|
1
|
Liu J, Liu T, Wang X and He A: Circles
reshaping the RNA world: From waste to treasure. Mol Cancer.
16:582017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
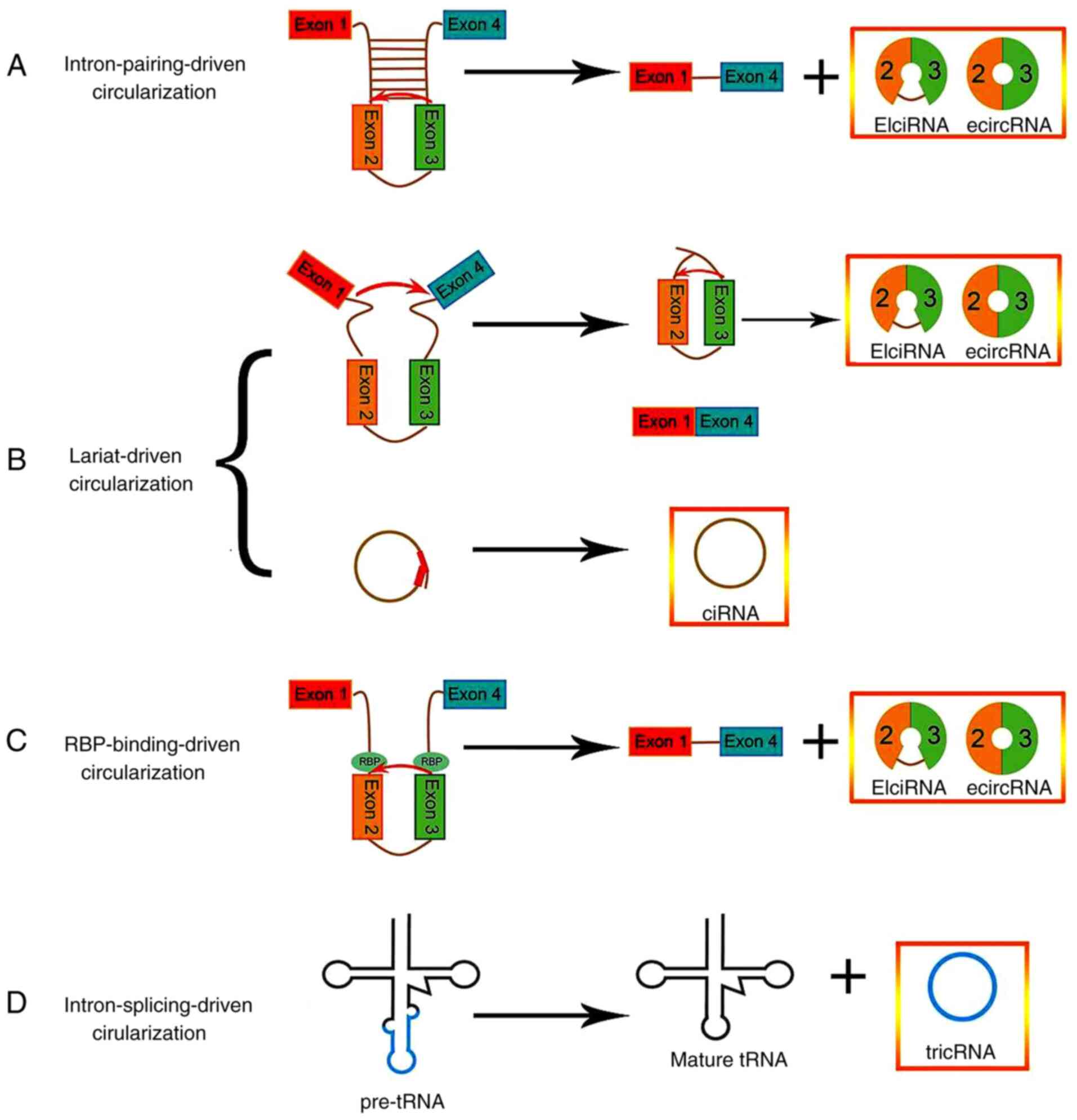
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
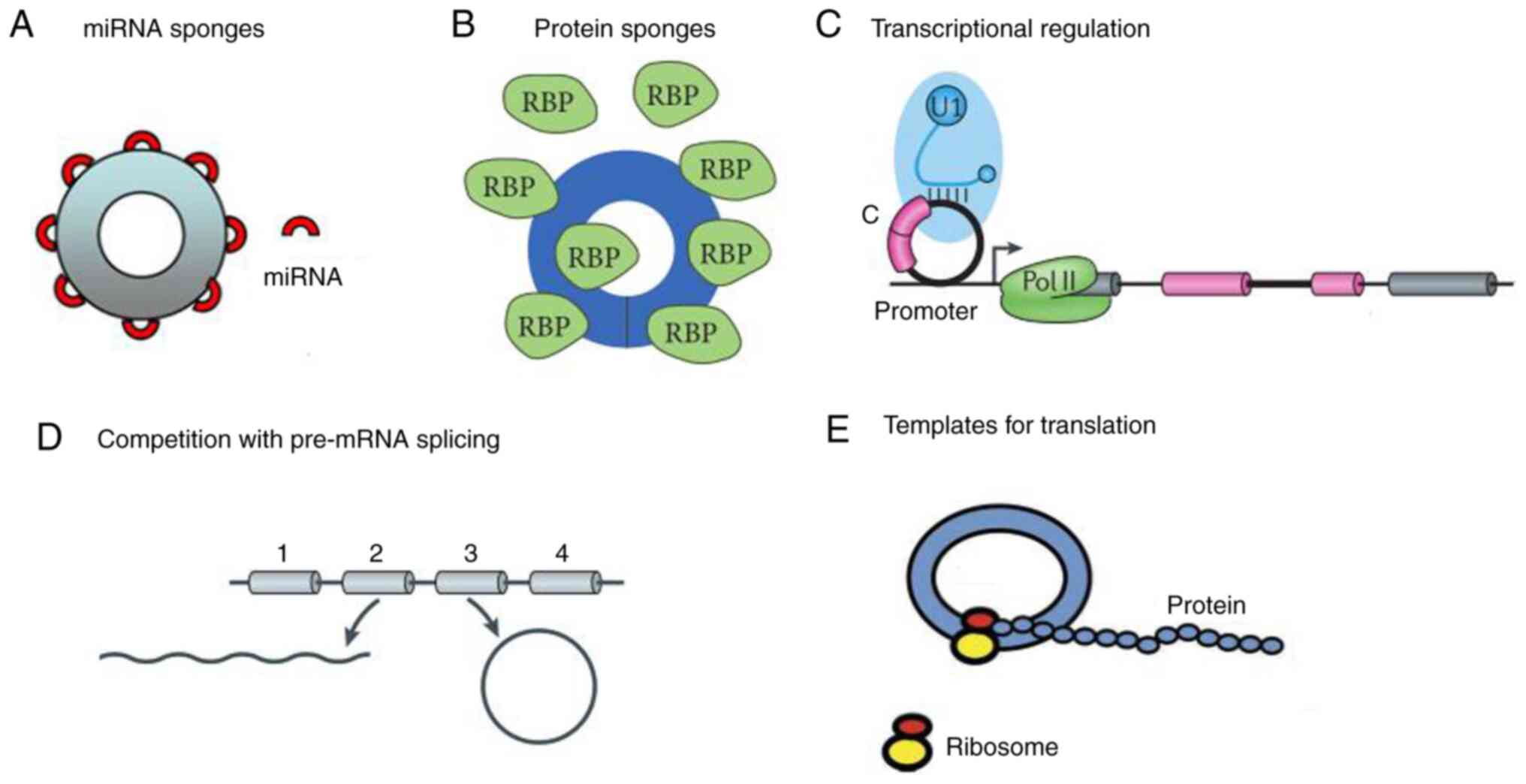
6
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang W, Qin P, Gong X, Huang L, Wang C,
Chen G, Chen J, Wang L and Lv Z: Identification of circRNAs in the
liver of whitespotted bamboo shark (Chiloscyllium
plagiosum). Front Genet. 11:5963082020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang J, Liu R, Zhu Y, Gong J, Yin S, Sun
P, Feng H, Wang Q, Zhao S, Wang Z and Li G: Identification and
characterization of circRNAs responsive to methyl jasmonate in
Arabidopsis thaliana. Int J Mol Sci. 21:7922020. View Article : Google Scholar
