Introduction
The adenosine monophosphate-activated protein kinase
(AMPK) is a highly conserved serine/threonine protein kinase
present in almost all eukaryotic cells. It plays a key role in
maintaining the balance of energy metabolism by maintaining
glycogen reserves and efficient mitochondrial oxidative metabolism
(1–3). Because disorders of energy balance
lead to various metabolic diseases in humans, such as type-2
diabetes, inflammatory disorders, and cancer, there has been
increasing interest in the development of pharmacological
activators of AMPK (4–7).
According to previous structural analyses, AMPK is a
heterotrimeric complex containing α, β, and γ subunits (8–11). The
α subunit has two subtypes, α1 and α2, which are encoded by two
separate genes and contain a catalytic serine/threonine protein
kinase domain. The key phosphorylation site, Thr172, is located in
this subunit. The β subunit serves as a bridge between the α and γ
subunits and has two subtypes, β1 and β2, which are encoded by two
separate genes. It is a regulatory unit, and the cleft between its
central carbohydrate-binding module and the kinase domain in the α
subunit is a critical binding site for direct activation. The γ
subunit is the allosteric activation site for AMP/ADP binding. It
also plays a regulatory role based on AMP/ATP and ADP/ATP ratio,
and has three subtypes, γ1, γ2 and γ3, which are encoded by three
different genes.
The activation of AMPK involves multiple mechanisms.
Firstly, AMPK is typically activated by the phosphorylation of
Thr172 in the catalytic domain of the α subunit. This process is
catalyzed by multiple protein kinases such as serine threonine
kinase 11 calcium/calmodulin dependent protein kinase kinase 2, and
MAP3K7 (12). Secondly, allosteric
activation associated with increased AMP/ATP and ADP/ATP ratios is
another mechanism for AMPK activation. AMP and its mimic ADP
binding to the γ subunit of AMPK not only cause allosteric effects
but also protect phosphorylated Thr172 from dephosphorylation
(13). Therefore, any agent that
can affect the binding of AMP/ADP to the γ subunit or inhibit
mitochondrial ATP production would indirectly activate the
phosphorylated AMPK. Thirdly, some exogenous compounds, called
direct AMPK modulators, can allosterically activate AMPK through an
AMP-independent pathway. The binding site of these activators is
located in the cleft between the β subunit central
carbohydrate-binding module and the α subunit kinase domain
(14,15).
Natural products are useful sources for drug
development. Indeed, natural products coming from various medicinal
plants can provide abundant drug screening entities with
high-molecular diversity (16). Our
previous studies examined the separation and identification of
natural products from traditional Chinese herbs (17–20).
Based on our previous studies, a natural product library containing
1,235 compounds derived from traditional Chinese herbs with
anti-metabolic effect was developed. Several of these compounds
have been shown to promote AMPK activation, as curcumin,
resveratrol, berberine, quercetin, and arctigenin (21–26).
Moreover, whereas some of these AMPK activators can directly
activate AMPK, others can also inhibit the respiratory chain and
indirectly activate AMPK. Therefore, identifying the specific
mechanisms of these natural activators may be beneficial for
further drug development. The aim of the present study was to
identify direct AMPK activators from our natural product
library.
Materials and methods
Materials
Liquiritin, ononin,
5,7-dihydroxy-3′,4′,6-trimethoxyflavone, leonurine, (−)-catechin,
luteolin, licochalcone-A were obtained from the National Institute
for Food and Drug Control (Beijing, China). The HepG2 liver cancer
cell line was obtained from the Cell Resource Center of Institute
of Basic Medical Sciences, Chinese Academy of Medical Sciences
& Peking Union Medical College. FBS was purchased from
Biological Industries. Modified Eagle's medium (MEM), non-essential
amino acids (NEAA), and penicillin/streptomycin were obtained from
Thermo Fisher Scientific, Inc. The triglyceride (TG) assay kit
(cat. no. 0220/0221/0222) was purchased from BioSino Bio-technology
and Science Inc. Sodium oleate and orlistat were obtained from
Sigma-Aldrich (Merck KGaA). Rabbit anti-AMPK (cat. no. 2532) and
anti-β-actin (cat. no. 4970) antibodies were from Cell Signaling
Technology, Inc. Rabbit anti-phosphorylated (p)-AMPK antibody (cat.
no. ab133448) and HRP-conjugated secondary antibodies (cat. nos.
ab6721 and ab6728) were from Abcam.
Data acquisition for protein and small
molecules
The 3D structure (PDB no., 4CFE) of AMPK
heterotrimeric complex (14) with
three types of ligands ([staurosporine,
5-[[6-chloranyl-5-(1-methylindol-5-yl)-1H-benzimidazol-2-yl]oxy]-2-methyl-benzoic
acid, and AMP) was downloaded from the Research Collaboratory for
Structural Bioinformatics Protein Data Bank (http://www.rcsb.org/pdb/home). The protein structure
was analyzed using Chimera 1.8.1 software (www.cgl.ucsf.edu/chimera/) and Discovery Studio 4.5
software (www.discoverystudio.net). The initial activator
benzimidazole derivative 991 was extracted as a reference ligand.
All of the co-ligands and water molecules were removed from the
co-crystallized complex. Addition of missing hydrogen atoms and
elimination of unnecessary polymers were performed using the
macromolecule preparation protocol, in the Discovery Studio
software. Protein structure was further analyzed using a clean
protein module to correct incomplete amino acid residues and
alternate conformations. Small molecules were constructed using
ChemBiodraw ultra 11.0 (https://www.chemdraw.com.cn/) and employed a Chemistry
at Harvard Macromolecular Mechanics (CHARMM) force field in
Discovery Studio, then minimized under a Dreiding-like force field
and CHARMM force field successively.
The compounds selected for virtual screening were
separated and purified from traditional Chinese herbs that have
anti-metabolic effects, according to our previous study. All the
structures were determined by nuclear magnetic resonance
spectrometry, infrared spectrum, and mass spectrometry (17–20).
