Introduction
Pulmonary fibrosis is the end result of a major
category of pulmonary diseases characterized by fibroblast
proliferation, extracellular matrix (ECM) deposition and tissue
structure destruction. It is also characterized by scar formation
caused by abnormal repair of damaged alveolar tissue (1,2).
Fibroblasts are derived from epithelial cells that have undergone
epithelial-mesenchymal transformation (EMT), in which the markers
of the epithelial cells are depleted and the abilities of adhesion,
proliferation and migration are enhanced (3). The phenotypic dysregulation of the
alveolar epithelial cells, accompanied by excessive ECM deposition,
is a crucial component in the progression of pulmonary fibrosis
(4). Studies investigating
fibrogenesis have made progress in recent years (5,6);
however, further investigation is required to elucidate the
pathogenesis of pulmonary fibrosis to facilitate the development of
effective therapeutic strategies.
Numerous studies have demonstrated that microRNAs
(miRNA/miR) act as a class of non-coding single-stranded RNA
molecules, which are ~22 nucleotides in length and encoded by
endogenous genes, and are involved in a series of biological and
pathological processes by binding and degrading the target mRNAs
(7,8). Recently, the regulatory functions of
miRNAs in pulmonary fibrosis have been increasingly discovered.
Using Affymetrix miRNA microarrays, miR-455-3p was found to be
downregulated in the lung tissues of patients with idiopathic
pulmonary fibrosis (9). Wei et
al (10) reported that
miR-455-3p was decreased in the fibrotic liver, and overexpression
of miR-455-3p could inhibit the expression levels of profibrotic
markers in hepatic stellate cells. Furthermore, miR-455-3p was also
involved in the accumulation of ECM and the expression of
fibrosis-related proteins in diabetic nephropathy (11).
In addition to miRNAs, another type of non-coding
RNA, which have lengths of >200 nucleotides, termed long
non-coding RNAs (lncRNAs), have been reported to regulate gene or
protein expression levels in the course of fibrogenesis. The lncRNA
nuclear enriched abundant transcript 1 (NEAT1) was highly expressed
in murine fibrotic livers and promoted liver fibrosis by regulating
miRNA-122 and Kruppel-like factor 6 (12). Huang et al (13) revealed that the downregulation of
NEAT1 repressed the proliferation of mesangial cells and fibrosis
in diabetic nephropathy by inactivating the Akt/mTOR signaling
pathway. However, the expression of NEAT1 in lung epithelial cells
and the regulatory mechanism involved remains largely unknown.
Numerous studies have described that lncRNA
function, as a competing endogenous RNA, could mediate miRNAs and
the targets of miRNAs (14,15). Binding sites between the sequences
of NEAT1 and miR-455-3p were predicted using bioinformatics
analysis. The present study aimed to determine the function of
NEAT1 in TGF-β1-treated human alveolar epithelial and bronchial
epithelial cell lines, and investigate its potential association
with miR-455-3p.
Materials and methods
Cell culture and transfection
The human HPAEpiC alveolar epithelial cells
(ScienCell Research Laboratories, Inc.) were cultured in DMEM/F12
(Hyclone; Cytiva) containing 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in a humidified incubator with 5%
CO2. The BEAS-2B human bronchial epithelial cells
(American Type Culture Collection) were cultured in complete
bronchial epithelial growth medium (Lonza Group, Ltd.) at 37°C in a
humidified incubator with 5% CO2.
TGF-β1 (R&D Systems China Co., Ltd.) is a potent
pro-fibrotic factor, and 10 ng/ml was used to stimulate the two
types of epithelial cells for 48 h. Two interfering sequences of
short hairpin NEAT1 (shRNA-NEAT1-1, 5′-GTGAGAAGTTGCTTAGAAA-3′; and
shRNA-NEAT1-2, 5′-TGGTAATGGTGGAGGAAGA-3′) were ligated into the
pGPH6/Neo vector (50 nM; Shanghai GenePharma Co., Ltd.). miR-455-3p
mimic (50 nM; 5′-GCAGUCCAUGGGCAUAUACAC-3′) and inhibitor (50 nM;
5′-GUGUAUAUGCCCAUGGACUGC-3′), as well as their corresponding
negative controls [50 nM; NC; mimic-NC;
5′-UUCUCCGAACGUGUCACGUTT-3′) and inhibitor-NC (50 nM;
5′-CAGUACUUUUGUGUAGUACAA-3′)] were also purchased from Shanghai
GenePharma Co., Ltd. The empty vector plasmid (50 nM; pcDNA3.1) and
pcDNA3.1-SMAD3 (50 nM; 5′-GAATCGCCACCATGTCGTCCATCCTGCCCTTC-3′ and
5′-CTCGAGCCTGGGGTTTTCTTCTGTGGTC-3′) were constructed by Sangon
Biotech Co., Ltd. Briefly, cells were seeded (5×105)
into 12-well plates and grown to 80% confluence. Subsequently,
cells were transfected with shRNA or mimic using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) for
48 h, according to the manufacturer's instructions. At 48 h
post-transfection, transfection efficacy was evaluated using
reverse transcription-quantitative PCR (RT-qPCR).
Prediction of target genes
The Encyclopedia of RNA Interactomes (ENCORI,
http://starbase.sysu.edu.cn). ENCORI is
an open-source platform for studying the miRNA-ncRNA, miRNA-mRNA,
ncRNA-RNA, RNA-RNA, RBP-ncRNA and RBP-mRNA interactions from
CLIP-seq, degradome-seq and RNA-RNA interactome data.
RT-qPCR
Total RNA was extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.), while the
PrimeScript™ RT reagent kit (Takara Bio, Inc.; 16°C for 30 min,
42°C for 30 min and 85°C for 5 min) and SYBR Green qPCR kit (Thermo
Fisher Scientific, Inc.) were used for reverse transcription and
qPCR, respectively. The following thermocycling conditions were
used for qPCR: Initial denaturation at 95°C for 5 min; followed by
40 cycles of denaturation at 95°C for 20 sec, annealing at 60°C for
30 sec and extension at 72°C for 20 sec. β-actin was used as the
endogenous control of lncRNA and mRNA. U6 was used to normalize the
relative expression levels of miRNA. Relative expression levels of
miRNA and mRNA were determined using the 2−ΔΔCq method
(16). The primer sequences used
are stated in Table I.
 | Table I.Primers used in the present
study. |
Table I.