|
|
9
|
Li L, Sun D, Li X, Yang B and Zhang W:
Identification of key circRNAs in non-small cell lung cancer. Am J
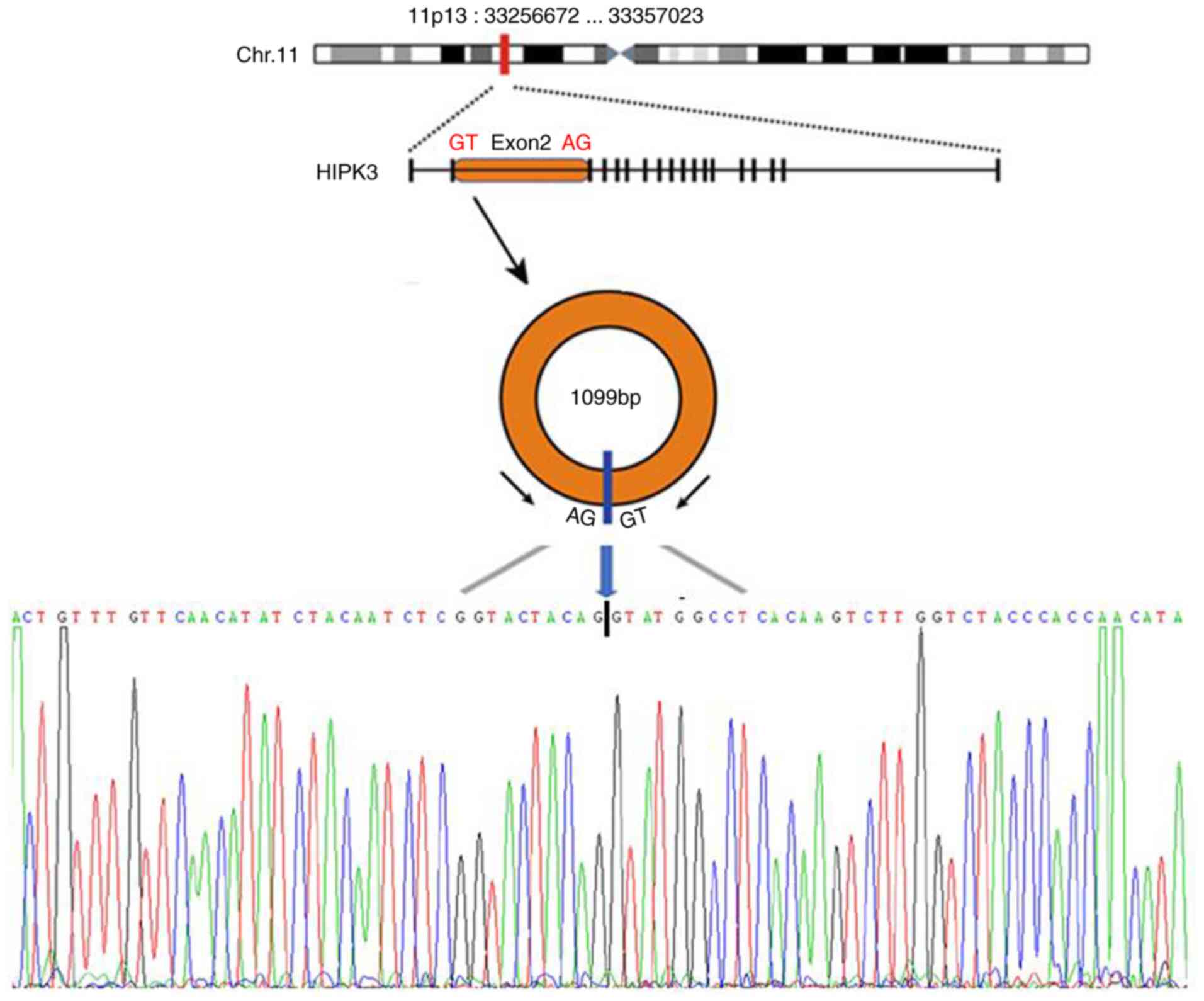
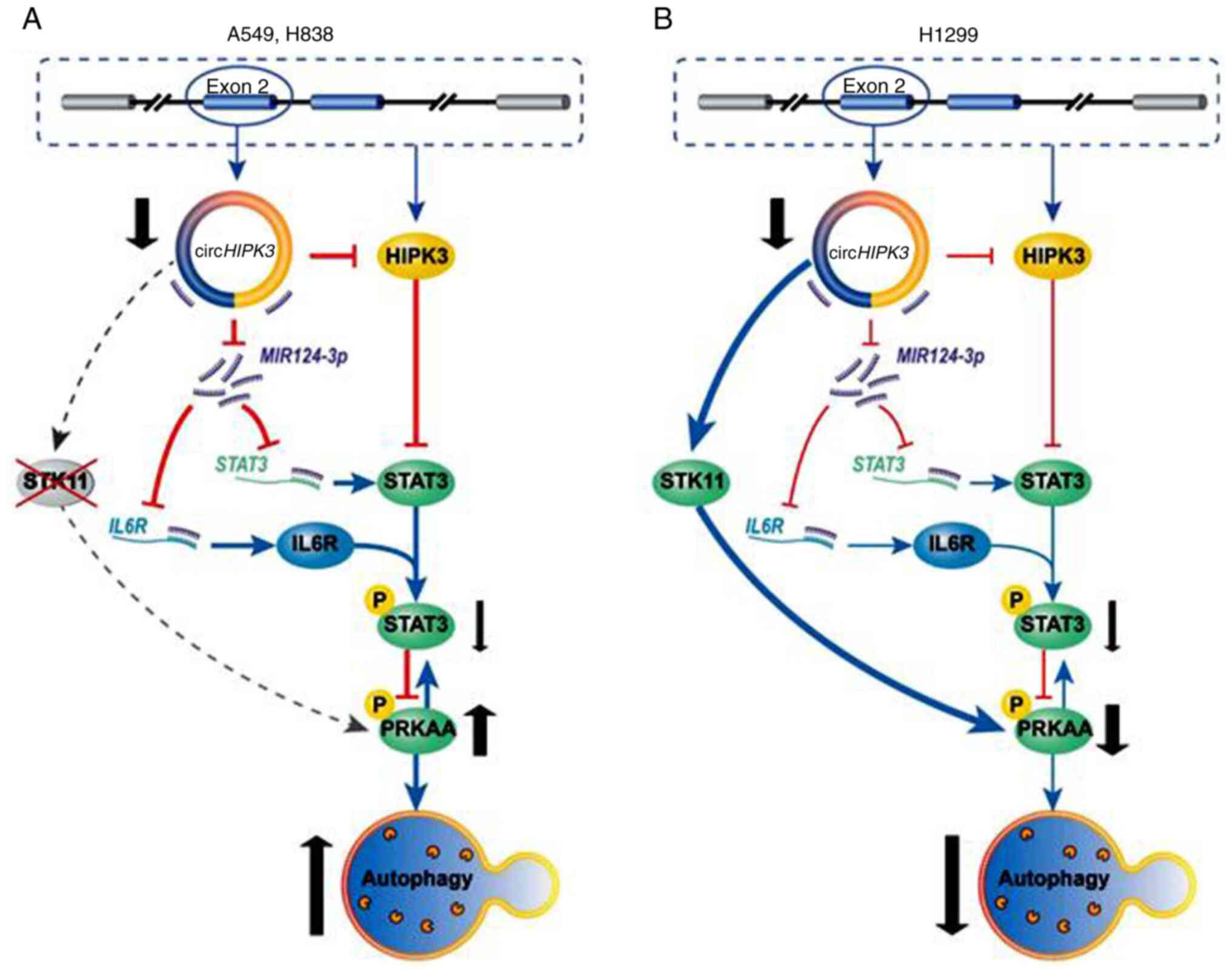
Med Sci. 361:98–105. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Geng X, Jia Y, Zhang Y, Shi L, Li Q, Zang
A and Wang H: Circular RNA: Biogenesis, degradation, functions and
potential roles in mediating resistance to anticarcinogens.
Epigenomics. 12:267–283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Salzman J: Circular RNA expression: Its
potential regulation and function. Trends Genet. 32:309–316. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wan B, Liu B and Lv C: Progress of
research into circular RNAs in urinary neoplasms. PeerJ.
8:e86662020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jahani S, Nazeri E, Majidzadeh-A K, Jahani