Molecular docking
A molecular docking analysis was conducted using the
CDOCKER module in Discovery Studio 4.5. CDOCKER is a
structure-based docking method by grid calculation under the CHARMM
force field (27). It is a typical
semi-flexible docking protocol. The interactions between different
conformations of ligands and macromolecules were collected and
analyzed using a scoring algorithm. According to pre-experimental
results, docking parameters were set for optimization. The binding
site sphere was defined around the activated ligand 991 with a
radius of 9.0 Å. Pose cluster radius was set as 0.1 Å, the number
of allowed random conformations was set to 10, the number of
allowed orientations was set to 10, and electrostatic interactions
were included. The rest of the parameters were set as default. The
top 10 conformations were saved for each ligand based on scoring
function value (−CDOCKER interaction energy). To estimate whether
the binding site model was sufficient for the AMPK docking system,
a re-docking process was performed by evaluating the root mean
squared error (RMSD) value between the initial and virtual docking
conformations.
Pharmacophore model generation
The pharmacophore-based virtual screening is a
ligand-based virtual screening generated from a set of active small
molecules. The Common Feature Pharmacophore Generation module
(HipHop) in Discovery Studio is an appropriate application for
pharmacophore model generation, especially for those ligands that
lack of uniform standard activity data (28). Based on published reports (Table SI; Fig. S1), 28 compounds with AMPK agonist
activity were collected and divided into two sets, of which 18
compounds were in the training set, and the remaining were in the
text set. The Feature Mapping function was used to identify key
chemical features of the training set for predicting the considered
features in the pharmacophore generation process. The principal
value was set as 2, and MaxOmitFeat value was set as 0 for
compounds 1–4. The principal value and MaxOmitFeat value was set as
1 for other training set compounds. Minimum Interfeature Distance
was set to 2.97 Å, Minimum Features was set to 3, Feature Misses
and Complete Misses was set to 0, Conformation Generation was set
to ‘none’. The rest of the parameters were set as default. The text
set was prepared for the validation of the pharmacophore model.
Virtual screening based on
pharmacophore
All the compounds that were optimized under the
CHARMM force field were used as a virtual screening library in the
Build 3D database module in Discovery Studio software. The
screening based on pharmacophore was conducted with the Search 3D
database function in Discovery Studio software. The search method
was set as ‘best’. The rest of the parameters were set as default.
Hit compounds were used in subsequent molecular docking
screening.
Cell culture
HepG2 cells were maintained in MEM supplemented with
1% NEAA, 10% FBS and 1% P/S, and kept in a humidified atmosphere of
5% CO2 at 37°C. Cells were grown to 70–80% confluence,
then seeded in multi-well plates containing serum-free medium and
incubated for 24 h before treatment at 37°C with 5%
CO2.
TG accumulation inhibitory effects
assay
To induce an intracellular lipid accumulation model,
sodium oleate was used as previously described (29). Briefly, after seeding in 48-well
plates in FBS-free medium for 24 h, HepG2 cells were exposed to 200
µM sodium oleate in the presence or absence of different compounds
(3 or 30 µM), or orlistat (5 µM) as a positive control, for another
48 h. The intracellular TG content was finally examined using a
commercial assay kit at a wavelength of 492 nm after cells were
washed with PBS. Protein concentrations were determined using a BCA
protein assay kit at a wavelength of 562 nm. TG concentrations were
normalized to the total protein content.
Western blot analysis
Protein isolation and western blotting were
performed as described previously (29). Cells were homogenized in ice-cold
RIPA lysis buffer for 30 min on ice to yield protein samples. The
insoluble protein solution was removed by centrifugation at 10,000
× g, 4°C for 10 min. The supernatant was collected from the
lysates, and protein concentrations were determined using the BCA
protein assay kit following the manufacturer's instructions. Equal
amounts of proteins (40 µg) were resolved by SDS-PAGE on 8% gels
and transferred onto PVDF membranes. The membranes were blocked
with 5% non-fat dry milk in TBST buffer for 1.5 h at room
temperature. The membranes were incubated overnight at 4°C with
primary antibodies (1:500). The blots were rinsed five times with
TBST buffer for 6 min each. The washed blots were incubated with a
HRP-conjugated secondary antibody (1:5,000) for 1 h at room
temperature, then washed five times with TBST buffer. The protein
bands were visualized with an enhanced chemiluminescence detection
kit (EMD Millipore). The optical density of each band was
quantified using ImageJ software v1.53c (National Institutes of
Health) and expressed as arbitrary units.
Statistical analysis
Statistical analysis was performed using one-way
ANOVA followed by Tukey's post hoc test for multiple comparisons.
All data are expressed as the mean ± SEM, n=5. P<0.05 was
considered to indicate a statistically significant difference.
Results and Discussion
Pharmacophore model generation and
validation
A total of ten pharmacophore models were generated
based on the spatial arrangement of key chemical features from the
training set (details in supporting information Table SII). Each of these models contained
at least three features.