Primers used in the present
study.
| Primer | Sequence
(5′→3′) |
|---|
| U6 snRNA | F:
GCGCGTCGTGAAGCGTTC |
|
| R:
GTGCAGGGTCCGAGGT |
| miR-455-3p | F:
ACACTCCAGCTGGGGCAGTCCACGGGCATATACAC |
|
| R:
GTGCAGGGTCCGAGGT |
| β-actin | F:
CAGAGCAAGAGAGGCATCC |
|
| R:
CTGGGGTGTTGAAGGTCTC |
| NEAT1 | F:
CAGGGTGTCCTCCACCTTTA |
|
| R:
AAACCAGCAGACCCCTTTTT |
| SMAD3 | F:
AAACTAGTGTCACAGTCCAACCAGAAAC |
|
| R:
GGAAGCTTTTGTACCAAGCCTGCAATT |
Wound healing assay
Cells were seeded (3×105 cells/well) into
a 6-well plate and incubated at 37°C in 5% CO2. At 80%
confluence, the scratches were created using a sterile 200 µl
pipette tip. Then, cells were washed gently to remove the floating
cells and the medium was replaced with serum-free medium for 24 h.
Images of the cells that had migrated into the wound were captured
under a light microscope (magnification, ×100; Zeiss AG).
Quantitative analysis of the wound healing area was performed using
ImageJ software (version 1.52r; National Institutes of Health).
Transwell invasion assay
A Transwell invasion assay was used to analyze the
invasive rate of cells. Briefly, AMC-HN-8 cells in 100 µl
(2×105) serum-free medium (Thermo Fisher Scientific,
Inc.) were plated into the upper chambers of an 8-µm Transwell
plate (Corning, Inc.) precoated with Matrigel (BD Biosciences) at
24 h (37°C) after transfection. DMEM/F12 (Hyclone; Cytiva)
containing 20% FBS was plated in the lower chamber to serve as a
chemoattractant. Following the incubation (37°C, 24 h), the
invading cells on the bottom surface of the filter were fixed with
methanol (100%, 4°C) for 30 min and stained with hematoxylin at
room temperature for 20 min. Cell invasion was analyzed in three
randomly selected fields under a fluorescent microscope
(magnification, ×20).
Western blot analysis
Total protein in the cells was extracted using a
RIPA lysis buffer (Beyotime Institute of Biotechnology). After
quantification using a bicinchoninic acid protein assay kit
(Beyotime Institute of Biotechnology), protein (40 µg/lane) in the
different experimental groups were separated via 10% SDS-PAGE, and
then transferred onto PVDF membranes using electrophoresis.
Following blocking with 5% skimmed milk for 1.5 h at room
temperature, the PVDF membranes were subsequently incubated with
primary antibodies overnight at 4°C. Next, the primary antibodies
were removed and the membrane was incubated with the secondary
antibodies for 1 h at room temperature. The primary antibodies
against α smooth muscle actin (α-SMA; cat. no. ab7817; 1:1,000),
collagen I (cat. no. ab34710; 1:1,000), collagen III (cat. no.
ab184993; 1:1,000) and E-cadherin (cat. no. ab1416; 1:1,000) were
obtained from Abcam, while the antibodies against SMAD3 (cat. no.
9523T; 1:1,000), fibronectin1 (cat. no. 26836S; 1:1,000) and
β-actin (cat. no. 4970T; 1:1,000) were obtained from Cell Signaling
Technology, Inc. The secondary antibodies (cat. nos. ab7090 and
ab97040; 1:5,000) were purchased from Abcam.
Luciferase reporter gene
The 3′untranslated region (UTR) of miR-455-3p,
containing NEAT1 wild-type (WT) or mutant (MUT) sites which were
amplified by Shanghai GenePharma Co., Ltd., were subcloned into the
pmirGLO vector (Promega Corporation). The cells were co-transfected
with miR-455-3p mimic or NC-mimic, and with a vector containing
NEAT1 WT or MUT sites using Lipofectamine 3000. The 3′UTR of
miR-455-3p containing SMAD3 WT or MUT sites were subcloned into the
pmirGLO vector (Promega Corporation). The cells were co-transfected
with miR-455-3p mimic or NC-mimic and with a vector containing
SMAD3 WT or MUT sites using Lipofectamine 3000. Relative luciferase
activity was measured by normalizing firefly luciferase activity to
Renilla luciferase activity at 48-h post-transfection using
a Dual-Luciferase Reporter assay (Promega Corporation).
Statistical analysis
The data are presented as the mean ± standard
deviation, and were analyzed using GraphPad Prism v6.0 statistical
software (GraphPad Software, Inc.). A Student's t-test was used for
comparisons between two groups, while ANOVA followed by the Tukey's
post hoc test was used for comparisons among multiple groups. All
experiments were performed in triplicate. P<0.05 was considered
to indicate a statistically significant difference.
Results
Knockdown of NEAT1 inhibits migration,
EMT and collagen generation of epithelial cells
The mRNA expression level of NEAT1 was significantly
increased in TGF-β1-treated HPAEpiC and BEAS-2B cells compared with
that in the control group (Fig. 1A and
B). The HPAEpiC and BEAS-2B cell lines were transfected with
shRNA-NEAT1 (Fig. 1C and D), and
shRNA-NEAT1-1 was selected for the further experiments due to lower
NEAT1 expression levels induced by shRNA-NEAT1-1 compared with
shRNA-NEAT1-2. The migratory and invasive abilities of the
TGF-β1-induced HPAEpiC cells were weakened by silencing NEAT1
(Fig. 1E-H), and similar results
were also found in the BEAS-2B cell line (Fig. 1I-L). The epithelial cell marker,
E-cadherin, was decreased in the HPAEpiC and BEAS-2B cell lines
treated with TGF-β1, whereas transfection with shRNA-NEAT1-1
reversed the effect of TGF-β1 (Fig.
1M). Fibronectin1 and α-SMA act as markers of mesenchymal cells
and were upregulated in TGF-β1-treated HPAEpiC and BEAS-2B cell
lines, while knockdown of NEAT1 reduced the protein expression
level of fibronectin1 and α-SMA. Similarly, shRNA-NEAT1-1 partially
abrogated the promotional effects of TGF-β1 on the protein
expression levels of collagen I and III (Fig. 1M). Collectively, these findings
illustrated the important roles of NEAT1 in cell migration, EMT and
collagen production of epithelial cells.