M and Esmaeili R: Circular RNA; a new biomarker for breast cancer:
A systematic review. J Cell Physiol. 235:5501–5510. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang G, Li S, Yang N, Zou Y, Zheng D and
Xiao T: Recent progress in circular RNAs in human cancers. Cancer
Lett. 404:8–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee ECS, Elhassan SAM, Lim GPL, Kok WH,
Tan SW, Leong EN, Tan SH, Chan EWL, Bhattamisra SK, Rajendran R and
Candasamy M: The roles of circular RNAs in human development and
diseases. Biomed Pharmacother. 111:198–208. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lei K, Bai H, Wei Z, Xie C, Wang J, Li J
and Chen Q: The mechanism and function of circular RNAs in human
diseases. Exp Cell Res. 368:147–158. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Soghli N, Qujeq D, Yousefi T and Soghli N:
The regulatory functions of circular RNAs in osteosarcoma.
Genomics. 112:2845–2856. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Conte A and Pierantoni GM: Update on the
regulation of HIPK1, HIPK2 and HIPK3 protein kinases by microRNAs.
Microrna. 7:178–186. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: circHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wen Y, Li B, He M, Teng S, Sun Y and Wang
G: circHIPK3 promotes proliferation and migration and invasion via
regulation of miR-637/HDAC4 signaling in osteosarcoma cells. Oncol
Rep. 45:169–179. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Y, Liu Q and Liao Q: circHIPK3: A
promising cancer-related circular RNA. Am J Transl Res.
12:6694–6704. 2020.PubMed/NCBI
|
|
24
|
Wang J, Zhu M, Pan J, Chen C, Xia S and
Song Y: Circular RNAs: A rising star in respiratory diseases.
Respir Res. 20:32019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie Y, Yuan X, Zhou W, Kosiba AA, Shi H,
Gu J and Qin Z: The circular RNA HIPK3 (circHIPK3) and its
regulation in cancer progression: Review. Life Sci. 254:1172522020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ragan C, Goodall GJ, Shirokikh NE and
Preiss T: Insights into the biogenesis and potential functions of
exonic circular RNA. Sci Rep. 9:20482019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang C, Ma L, Niu Y, Wang Z, Xu X, Li Y
and Yu Y: Circular RNA in lung cancer research: Biogenesis,
functions, and roles. Int J Biol Sci. 16:803–814. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Panda AC, Grammatikakis I, Munk R, Gorospe
M and Abdelmohsen K: Emerging roles and context of circular RNAs.