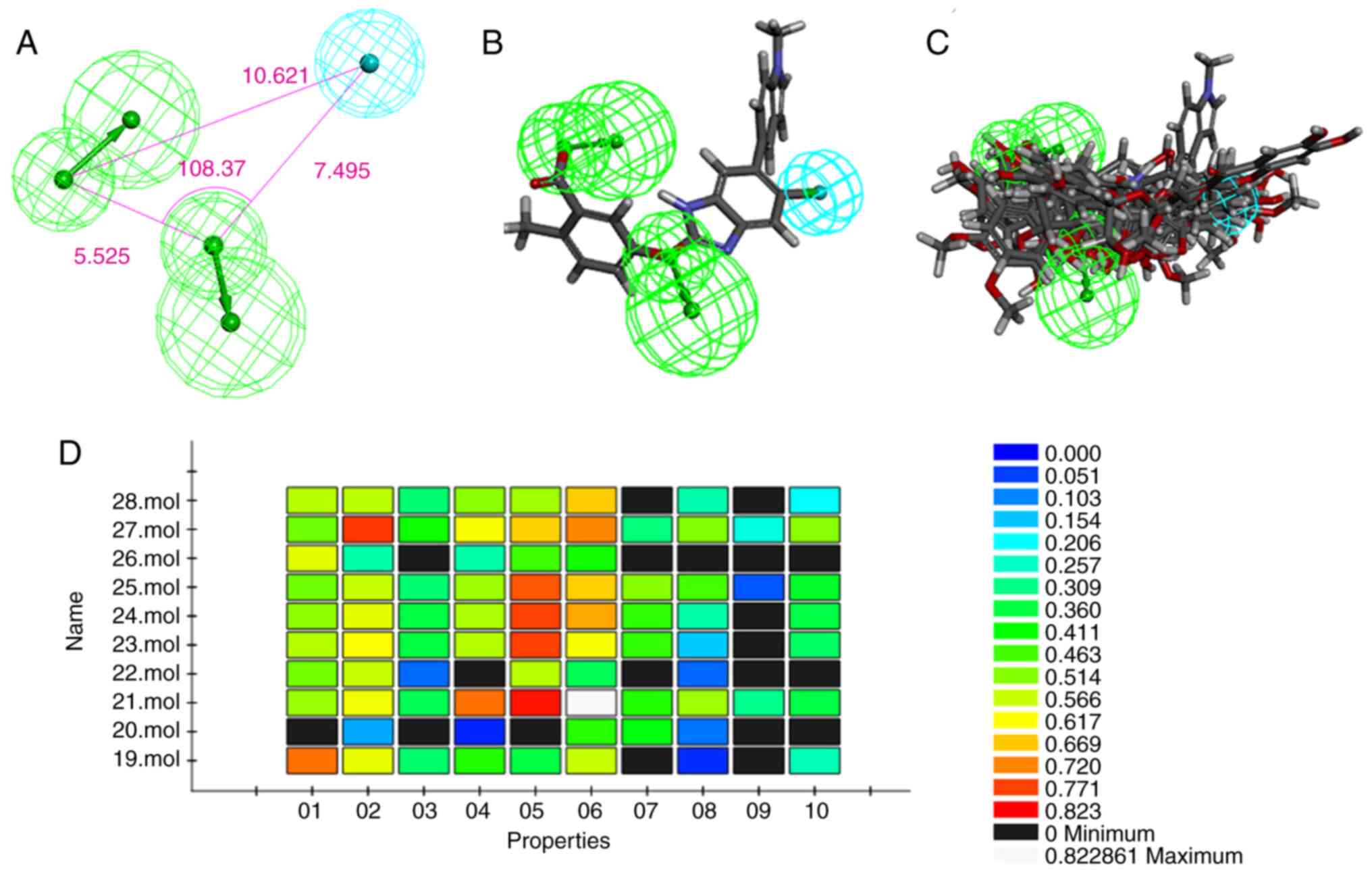
The top ranked model (HipHop1) was chosen as the
best hypothesis containing one hydrophobic region feature and two
hydrogen bond acceptor lipid (Ali1 and Ali2) features (Fig. 1A). The hydrophobic region feature
was farther from the other two hydrogen bond acceptor lipid
features. The pharmacophore hypotheses matched well with those
potent ligands, especially with initial ligand 991. The
pharmacophore model was also validated using the Ligand profiler
method from the Discovery Studio software.
Virtual screening based on
pharmacophore
The natural product library, consisting of 1,235
natural compounds, was constructed using Build 3D database modular.
The screening based on pharmacophore was conducted using the Search
3D database. All the hit compounds were aligned in the
pharmacophore model in Fig. 2.
Compared with the binding site structure, the distance between the
hydrophobic region feature and the other two hydrogen bond acceptor
lipid features facilitated hydrogen bond interactions between
ligands and amino acid residues B:ASN111, A:LYS29, B:THR106, and
B:phosphoserine (SER)108 in the AMPK activation site and was also
beneficial for hydrophobic interactions between ligands and the
hydrophobic region of the receptor.
Molecular docking
Benzimidazole derivative 991 is a well-characterized
direct AMPK activator binding in the specific binding site of AMPK.
The 3D crystal structure of AMPK-ligand (991) complex draws a
distinct image of this specific binding site, and paves a
convenient way for screening of direct AMPK activators (30,31).
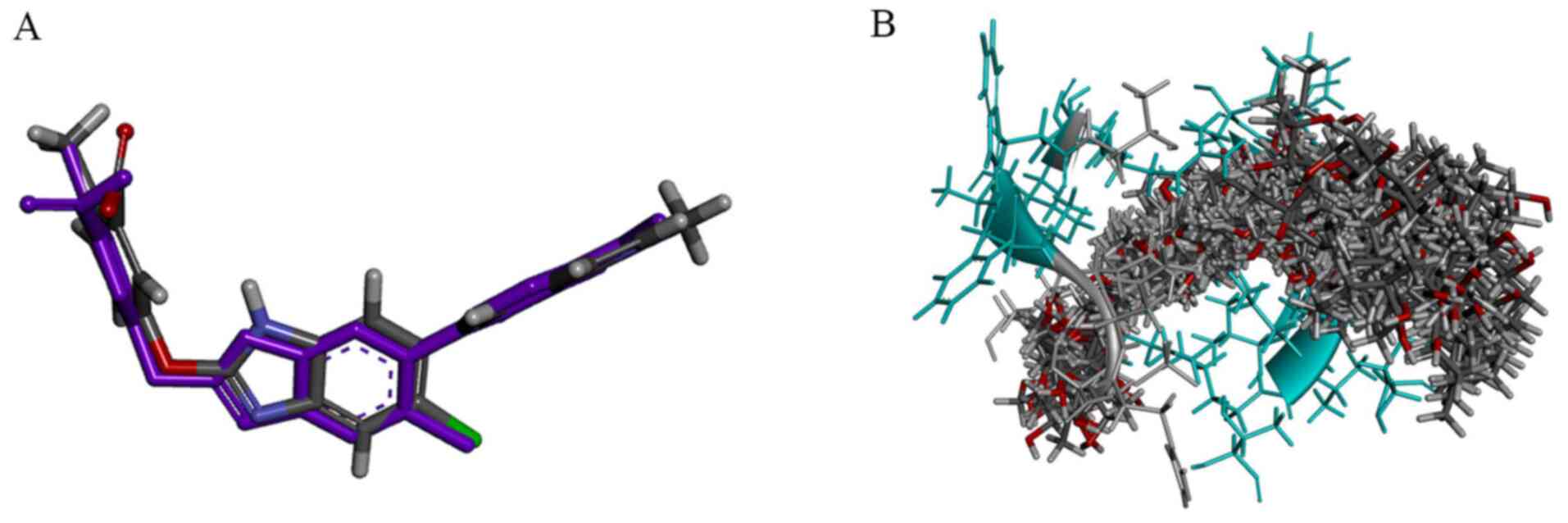
To verify the applicability of the binding site model for AMPK
docking screening, a redocking process was utilized, starting from
a random conformation of the initial ligand 991. The RMSD value
between the initial conformation and virtual docking conformation
was 1.4035 Å (Fig. 3A). This result
illustrated that this binding site model could simulate the real
ligand binding model (real crystal structure of Benzimidazole
derivative 991 binding to the active site of AMPK) and could be
appropriate for further docking studies.