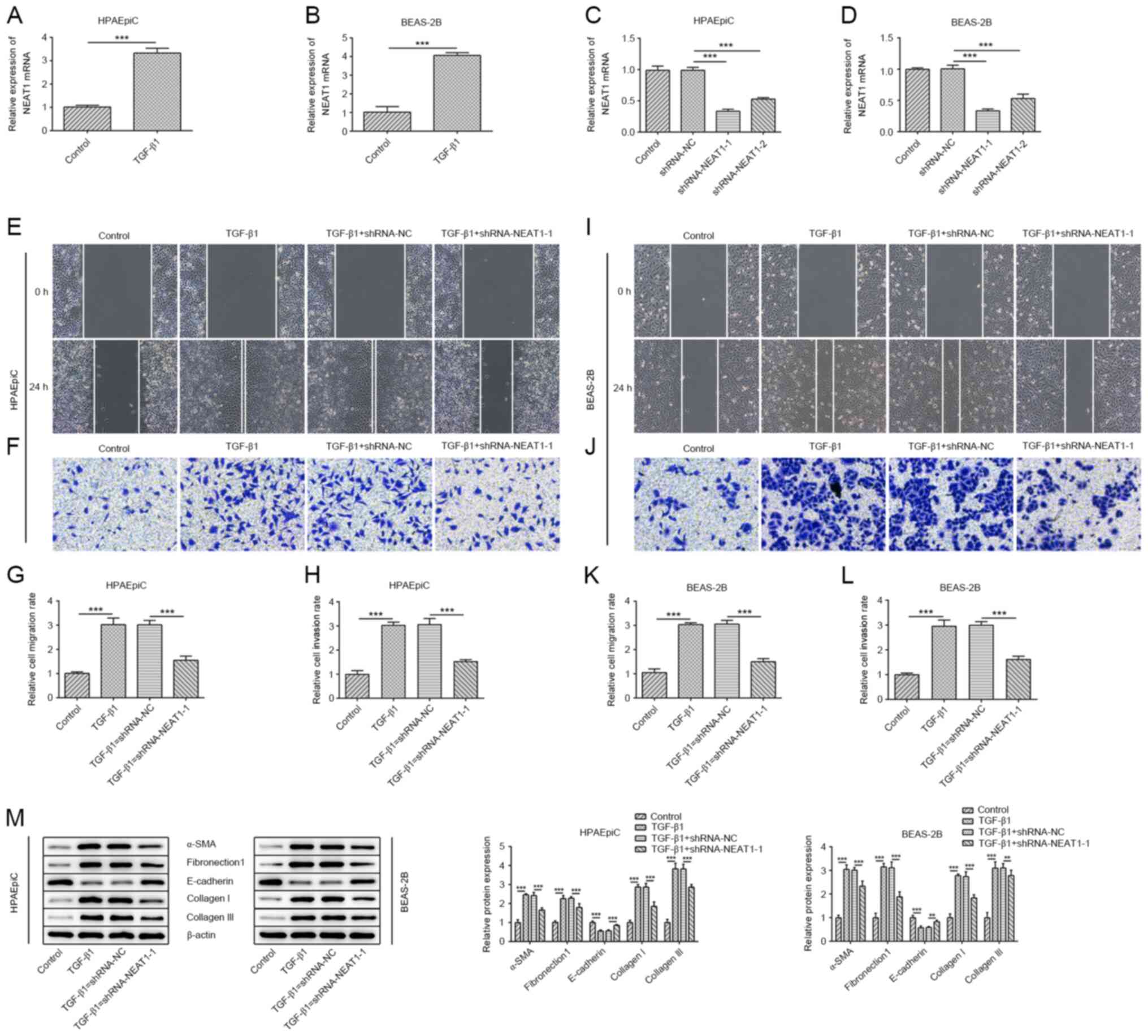 | Figure 1.NEAT1 knockdown inhibits epithelial
cell migration, EMT and collagen generation. (A) HPAEpiC and (B)
BEAS-2B cells were treated with 10 ng/ml TGF-β1 for 48 h then, the
mRNA expression of NEAT1 was determined using RT-qPCR. Transfection
efficiency of shRNA-NEAT1 was validated using RT-qPCR in (C)
HPAEpiC and (D) BEAS-2B cells. A wound healing assay was performed
to assess the migratory ability of the (E) HPAEpiC and (I) BEAS-2B
cell lines (magnification, ×100). The migration rate of (G) HPAEpiC
and (K) BEAS-2B cells. A Transwell assay was performed to assess
the invasive ability of the (F) HPAEpiC and (J) BEAS-2B cell lines
(magnification, ×100). The invasion rate of (H) HPAEpiC and (L)
BEAS-2B cell lines. (M) The protein expression levels of
E-cadherin, α-SMA, collagen I, collagen III and fibronectin1 in the
HPAEpiC and BEAS-2B cell lines were evaluated using western blot
analysis. **P<0.01, ***P<0.001. sh, short hairpin; RT-qPCR,
reverse transcription-quantitative PCR; α-SMA, α smooth muscle
actin; NEAT1, nuclear enriched abundant transcript 1; NC, negative
control. |
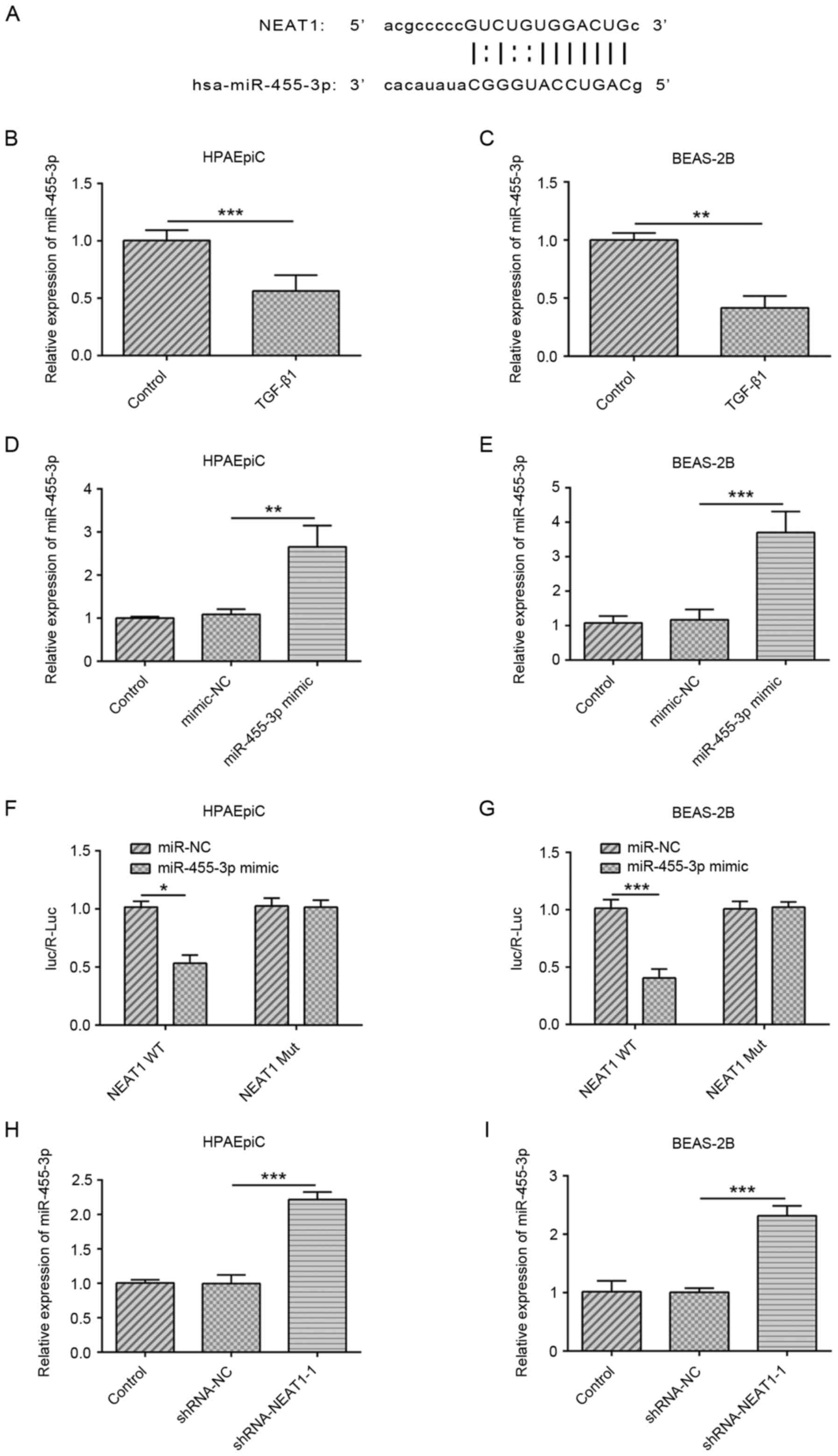
NEAT1 modulates the expression of
miR-455-3p
NEAT1 was predicted to be a sponge of miR-455-3p
using the StarBase software (Fig.
2A). miR-455-3p expression in TGF-β1-treated HPAEpiC and
BEAS-2B cell lines were decreased (Fig.
2B and C). The transfection efficiency of the miR-455-3p mimic
is shown in Fig. 2D and E. A
luciferase reporter assay was performed to validate the interaction
between miR-455-3p and NEAT1, and the luciferase activity was found
to be downregulated in cells co-transfected with NEAT1-WT and
miR-455-3p mimic, suggesting that miR-455-3p could bind to NEAT1
(Fig. 2F and G). Furthermore, the