Wiley Interdiscip Rev RNA. 8:10.1002/wrna.1386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kelly S, Greenman C, Cook PR and
Papantonis A: Exon skipping is correlated with exon
circularization. J Mol Biol. 427:2414–2417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stagsted LVW, O'Leary ET, Ebbesen KK and
Hansen TB: The RNA-binding protein SFPQ preserves long-intron
splicing and regulates circRNA biogenesis in mammals. Elife.
10:e630882021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schmidt CA, Giusto JD, Bao A, Hopper AK
and Matera AG: Molecular determinants of metazoan tricRNA
biogenesis. Nucleic Acids Res. 47:6452–6465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wen J, Liao J, Liang J, Chen XP, Zhang B
and Chu L: Circular RNA HIPK3: A key circular RNA in a variety of
human cancers. Front Oncol. 10:7732020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer
A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J
and Califano A: An extensive microRNA-mediated network of RNA-RNA
interactions regulates established oncogenic pathways in
glioblastoma. Cell. 147:370–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Capel B, Swain A, Nicolis S, Hacker A,
Walter M, Koopman P, Goodfellow P and Lovell-Badge R: Circular
transcripts of the testis-determining gene Sry in adult mouse
testis. Cell. 73:1019–1030. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: CircNT5E acts as a sponge of miR-422a to
promote glioblastoma tumorigenesis. Cancer Res. 78:4812–4825. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang
L, Ma J, Li X, Zeng Y, Yang Z, et al: A circular RNA promotes
tumorigenesis by inducing c-myc nuclear translocation. Cell Death
Differ. 24:1609–1620. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Barrett SP and Salzman J: Circular RNAs:
Analysis, expression and potential functions. Development.
143:1838–1847. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Joassard OR, Bélanger G, Karmouch J, Lunde
JA, Shukla AH, Chopard A, Legay C and Jasmin BJ: HuR mediates
changes in the stability of AChR β-subunit mRNAs after skeletal
muscle denervation. J Neurosci. 35:10949–10962. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by circPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fang L, Du WW, Awan FM, Dong J and Yang
BB: The circular RNA circ-Ccnb1 dissociates Ccnb1/Cdk1 complex
suppressing cell invasion and tumorigenesis. Cancer Lett.
459:216–226. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chao CW, Chan DC, Kuo A and Leder P: The
mouse formin (Fmn) gene: Abundant circular RNA transcripts and
gene-targeted deletion analysis. Mol Med. 4:614–628. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Conn VM, Hugouvieux V, Nayak A, Conos SA,
Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta
C and Conn SJ: A circRNA from SEPALLATA3 regulates splicing of its
cognate mRNA through R-loop formation. Nat Plants. 3:170532017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li Y, Ge YZ, Xu L and Jia R: Circular RNA
ITCH: A novel tumor suppressor in multiple cancers. Life Sci.
254:1171762020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of CircRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen CY and Sarnow P: Initiation of
protein synthesis by the eukaryotic translational apparatus on
circular RNAs. Science. 268:415–417. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar
|
|
55
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei
P, Liu H, Xu J, Xiao F, Zhou H, et al: A peptide encoded by
circular form of LINC-PINT suppresses oncogenic transcriptional
elongation in glioblastoma. Nat Commun. 9:44752018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Paschalis A and de Bono JS: Prostate
cancer 2020: ‘The times they are a'changing’. Cancer Cell.
38:25–27. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C
and Sun F: circHIPK3 overexpression accelerates the proliferation
and invasion of prostate cancer cells through regulating
miRNA-338-3p. Onco Targets Ther. 12:3363–3372. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen D, Lu X, Yang F and Xing N: Circular
RNA circHIPK3 promotes cell proliferation and invasion of prostate
cancer by sponging miR-193a-3p and regulating MCL1 expression.