The hit compounds from the pharmacophore screening
were subjected to the CDOCKER docking process, resulting in 284 hit
compounds for in vitro activity studies. The hit compounds
were aligned in Fig. 3B, and all
hit compounds from the docking study displayed similar spatial
conformations. The top 54 hit compounds (score >50 kcal/mol) are
listed in Table I (details of these
compounds Table SIII and Fig. S2).
 | Table I.Top list of hit compounds (score value
>50 kcal/mol). |
Table I.
Top list of hit compounds (score value
>50 kcal/mol).
| Compound no. | Compound name | Scoreb, kcal/mol | PubChem CID/CAS
no.c |
|---|
|
Referencea | 991 | 64.08 | 45256693 |
| 152 | Acotarin D | 64.2332 | 2306174-08-1 |
| 803 |
(E,E)-terrestribisamide | 63.872 | 5321825 |
| 804 |
N-p-coumaroyl-N′-feruloylputrescine | 60.2194 | 44241259 |
| 656 | 6′-benzoate
phloroglucinol 1-β-D-glucopyranoside | 59.2064 | 2407648-32-0 |
| 1211 |
Demethoxycurcumin | 58.5833 | 5469424 |
| 1145 | Liquiritin | 57.6091 | 503737 |
| 1209 |
5-hydroxy-7-(4-hydroxy-3-methoxyphenyl)-1-phenyl-3-heptanone | 57.2732 | 5318228 |
| 545 | Ononin | 56.276 | 442813 |
| 162 |
7S,8R-erythro-4,7,9,9′-tetrahydroxy-3,3′-dimethoxy-8-O-4′-neolignan | 56.1007 | 13893597 |
| 1148 | Licochalcone-A | 55.7685 | 5318998 |
| 374 | Epimedokoreanin
D | 55.6196 | 5315125 |
| 161 |
3-[2-(3,4-dimethoxyphenyl)-3-(hydroxymethyl)-7-
methoxy-2,3-dihydrobenzofuran-5-yl]-1-propanol | 55.1088 | 4639677 |
| 283 |
(−)-secoisolariciresinol | 55.0016 | 11552274 |
| 578 |
(7S,8R)-3,3′,5-trimethoxy-4′,7-epoxy-8,5′-neolignan-4,9,9′-triol | 54.7096 | 56838440 |
| 522 | (−)-Catechin | 54.2336 | 73160 |
| 540 | Malaferin C | 54.1877 | 71596193 |
| 1073 | Rosmarinic
acid | 54.0327 | 5281792 |
| 375 |
2-(3,4-dihydroxy-5-(3-methylbut-2-enyl)phenyl)-5,7-
dihydroxy-8-(2-hydroxy-3-methylbut-3-enyl)chromen-4-one | 53.9136 | 131676094 |
| 567 |
2-(4-hydroxy-3,5-dimethoxyphenyl)ethyl-β-D-glucopyranoside | 53.9011 | 76308955 |
| 615 | Methyl
5-O-caffeoylquinic acid methyl ester | 53.8888 | 6476139 |
| 441 |
5,7-dihydroxy-3′,4′,6-trimethoxyflavone | 53.8865 | 5273755 |
| 1214 | Arctigenin | 53.7069 | 64981 |
| 1011 |
p-hydroxybenzoyl-β-D-glucopyranoside | 53.6545 | 14132342 |
| 1195 | Curcumin | 53.6135 | 969516 |
| 395 |