expression of miR-455-3p was significantly higher in
shRNA-NEAT1-1-transfected cells compared with that in the shRNA-NC
group (Fig. 2H and I). These data
suggested that NEAT1 may bind to miR-455-3p and regulate the
expression levels of miR-455-3p.
Regulation of NEAT1 in migration,
collagen production and EMT depends on miR-455-3p
Subsequently, a miR-455-3p inhibitor was generated
and the transfection efficiency was determined (Fig. 3A and B). As shown in Fig. 3C-J, the results of the wound healing
and Transwell invasion assays indicated that the miR-455-3p
inhibitor promoted the migratory and invasive abilities of the
HPAEpiC and BEAS-2B cell lines. Furthermore, knockdown of NEAT1
counteracted the effects of TGF-β1 on the expression levels of
E-cadherin, whereas the miR-455-3p inhibitor further abolished the
function of shRNA-NEAT1-1. The protein expression levels of
fibronectin1, α-SMA, collagen I and collagen III showed the
opposite trend to E-cadherin (Fig.
3K).
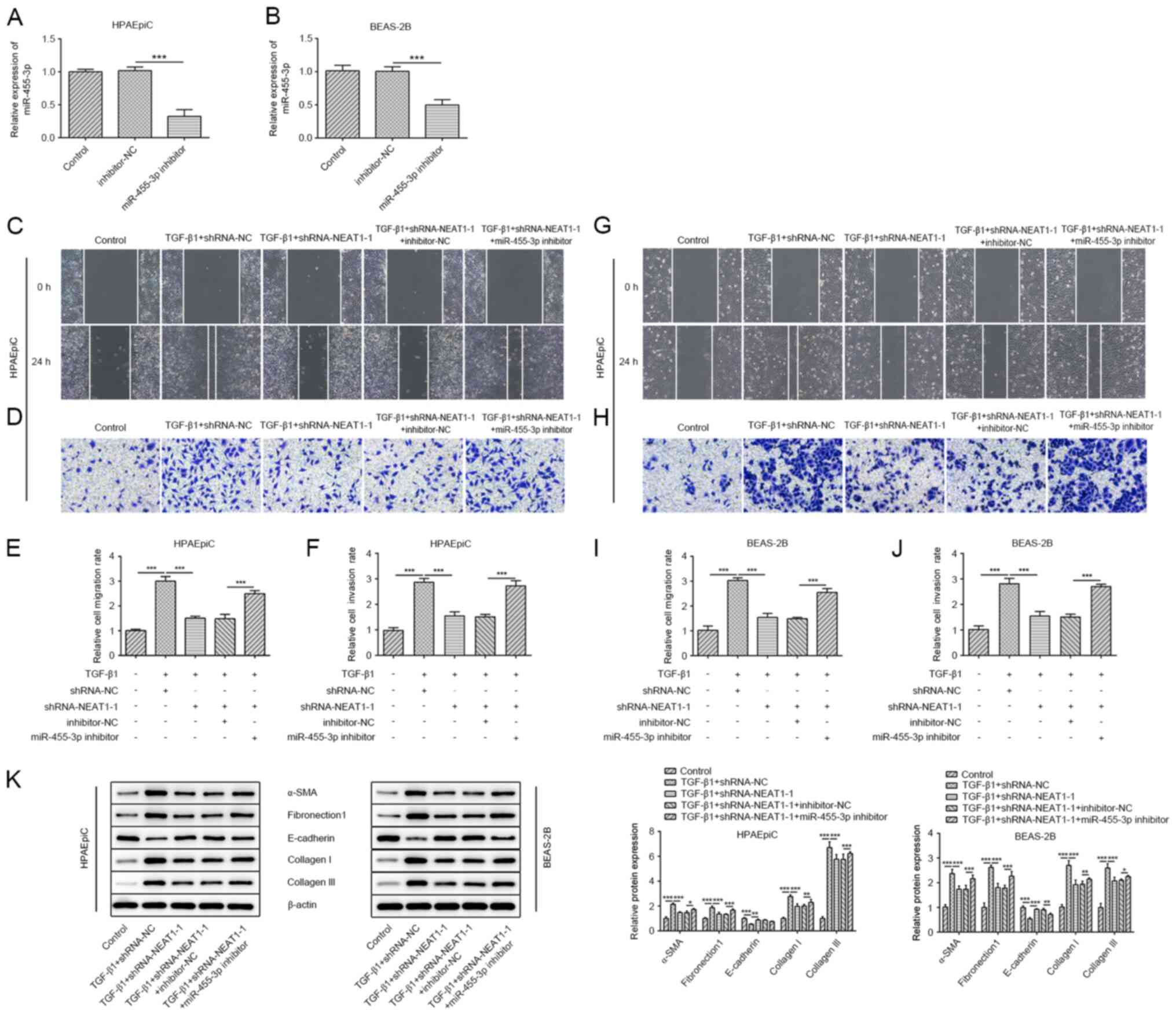 | Figure 3.Regulation of NEAT1 in migration,
collagen production and EMT depends on miR-455-3p. The mRNA
expression levels of miR-455-3p in the (A) HPAEpiC and (B) BEAS-2B
cell lines transfected with miR-455-3p inhibitor were determined
using RT-qPCR. A wound healing assay was performed to assess the
migratory ability of the (C) HPAEpiC and (G) BEAS-2B cell lines
(magnification, ×100). The migration rate of (E) HPAEpiC and (I)
BEAS-2B cells. A Transwell assay was performed to assess the
invasion ability of the (D) HPAEpiC and (H) BEAS-2B cell lines
(magnification, ×100). The invasion rate of (F) HPAEpiC and (J)
BEAS-2B cells. (K) The protein expression levels of E-cadherin,
α-SMA, collagen I, collagen III and fibronectin1 in the HPAEpiC and
BEAS-2B cell lines were evaluated using western blot analysis.