Cancer Manag Res. 11:1415–1423. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yu H, Chen Y and Jiang P: Circular RNA
HIPK3 exerts oncogenic properties through suppression of miR-124 in
lung cancer. Biochem Biophys Res Commun. 506:455–462. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lu H, Han X, Ren J, Ren K, Li Z and Sun Z:
Circular RNA HIPK3 induces cell proliferation and inhibits
apoptosis in non-small cell lung cancer through sponging miR-149.
Cancer Biol Ther. 21:113–121. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hong W, Zhang Y, Ding J, Yang Q, Xie H and
Gao X: circHIPK3 acts as competing endogenous RNA and promotes
non-small-cell lung cancer progression through the miR-107/BDNF
signaling pathway. Biomed Res Int. 2020:60759022020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Chen X, Mao R, Su W, Yang X, Geng Q, Guo
C, Wang Z, Wang J, Kresty LA, Beer DG, et al: Circular RNA
circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα
signaling in STK11 mutant lung cancer. Autophagy. 16:659–671. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Malvezzi M, Carioli G, Bertuccio P,
Boffetta P, Levi F, La Vecchia C and Negri E: European cancer
mortality predictions for the year 2018 with focus on colorectal
cancer. Ann Oncol. 29:1016–1022. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yan Y, Su M and Qin B: circHIPK3 promotes
colorectal cancer cells proliferation and metastasis via modulating
of miR-1207-5p/FMNL2 signal. Biochem Biophys Res Commun.
524:839–846. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ilson DH: Advances in the treatment of
gastric cancer: 2019. Curr Opin Gastroenterol. 35:551–554. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cheng J, Zhuo H, Xu M, Wang L, Xu H, Peng
J, Hou J, Lin L and Cai J: Regulatory network of circRNA-miRNA-mRNA
contributes to the histological classification and disease
progression in gastric cancer. J Transl Med. 16:2162018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Wei J, Xu H, Wei W, Wang Z, Zhang Q, De W
and Shu Y: circHIPK3 promotes cell proliferation and migration of
gastric cancer by sponging miR-107 and regulating BDNF expression.
Onco Targets Ther. 13:1613–1624. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F and Jiang G: circHIPK3 sponges
miR-558 to suppress heparanase expression in bladder cancer cells.
EMBO Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xie F, Zhao N, Zhang H and Xie D: Circular
RNA circHIPK3 promotes gemcitabine sensitivity in bladder cancer. J
Cancer. 11:1907–1912. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Akcay M, Etiz D, Celik O and Ozen A:
Evaluation of prognosis in nasopharyngeal cancer using machine
learning. Technol Cancer Res Treat. 19:15330338209098292020.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ke Z, Xie F, Zheng C and Chen D: circHIPK3
promotes proliferation and invasion in nasopharyngeal carcinoma by
abrogating miR-4288-induced ELF3 inhibition. J Cell Physiol.
234:1699–1706. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Sharma A, Sharma KL, Gupta A, Yadav A and
Kumar A: Gallbladder cancer epidemiology, pathogenesis and
molecular genetics: Recent update. World J Gastroenterol.
23:3978–3998. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kai D, Yannian L, Yitian C, Dinghao G, Xin
Z and Wu J: Circular RNA HIPK3 promotes gallbladder cancer cell
growth by sponging microRNA-124. Biochem Biophys Res Commun.
503:863–869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Chen G, Shi Y, Liu M and Sun J: circHIPK3
regulates cell proliferation and migration by sponging miR-124 and
regulating AQP3 expression in hepatocellular carcinoma. Cell Death
Dis. 9:1752018. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wang Y, Zhang Y, Yang T, Zhao W, Wang N,
Li P, Zeng X and Zhang W: Long non-coding RNA MALAT1 for promoting
metastasis and proliferation by acting as a ceRNA of miR-144-3p in
osteosarcoma cells. Oncotarget. 8:59417–59434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Xiao-Long M, Kun-Peng Z and Chun-Lin Z:
Circular RNA circ_HIPK3 is down-regulated and suppresses cell
proliferation, migration and invasion in osteosarcoma. J Cancer.