3′-(2-hydroxy-3-methyl-3-butenyl)-4′,3,5,7-tetrahydroxy-5′-prenylflavone | 53.1348 | 1812887-75-4 |
| 1007 |
3,4-dimethoxycinnamyl-β-D-glucopyranoside | 52.7184 | 21581586 |
| 287 | 4-O-caffeoylquinic
acid methyl ester | 52.6425 | 71720840 |
| 396 |
3′-Prenylnaringenin | 52.5223 | 5315396 |
| 1012 |
4-methoxybenzyl-β-D-glucopyranoside | 52.4808 | 10685601 |
| 566 |
3-methoxyphenethyl-alcohol-4-O-β-D-glucopyranoside | 52.2909 | 23815379 |
| 284 | (−)-arctigenin | 52.1262 | 119205 |
| 867 | Salidreoside | 52.0402 | 159278 |
| 1147 | Glabridin | 52.023 | 124052 |
| 376 |
2-(3,4-dihydroxy-5-(3-methylbut-2-en-1-yl)phenyl)-5-hydroxy-8-(2-hydroxypropan-2-yl)-8,9-dihydro-4H-furo(2,3-h)chromen-4-one | 51.9827 | 2196202-19-2 |
| 553 |
(−)-Epicatechin | 51.8918 | 72276 |
| 143 | Leonurine | 51.8901 | 161464 |
| 1196 | Desmethoxy
curcumin | 51.6991 | 5469424 |
| 982 |
(2R,3R)-3,5,6,7,4′-Pentahydroxyflavanonol | 51.6525 | 101353295 |
| 160 |
Dihydrodehydrodiconiferyl alcohol | 51.4808 | 4365980 |
| 1205 |
(R)-5-hydroxy-1,7-diphenyl-3-heptanone | 51.4631 | 46213118 |
| 1140 | Hesperidin | 51.3406 | 72281 |
| 137 |
Phenethyl-β-D-glucopyranoside | 51.1543 | 11289099 |
| 669 |
5,6,7,3′,4′-pentamethoxyflavone | 51.1409 | 145659 |
| 560 | Isotachioside | 51.1143 | 15098566 |
| 166 | N-trans ferulic
acid casein amide | 51.0104 | 5280537 |
| 19 | Luteolin | 51.0005 | 5280445 |
| 19 | Luteolin | 51.0005 | 5280445 |
| 752 |
2-methyl-3-O-β-D-glucopyranosyl-pyran-4-one | 50.6335 | 5316639 |
| 1197 |
7-(4-hydroxy-3-methoxyphenyl)-1-phenyl-4E-heptene-3-one | 50.5732 | 5318278 |
| 371 |
3,6,7-trimethoxy-9,10-dihydrophenanthrene-2,5-diol | 50.5535 | 14135385 |
| 552 | (+)-Taxifolin | 50.2719 | 439533 |
| 900 |
3-O-trans-coumaroylquinic acid | 50.2104 | 11078262 |
| 665 | 2S-eriodictyol | 50.1198 | 440735 |
| 434 | Artemetin | 50.0462 | 5320351 |
| 382 | 4, 5-
Phenanthrenediol, 9, 10- dihydro- 2, 3, 6-
trimethoxy-9,10-Dihydrophenanthrene | 50.0364 | 25141334 |
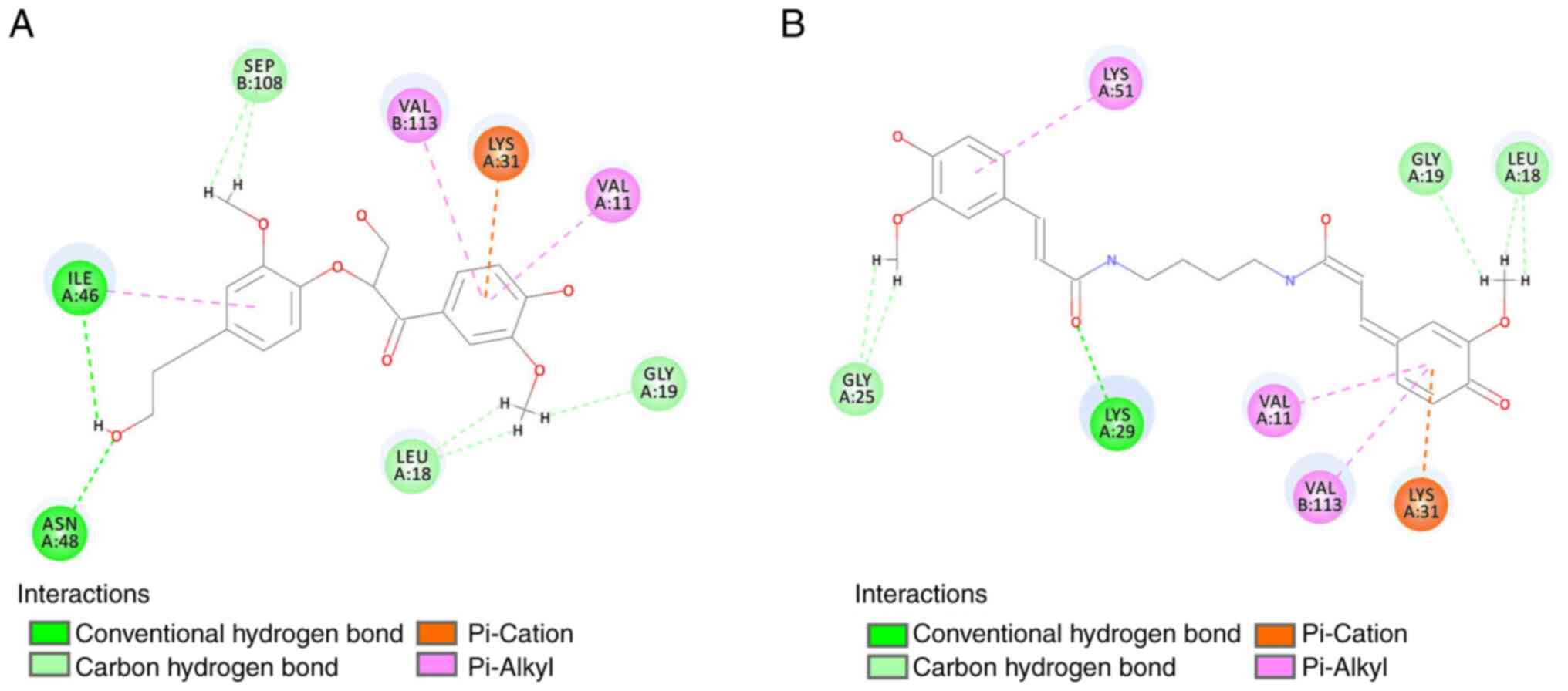
The 2D interaction diagram of the top two hits,
acotarin D and (E,E)-terrestribisamide, indicates that the residues
A:Gly19, A:Leu18, A:ILE46, A:ASN48, B:SEP108, A:Gly25, and A:Lys29
were involved in the formation of hydrogen bonds; the residues
A:Lys31, B:VAL113 and A:VAL11 were involved in the formation of
hydrophobic interactions (Fig.
4).