*P<0.05, **P<0.01, ***P<0.001. RT-qPCR, reverse
transcription-quantitative PCR; miR, microRNA; α-SMA, α smooth
muscle actin; NC, negative control; NEAT1, nuclear enriched
abundant transcript 1; sh, short hairpin. |
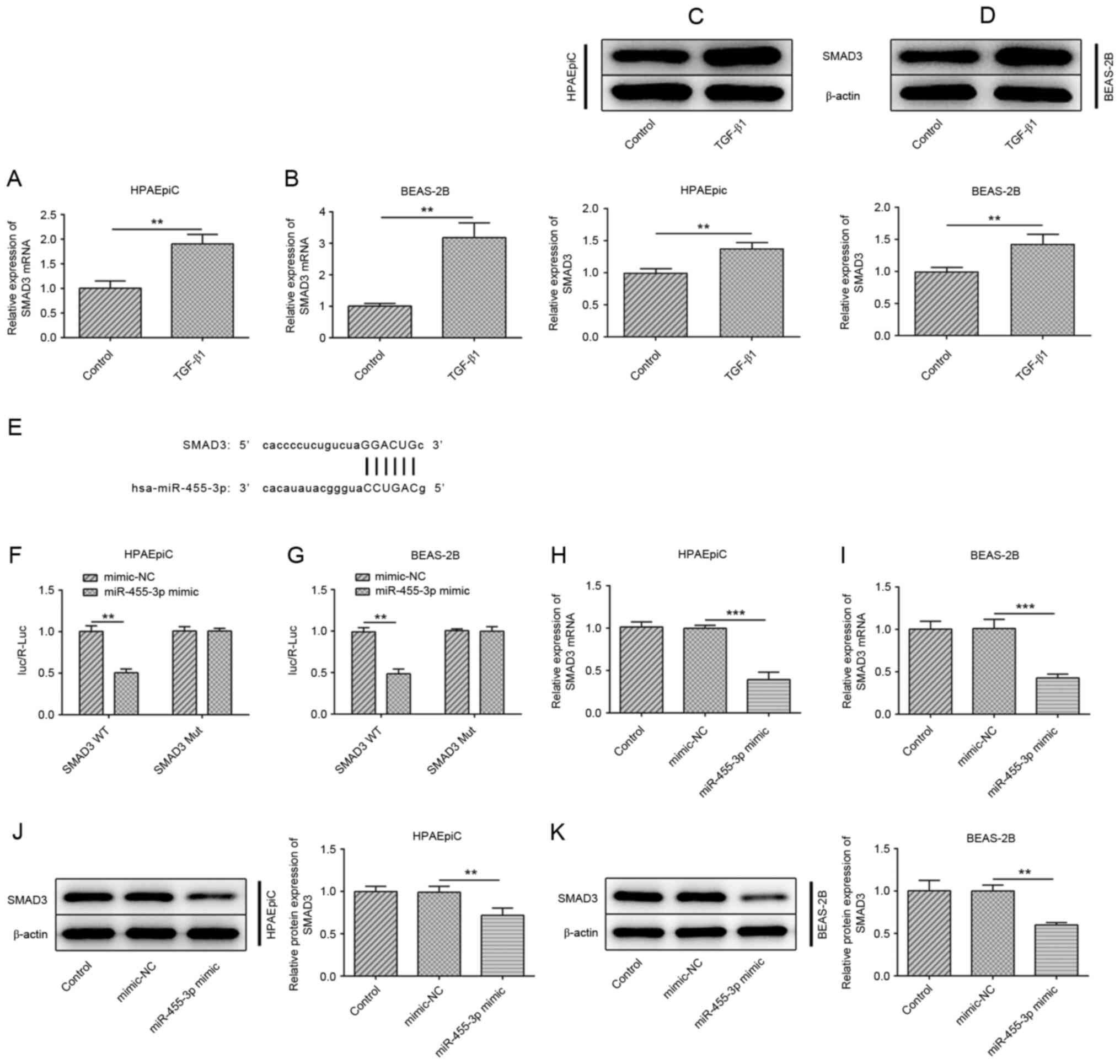
SMAD3 serves as a target of
miR-455-3p
SMAD3 has been identified to be an important
mediator in the progression of pulmonary fibrosis (17,18).
In the TGF-β1-treated HPAEpiC and BEAS-2B cell lines, the mRNA
expression levels of SMAD3 were increased compared with that in the
control group (Fig. 4A and B),
while SMAD3 was also elevated at the protein level (Fig. 4C and D). Notably, SMAD3 was
predicted as a target gene of miR-455-3p using the StarBase
software (Fig. 4E), and the results
from a luciferase reporter assay revealed decreased luciferase
activity in the group co-transfected with SMAD3 WT and miR-455-3p
mimic, demonstrating that there was an interaction between
miR-455-3p and SMAD3 (Fig. 4F and
G). Furthermore, the mRNA and protein expression levels of
SMAD3 were downregulated in the HPAEpiC or BEAS-2B cell lines
transfected with miR-455-3p mimic (Fig.