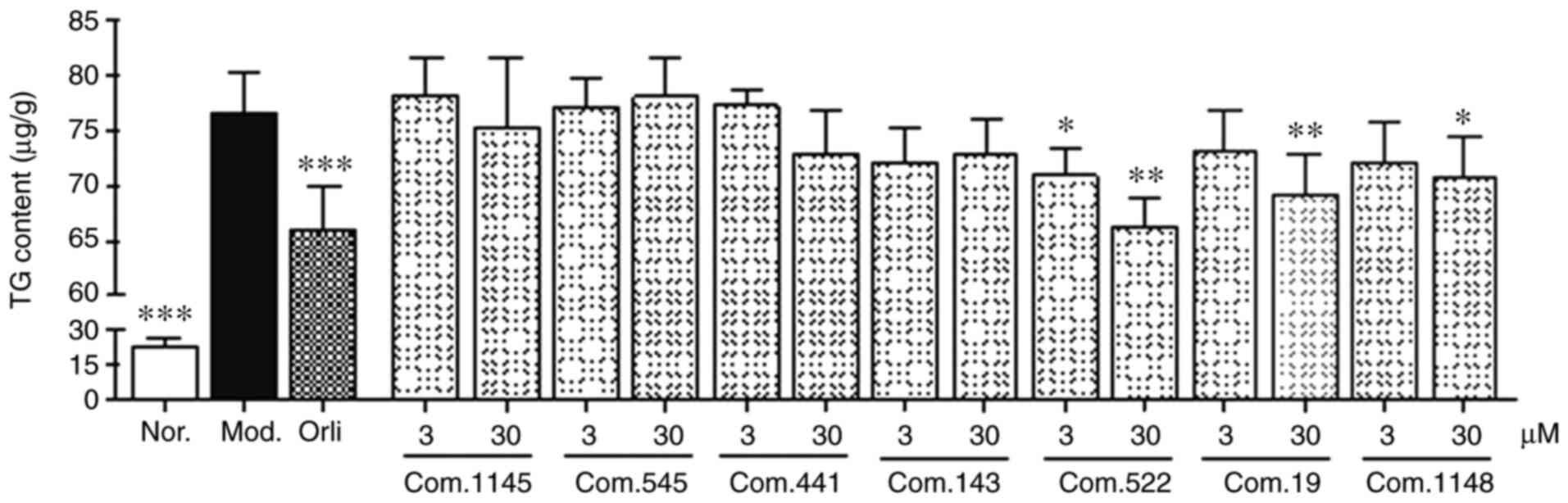
TG accumulation inhibitory effects of
selected compounds
AMPK is a key sensor that maintains the balance of
energy metabolism by regulating glucose and lipid metabolism.
Activation of AMPK leads to increases in fatty acid oxidation
though multiple pathways such as activation of malonyl CoA
decarboxylase and reduction of the inhibitory effect of malonyl CoA
to carnitine palmitoyl transterase-1 (1,2).
Therefore, inhibition of TG accumulation is a preliminary indicator
for AMPK activation (29).
TG accumulation inhibitory assays were carried out
for preliminary activity screening in sodium oleate-treated HepG2
cells. Considering docking score and structural diversity, seven
compounds (Table II) with a score
>50 kcal/mol were selected and evaluated for their inhibitory
effects on TG accumulation.
 | Table II.Selected compounds list for activity
verification. |
Table II.
Selected compounds list for activity
verification.
| Compound no. | Compound name |
|---|
| 1145 | Liquiritin |
| 545 | Ononin |
| 441 |
5,7-dihydroxy-3′,4′,6-trimethoxyflavone |
| 143 | Leonurine |
| 522 | (−)-Catechin |
| 19 | Luteolin |
| 1148 | Licochalcone A |
Following sodium oleate challenge, the intracellular
TG levels increased almost four-fold in the model group (Fig. 5), compared with the control.
However, this increase in TG levels induced by sodium oleate was
significantly reduced by ~13.8% following orlistat treatment.
Moreover, 30 µM (−)-catechin (compound 522), luteolin (compound
19), and licochalcone A (compound 1148) also significantly reduced
intracellular TG levels by 13.4, 9.7 and 7.7%, respectively. In
addition, (−)-catechin also exhibited a moderate inhibitory effect
on TG accumulation at a concentration of 3 µM. The other compounds
did not result in any detectable effects.
AMPK phosphorylation effect of
selected compounds in sodium oleate-induced HepG2 cells
The activation effects of (−)-catechin (compound
522), luteolin (compound 19), and licochalcone A (compound 1148) on
AMPK were further assessed. Immunoblotting analysis indicated that
AMPK phosphorylation levels significantly decreased after 200 µM
sodium oleate challenge in the model group (Fig. 6), compared with the control. Among
the tested compounds, compound 522 [(−)-catechin] and 1148
(licochalcone A) significantly increased AMPK phosphorylation
levels at a concentration of 30 µM, as evidenced by higher
p-AMPK/t-AMPK ratios. However, other selected compounds had no
apparent effect on AMPK phosphorylation.
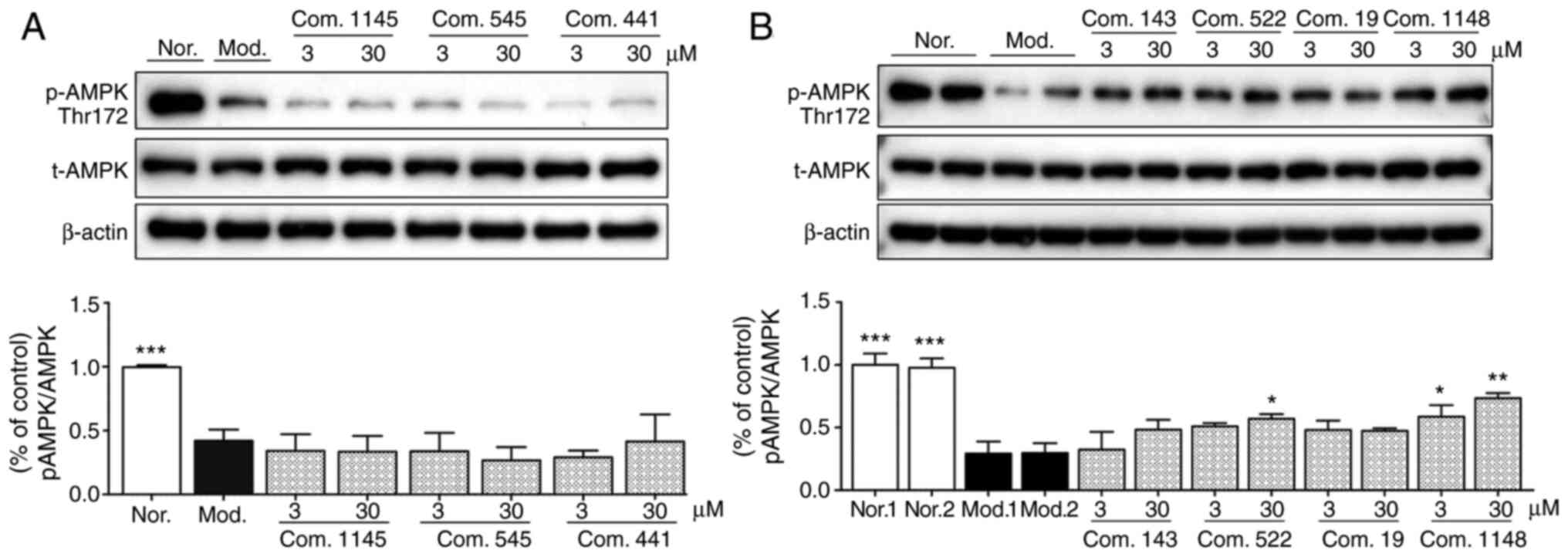 | Figure 6.AMPK activation assay. Data are
presented as the mean ± SEM. (A) Results for com. 1145, com. 545,
and com. 441. ***P<0.001 vs. Mod. (B) Results for com. 143, com.