4H-K).
NEAT1/miR-455-3p mediates migration,
EMT and collagen generation via SMAD3
Compared with the control group, knockdown of NEAT1
was found to significantly inhibit the expression of SMAD3, which
was rescued by the miR-455-3p inhibitor (Fig. 5A). As shown in Fig. 5B, the transfection efficiency of
pcDNA3.1-SMAD3 was validated using western blot analysis. It was
found that the effects of co-transfection with shRNA-NEAT1-1 and
miR-455-3p mimic could further inhibit the migratory and invasive
abilities of the epithelial cells compared with that in cells
transfected with shRNA-NEAT1-1 alone, following treatment with
TGF-β1; however, overexpression of SMAD3 weakened the synergistic
effect of shRNA-NEAT1-1 and miR-455-3p mimic (Fig. 5C-J). Similar results were observed
in EMT; miR-455-3p mimic enhanced the inhibition of shRNA-NEAT1-1
on the protein expression levels of α-SMA, collagen I, collagen III
and fibronectin1, but upregulated the expression of E-cadherin.
SMAD3 overexpression reversed the effect of shRNA-NEAT1-1 and
miR-455-3p mimic (Fig. 5K). Taken
together, these data demonstrated that the inhibitory effects of
shRNA-NEAT1-1 and miR-455-3p on migration, EMT and collagen
generation were abrogated by the overexpression of SMAD3.
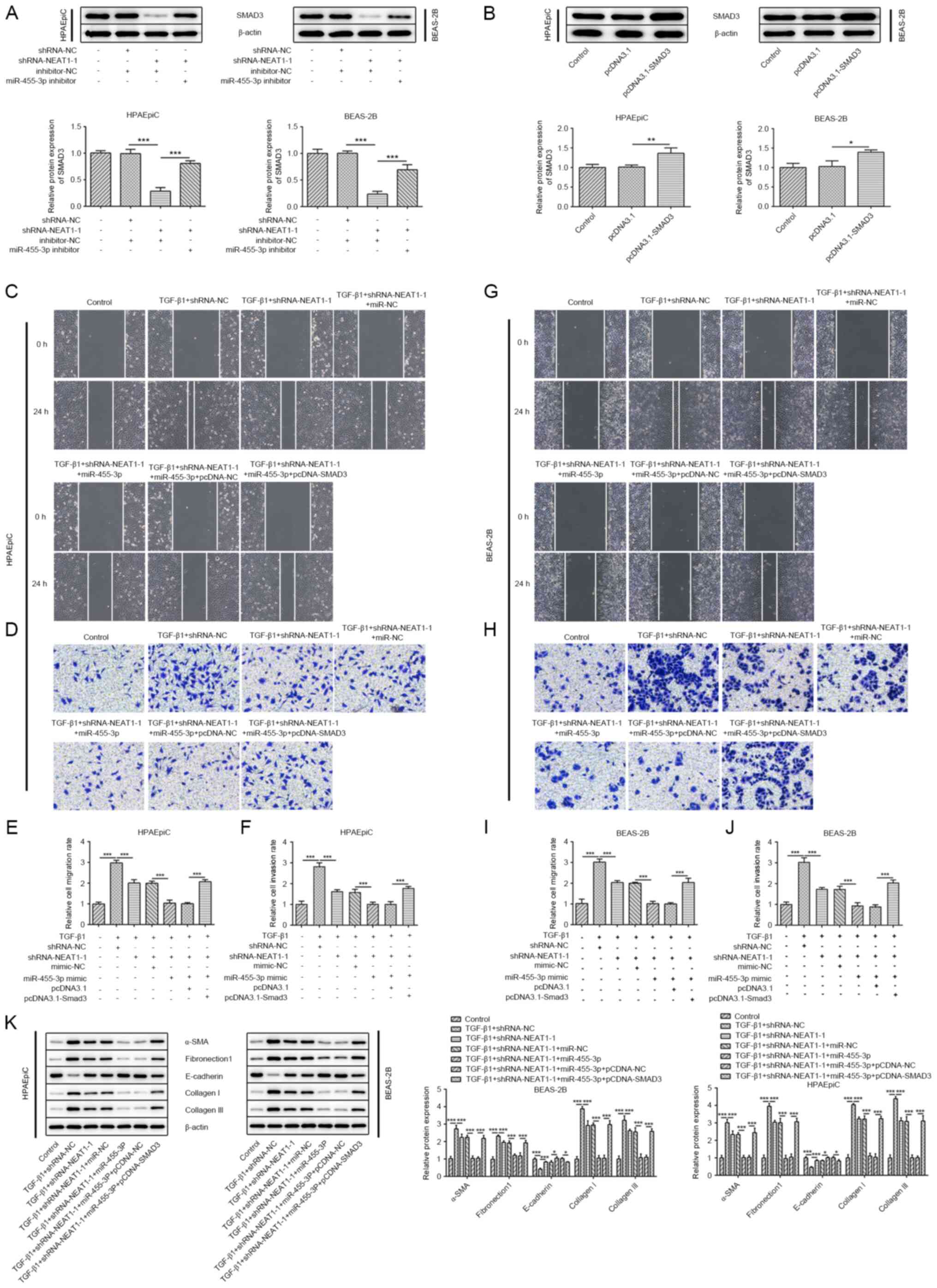 | Figure 5.NEAT1/miR-455-3p mediates migration,
EMT and collagen generation via SMAD3. (A) The protein expression
levels of SMAD3 in the HPAEpiC and BEAS-2B cell lines transfected
with shRNA-NEAT1-1 and/or miR-455-3p inhibitor were determined
using western blot analysis. (B) The protein expression levels of
SMAD3 in the HPAEpiC and BEAS-2B cell lines transfected with
pcDNA3.1-SMAD3. A wound healing assay was performed to assess the
migratory ability of (C) HPAEpiC and (G) BEAS-2B cell lines
(magnification, ×100). Migration rate of (E) HPAEpiC and (I)
BEAS-2B cells. A Transwell assay was performed to assess the
invasive ability of the (D) HPAEpiC and (H) BEAS-2B cell lines
(magnification, ×100). Invasion rate of (F) HPAEpiC and (J) BEAS-2B
cells. (K) The protein expression levels of E-cadherin, α-SMA,
collagen I, collagen III and fibronectin1 in the HPAEpiC and
BEAS-2B cell lines were evaluated using western blot analysis.