522, com. 19, and com. 1148. ***P<0.001, **P<0.01, *P<0.05
vs. Mod. 1. AMPK, adenosine monophosphate-activated protein kinase;
Com., compound; Mod., model; Nor., normal; p, phosphorylated; t,
total. |
Comparing the results of the TG accumulation and the
AMPK phosphorylation assays, the activity trends of these two tests
were consistent, and compound 522 [(−)-catechin] and 1148
(licochalcone A) performed well in both assays. Our findings agreed
with the results of previous studies indicating that both compounds
522 [(−)-catechin] and 1148 (licochalcone A) are potential AMPK
activators (32,33). According to the docking results,
these two natural products may directly modulate AMPK activity in
an AMP-independent way by binding to the specific binding site.
Conclusions
The present study followed on our research efforts
on the separation and purification of natural compounds from
traditional Chinese herbs. In this study, virtual screening for
direct natural AMPK activators was conducted using a combination of
ligand- and structure-based screening. The hit compounds displayed
similar spatial conformations and bound to the AMPK-specific
binding site through hydrogen bonds and hydrophobic interactions. A
total of seven hit compounds were chosen for subsequent activity
validation. (−)-Catechin (compound 522) and licochalcone A
(compound 1148) performed well in two activity tests and could be
valuable for further pharmacological evaluation. The present
findings suggest that these two natural products may directly
modulate AMPK activity in an AMP-independent manner, through the
specific binding site of AMPK. This study may provide insight into
the development of AMPK activators from natural resources and their
modulatory mechanisms.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This project was supported by the National Natural
Science Foundation (grant nos. 81673703 and 81803691), and the
Important Drug Development Fund, Ministry of Science and Technology
of China (grant nos. 2018ZX09735-002 and
2019ZX09201005-002-007).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
TW designed the experiment and was responsible for
the conception of the study. JH, ZY and LH performed the laboratory
experiments and wrote the manuscript. JL and YZ contributed to the
pharmacological experiments. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gruzman A, Babai G and Sasson S: Adenosine
monophosphate-activated protein kinase (AMPK) as a new target for
antidiabetic drugs: A review on metabolic, pharmacological and
chemical considerations. Rev Diabet Stud. 6:13–36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Daskalopoulos EP, Dufeys C, Bertrand L,
Beauloye C and Horman S: AMPK in cardiac fibrosis and repair:
Actions beyond metabolic regulation. J Mol Cell Cardiol.
91:188–200. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shirwany NA and Zou MH: AMPK: A cellular
metabolic and redox sensor. A minireview. Front Biosci (Landmark
Ed). 19:447–474. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gejjalagere Honnappa C and Mazhuvancherry
Kesavan U: A concise review on advances in development of small
molecule anti-inflammatory therapeutics emphasising AMPK: An
emerging target. Int J Immunopathol Pharmacol. 29:562–571. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim J, Yang G, Kim Y, Kim J and Ha J: AMPK
activators: Mechanisms of action and physiological activities. Exp
Mol Med. 48:e2242016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Plews RL, Mohd Yusof A, Wang C, Saji M,
Zhang X, Chen CS, Ringel MD and Phay JE: A novel dual AMPK
activator/mTOR inhibitor inhibits thyroid cancer cell growth. J
Clin Endocrinol Metab. 100:E748–E756. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Law BY, Mok SW, Chan WK, Xu SW, Wu AG, Yao
XJ, Wang JR, Liu L and Wong VK: Hernandezine, a novel AMPK
activator induces autophagic cell death in drug-resistant cancers.
Oncotarget. 7:8090–8104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grahame Hardie D: Regulation of
AMP-activated protein kinase by natural and synthetic activators.
Acta Pharm Sin B. 6:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Polekhina G, Gupta A, Michell BJ, van
Denderen B, Murthy S, Feil SC, Jennings IG, Campbell DJ, Witters
LA, Parker MW, et al: AMPK beta subunit targets metabolic stress
sensing to glycogen. Curr Biol. 13:867–871. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lefort N, St-Amand E, Morasse S, Côté CH
and Marette A: The alpha-subunit of AMPK is essential for
submaximal contraction-mediated glucose transport in skeletal
muscle in vitro. Am J Physiol Endocrinol Metab. 295:E1447–E1454.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Day P, Sharff A, Parra L, Cleasby A,
Williams M, Hörer S, Nar H, Redemann N, Tickle I and Yon J:
Structure of a CBS-domain pair from the regulatory gamma1 subunit
of human AMPK in complex with AMP and ZMP. Acta Crystallogr D Biol
Crystallogr. 63(Pt 5):587–596. 2007. View Article : Google Scholar
|
|
12
|
Hardie DG, Ross FA and Hawley SA: AMPK: A
nutrient and energy sensor that maintains energy homeostasis. Nat
Rev Mol Cell Biol. 13:251–262. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gowans GJ, Hawley SA, Ross FA and Hardie
DG: AMP is a true physiological regulator of AMP-activated protein
kinase by both allosteric activation and enhancing net
phosphorylation. Cell Metab. 18:556–566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao B, Sanders MJ, Carmena D, Bright NJ,
Haire LF, Underwood E, Patel BR, Heath RB, Walker PA, Hallen S, et
al: Structural basis of AMPK regulation by small molecule
activators. Nat Commun. 4:30172013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zadra G, Photopoulos C, Tyekucheva S,
Heidari P, Weng QP, Fedele G, Liu H, Scaglia N, Priolo C, Sicinska
E, et al: A novel direct activator of AMPK inhibits prostate cancer
growth by blocking lipogenesis. EMBO Mol Med. 6:519–538. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Newman DJ and Cragg GM: Natural products
as sources of new drugs over the nearly four decades from 01/1981
to 09/2019. J Nat Prod. 83:770–803. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang T, Ruan J, Li X, Chao L, Shi P, Han
L, Zhang Y and Wang T: Bioactive cyclolanstane-type saponins from
the stems of Astragalus membranaceus (Fisch.) Bge. var. mongholicus
(Bge.) Hsiao. J Nat Med. 70:198–206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Chao L, Ruan J, Zheng C, Yu H, Qu
L, Han L and Wang T: Bioactive constituents from the rhizomes of
Dioscorea septemloba, Thunb. Fitoterapia. 115:165–172. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Y, Nakamura S, Nakashima S, Wang T,
Yoshikawa M and Matsuda H: Chemical structures of constituents from
the seeds of Cassia auriculata. Tetrahedron. 71:6727–6732. 2015.
View Article : Google Scholar
|
|
20
|
Zhang Y, Han L, Ge D, Liu X, Liu E, Wu C,
Gao X and Wang T: Isolation, structural elucidation, MS profiling,
and evaluation of triglyceride accumulation inhibitory effects of
benzophenone C-glucosides from, leaves of mangifera indica L. J
Agric Food Chem. 61:1884–1895. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee YK, Lee WS, Hwang JT, Kwon DY, Surh YJ
and Park OJ: Curcumin exerts antidifferentiation effect through
AMPKalpha-PPAR-gamma in 3T3-L1 adipocytes and antiproliferatory
effect through AMPKalpha-COX-2 in cancer cells. J Agric Food Chem.
57:305–310. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Z, Cui C, Xu P, Dang R, Cai H, Liao D,
Yang M, Feng Q, Yan X and Jiang P: Curcumin activates AMPK pathway
and regulates lipid metabolism in rats following prolonged
clozapine exposure. Front Neurosci. 11:5582017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Price NL, Gomes AP, Ling AJ, Duarte FV,
Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro
JS, et al: SIRT1 is required for AMPK activation and the beneficial
effects of resveratrol on mitochondrial function. Cell Metab.
15:675–690. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin Y, Liu S, Ma Q, Xiao D and Chen L:
Berberine enhances the AMPK activation and autophagy and mitigates
high glucose-induced apoptosis of mouse podocytes. Eur J Pharmacol.
794:106–114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim SG, Kim JR and Choi HC:
Quercetin-induced AMP-activated protein kinase activation
attenuates vasoconstriction through LKB1-AMPK signaling pathway. J
Med Food. 21:146–153. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han YH, Kee JY, Park J, Kim HL, Jeong MY,
Kim DS, Jeon YD, Jung Y, Youn DH, Kang J, et al: Arctigenin
inhibits adipogenesis by inducing AMPK activation and reduces
weight gain in high-fat diet-induced obese mice. J Cell Biochem.
117:2067–2077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu G, Robertson DH, Brooks CL III and
Vieth M: Detailed analysis of grid-based molecular docking: A case
study of CDOCKER-A CHARMm-based MD docking algorithm. J Comput
Chem. 24:1549–1562. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Patel Y, Gillet VJ, Bravi G and Leach AR:
A comparison of the pharmacophore identification programs:
Catalyst, DISCO and GASP. J Comput Aided Mol Des. 16:653–681. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Liu X, Han L, Gao X, Liu E and
Wang T: Regulation of lipid and glucose homeostasis by mango tree
leaf extract is mediated by AMPK and PI3K/AKT signaling pathways.
Food Chem. 141:2896–2905. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Potunuru UR, Priya KV, Varsha MKNS, Mehta
N, Chandel S, Manoj N, Raman T, Ramar M, Gromiha MM and Dixit M:
Amarogentin, a secoiridoid glycoside, activates AMP-activated
protein kinase (AMPK) to exert beneficial vasculo-metabolic
effects. Biochim Biophys Acta Gen Subj. 1863:1270–1282. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mok SWF, Zeng W, Niu Y, Coghi P, Wu Y, Sin
WM, Ng SI, Gordillo-Martínez F, Gao JY, Law BYK, et al: A method
for rapid screening of anilide-containing AMPK modulators based on
computational docking and biological validation. Front Pharmacol.
9:7102018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hwang JT, Park OJ, Lee YK, Sung MJ, Hur
HJ, Kim MS, Ha JH and Kwon DY: Anti-tumor effect of luteolin is
accompanied by AMP-activated protein kinase and nuclear factor-KB
modulation in HepG2 hepatocarcinoma cells. Int J Mol Med. 28:25–31.
2011.PubMed/NCBI
|
|
33
|
Bae UJ, Park J, Park IW, Chae BM, Oh MR,
Jung SJ, Ryu GS, Chae SW and Park BH:
Epigallocatechin-3-Gallate-rich green tea extract ameliorates fatty
liver and weight gain in mice fed a high fat diet by activating the
sirtuin 1 and AMP activating protein kinase pathway. Am J Chin Med.
46:617–632. 2018. View Article : Google Scholar : PubMed/NCBI
|