*P<0.05, **P<0.01, ***P<0.001. NEAT1, nuclear enriched
abundant transcript 1; sh, short hairpin; miR, microRNA; NC,
negative control; α-SMA, α smooth muscle actin. |
Discussion
Numerous studies have reported the emerging role of
lncRNAs in the pathogenesis of pulmonary fibrosis (19–21). A
previous study found that NEAT1 was increased in human fibrotic
liver samples and murine fibrotic livers (12), while Huang et al (13) reported that NEAT1 accelerated
fibrosis in diabetic nephropathy. Non-coding RNA-NEAT1 has been
demonstrated to play a role in the differentiation and regulation
of the development of various diseases (22,23).
For example, NEAT1 was reported to be highly expressed in
Parkinson's disease, and NEAT1 knockdown inhibited PD progression
via regulating the miR-212-3p/axin 1 signaling pathway (24). NEAT1 also accelerated apoptosis and
inflammation in lipopolysaccharide-induced sepsis models by
targeting miR-590-3p (25). In the
present study, NEAT1 was found to be upregulated in the
TGF-β1-treated HPAEpiC and BEAS-2B cell lines, suggesting that the
high expression level of NEAT1 could be associated with pulmonary
fibrosis. TGF-β1 is commonly considered to be a potent pro-fibrotic
factor, which could stimulate epithelial-derived fibroblasts to
produce fibronectin and collagen, the main components of the ECM
(26,27). Knockdown of NEAT1 weakened the
abilities of EMT and collagen production in epithelial cells in the
present study. A previous study demonstrated that downregulation of
NEAT1 reduced the expression levels of collagen I and fibronectin
in mouse mesangial cells (28).
Jin et al (29) found that NEAT1 promoted liver
fibrosis by mediating miR-506 and transcriptional activator GLI3.
In addition, NEAT1 has been found to impair lung function through
the interaction with miR-124 (30).
Understanding the crosstalk between lncRNA, miRNA and mRNA, and
their regulatory pattern could provide a novel perspective for the
therapy of pulmonary fibrosis. In the present study, NEAT1 was
identified to be a sponge of miR-455-3p. miR-455-3p is lowly
expressed in various tumors, including prostate (31), colorectal (32), pancreatic (33) and breast cancer (34). miR-455-3p serves as an important
regulator in organ fibrosis, including pulmonary fibrosis (10,35,36).
The present study found that the miR-455-3p inhibitor partially
reversed the regulatory effects of shRNA-NEAT1-1 on EMT and
collagen generation. SMAD3 is commonly known as a downstream
intracellular effector of TGF-β1 (37); in addition, SMAD3 was identified as
a target mRNA of miR-455-3p and overexpression of SMAD3 abolished
the effects of NEAT1/miR-455-3p on cell fibrosis in the present
study.
In summary, the present study illustrated that NEAT1
knockdown alleviated TGF-β1-induced epithelial cell migration, EMT
and collagen production by regulating the miR-455-3p/SMAD3 axis.
These findings suggested that NEAT1 and miR-455-3p may be potential
targets for treatment of pulmonary fibrosis. However, the use of
only in vitro methods was a limitation of the present study.
Therefore, in subsequent research, an in vivo pulmonary
fibrosis model will be established to determine the expression
levels of NEAT1, miR-455-3p or SMAD3 in fibrotic lung tissues,
furthermore the role of NEAT1 in pulmonary fibrosis requires
validation by interfering with the expression of NEAT in lung
tissue.
Acknowledgements
Not applicable.
Funding
This work was supported by National Natural Science
Foundation of China (no. 81960304) and Natural Science Foundation
of Inner Mongolia [no. 2017MS (LH) 0812].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YL, FAL, LW and CFW searched the literature,
designed the experiments and performed the experiments. FAL and YFW
analyzed and interpreted the data. YFW and CFW wrote the
manuscript. CFW revised the manuscript. YL and CFW confirmed the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ley B, Collard HR and King TE Jr: Clinical
course and prediction of survival in idiopathic pulmonary fibrosis.
Am J Respir Crit Care Med. 183:431–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Povedano JM, Martinez P, Flores JM, Mulero
F and Blasco MA: Mice with Pulmonary Fibrosis Driven by Telomere
Dysfunction. Cell Rep. 12:286–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Selman M and Pardo A: Role of epithelial
cells in idiopathic pulmonary fibrosis: From innocent targets to
serial killers. Proc Am Thorac Soc. 3:364–372. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Willis BC, Liebler JM, Luby-Phelps K,
Nicholson AG, Crandall ED, du Bois RM and Borok Z: Induction of
epithelial-mesenchymal transition in alveolar epithelial cells by
transforming growth factor-beta1: Potential role in idiopathic
pulmonary fibrosis. Am J Pathol. 166:1321–1332. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hou J, Shi J, Chen L, Lv Z, Chen X, Cao H,
Xiang Z and Han X: M2 macrophages promote myofibroblast
differentiation of LR-MSCs and are associated with pulmonary
fibrogenesis. Cell Commun Signal. 16:892018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saito S, Zhuang Y, Shan B, Danchuk S, Luo
F, Korfei M, Guenther A and Lasky JA: Tubastatin ameliorates
pulmonary fibrosis by targeting the TGFβ-PI3K-Akt pathway. PLoS
One. 12:e01866152017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li S, Geng J, Xu X, Huang X, Leng D, Jiang
D, Liang J, Wang C, Jiang D and Dai H: miR-130b-3p Modulates
Epithelial-Mesenchymal Crosstalk in Lung Fibrosis by Targeting
IGF-1. PLoS One. 11:e01504182016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wei S, Wang Q, Zhou H, Qiu J, Li C, Shi C,
Zhou S, Liu R and Lu L: miR-455-3p Alleviates Hepatic Stellate Cell
Activation and Liver Fibrosis by Suppressing HSF1 Expression. Mol
Ther Nucleic Acids. 16:758–769. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu XJ, Gong Z, Li SJ, Jia HP and Li DL:
Long non-coding RNA Hottip modulates high-glucose-induced
inflammation and ECM accumulation through miR-455-3p/WNT2B in mouse
mesangial cells. Int J Clin Exp Pathol. 12:2435–2445.
2019.PubMed/NCBI
|
|
12
|
Yu F, Jiang Z, Chen B, Dong P and Zheng J:
NEAT1 accelerates the progression of liver fibrosis via regulation
of microRNA-122 and Kruppel-like factor 6. J Mol Med (Berl).
95:1191–1202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang S, Xu Y, Ge X, Xu B, Peng W, Jiang
X, Shen L and Xia L: Long noncoding RNA NEAT1 accelerates the
proliferation and fibrosis in diabetic nephropathy through
activating Akt/mTOR signaling pathway. J Cell Physiol.
234:11200–11207. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yao W, Li Y, Han L, Ji X, Pan H, Liu Y,
Yuan J, Yan W and Ni C: The CDR1as/miR-7/TGFBR2 Axis Modulates EMT
in Silica-Induced Pulmonary Fibrosis. Toxicol Sci. 166:465–478.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao X, Sun J, Chen Y, Su W, Shan H, Li Y,
Wang Y, Zheng N, Shan H and Liang H: lncRNA PFAR Promotes Lung
Fibroblast Activation and Fibrosis by Targeting miR-138 to Regulate
the YAP1-Twist Axis. Mol Ther. 26:2206–2217. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang X, Cheng Z, Dai L, Jiang T, Jia L,
Jing X, An L, Wang H and Liu M: Knockdown of Long Noncoding RNA H19
Represses the Progress of Pulmonary Fibrosis through the
Transforming Growth Factor β/Smad3 Pathway by Regulating MicroRNA
140. Mol Cell Biol. 39:e00143–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu Q, Han L, Yan W, Ji X, Han R, Yang J,
Yuan J and Ni C: miR-489 inhibits silica-induced pulmonary fibrosis
by targeting MyD88 and Smad3 and is negatively regulated by lncRNA
CHRF. Sci Rep. 6:309212016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun J, Su W, Zhao X, Shan T, Jin T, Guo Y,
Li C, Li R, Zhou Y, Shan H, et al: lncRNA PFAR contributes to
fibrogenesis in lung fibroblasts through competitively binding to
miR-15a. Biosci Rep. 39:BSR201902802019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao X, Sun J, Chen Y, Su W, Shan H, Li Y,
Wang Y, Zheng N, Shan H and Liang H: lncRNA PFAR Promotes Lung
Fibroblast Activation and Fibrosis by Targeting miR-138 to Regulate
the YAP1-Twist Axis. Mol Ther. 26:2206–2217. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hao X, Du Y, Qian L, Li D and Liu X:
Upregulation of long noncoding RNA AP003419.16 predicts high risk
of aging associated idiopathic pulmonary fibrosis. Mol Med Rep.
16:8085–8091. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li JH, Zhang SQ, Qiu XG, Zhang SJ, Zheng
SH and Zhang DH: Long non-coding RNA NEAT1 promotes malignant
progression of thyroid carcinoma by regulating miRNA-214. Int J
Oncol. 50:708–716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, Wang S, Li Z, Long X, Guo Z, Zhang
G, Zu J, Chen Y and Wen L: Retracted: NEAT1 induces
epithelial-mesenchymal transition and 5-FU resistance through the
miR-129/ZEB2 axis in breast cancer. FEBS Lett. Nov 1–2016.(Epub
ahead of print). doi: 10.1002/1873-3468.12474.
|
|
24
|
Liu T, Zhang Y, Liu W and Zhao J: lncRNA
NEAT1 Regulates the Development of Parkinson's Disease by Targeting
AXIN1 Via Sponging miR-212-3p. Neurochem Res. Nov 26–2020.(Epub
ahead of print). doi: 10.1007/s11064-020-03157-1. View Article : Google Scholar
|
|
25
|
Liu L, Liu F, Sun Z, Peng Z, You T and Yu
Z: lncRNA NEAT1 promotes apoptosis and inflammation in LPS-induced
sepsis models by targeting miR-590-3p. Exp Ther Med. 20:3290–3300.
2020.PubMed/NCBI
|
|
26
|
Dong C, Gongora R, Sosulski ML, Luo F and
Sanchez CG: Regulation of transforming growth factor-beta1
(TGF-β1)-induced pro-fibrotic activities by circadian clock gene
BMAL1. Respir Res. 17:42016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshida M, Romberger DJ, Illig MG,
Takizawa H, Sacco O, Spurzem JR, Sisson JH, Rennard SI and Beckmann
JD: Transforming growth factor-beta stimulates the expression of
desmosomal proteins in bronchial epithelial cells. Am J Respir Cell
Mol Biol. 6:439–445. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma J, Zhao N, Du L and Wang Y:
Downregulation of lncRNA NEAT1 inhibits mouse mesangial cell
proliferation, fibrosis, and inflammation but promotes apoptosis in
diabetic nephropathy. Int J Clin Exp Pathol. 12:1174–1183.
2019.PubMed/NCBI
|
|
29
|
Jin SS, Lin XF, Zheng JZ, Wang Q and Guan
HQ: lncRNA NEAT1 regulates fibrosis and inflammatory response
induced by nonalcoholic fatty liver by regulating miR-506/GLI3. Eur
Cytokine Netw. 30:98–106. 2019.PubMed/NCBI
|
|
30
|
Li X, Ye S and Lu Y: Long non-coding RNA
NEAT1 overexpression associates with increased exacerbation risk,
severity, and inflammation, as well as decreased lung function
through the interaction with microRNA-124 in asthma. J Clin Lab
Anal. 34:e230232020.PubMed/NCBI
|
|
31
|
Zhao Y, Yan M, Yun Y, Zhang J, Zhang R, Li
Y, Wu X, Liu Q, Miao W and Jiang H: MicroRNA-455-3p functions as a
tumor suppressor by targeting eIF4E in prostate cancer. Oncol Rep.
37:2449–2458. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng J, Lin Z, Zhang L and Chen H:
MicroRNA-455-3p Inhibits Tumor Cell Proliferation and Induces
Apoptosis in HCT116 Human Colon Cancer Cells. Med Sci Monit.
22:4431–4437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhan T, Huang X, Tian X, Chen X, Ding Y,
Luo H and Zhang Y: Downregulation of MicroRNA-455-3p Links to
Proliferation and Drug Resistance of Pancreatic Cancer Cells via
Targeting TAZ. Mol Ther Nucleic Acids. 10:215–226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guo J, Liu C, Wang W, Liu Y, He H, Chen C,
Xiang R and Luo Y: Identification of serum miR-1915-3p and
miR-455-3p as biomarkers for breast cancer. PLoS One.
13:e02007162018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou Y and Chai X: Protective effect of
bicyclol against pulmonary fibrosis via regulation of
microRNA-455-3p in rats. J Cell Biochem. 121:651–660. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu J, Liu J, Ding Y, Zhu M, Lu K, Zhou J,
Xie X, Xu Y, Shen X, Chen Y, et al: MiR-455-3p suppresses renal
fibrosis through repression of ROCK2 expression in diabetic
nephropathy. Biochem Biophys Res Commun. 503:977–983. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|