9:1856–1862. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Quartuccio N, Laudicella R, Vento A,
Pignata S, Mattoli MV, Filice R, Comis AD, Arnone A, Baldari S,
Cabria M and Cistaro A: The additional value of 18F-FDG
PET and MRI in patients with glioma: A review of the literature
from 2015 to 2020. Diagnostics (Basel). 10:3572020. View Article : Google Scholar
|
|
83
|
Hu D and Zhang Y: Circular RNA HIPK3
promotes glioma progression by binding to miR-124-3p. Gene.
690:81–89. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jin P, Huang Y, Zhu P, Zou Y, Shao T and
Wang O: CircRNA circHIPK3 serves as a prognostic marker to promote
glioma progression by regulating miR-654/IGF2BP3 signaling. Biochem
Biophys Res Commun. 503:1570–1574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Cai Z and Liu Q: Understanding the global
cancer statistics 2018: Implications for cancer control. Sci China
Life Sci. Aug 26–2019.(Epub ahead of print). doi:
10.1007/s11427-019-9816-1. View Article : Google Scholar
|
|
86
|
Teng F, Xu J, Zhang M, Liu S, Gu Y, Zhang
M, Wang X, Ni J, Qian B, Shen R and Jia X: Comprehensive circular
RNA expression profiles and the tumor-suppressive function of
circHIPK3 in ovarian cancer. Int J Biochem Cell Biol. 112:8–17.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liang S, Zhang S, Wang P, Yang C, Shang C,
Yang J and Wang J: LncRNA, TUG1 regulates the oral squamous cell
carcinoma progression possibly via interacting with Wnt/β-catenin
signaling. Gene. 608:49–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang J, Zhao SY, Ouyang SS, Huang ZK, Luo
Q and Liao L: Circular RNA circHIPK3 acts as the sponge of
microRNA-124 to promote human oral squamous cell carcinoma cells
proliferation. Zhonghua Kou Qiang Yi Xue Za Zhi. 53:546–551.
2018.(In Chinese). PubMed/NCBI
|
|
89
|
Minciacchi VR, Kumar R and Krause DS:
Chronic myeloid leukemia: A model disease of the past, present and
future. Cells. 10:1172021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Feng XQ, Nie SM, Huang JX, Li TL, Zhou JJ,
Wang W, Zhuang LK and Meng FJ: Circular RNA circHIPK3 serves as a
prognostic marker to promote chronic myeloid leukemia progression.
Neoplasma. 67:171–177. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Congdon NG, Friedman DS and Lietman T:
Important causes of visual impairment in the world today. JAMA.
290:2057–2060. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Liu X, Liu B, Zhou M, Fan F, Yu M, Gao C,
Lu Y and Luo Y: Circular RNA HIPK3 regulates human lens epithelial
cells proliferation and apoptosis by targeting the miR-193a/CRYAA
axis. Biochem Biophys Res Commun. 503:2277–2285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Gathiram P and Moodley J: Pre-eclampsia:
Its pathogenesis and pathophysiolgy. Cardiovasc J Afr. 27:71–78.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang Y, Cao L, Jia J, Ye L, Wang Y, Zhou
B and Zhou R: circHIPK3 is decreased in preeclampsia and affects
migration, invasion, proliferation, and tube formation of human
trophoblast cells. Placenta. 85:1–8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Vera R, Dotor E, Feliu J, González E,
Laquente B, Macarulla T, Martínez E, Maurel J, Salgado M and
Manzano JL: SEOM clinical guideline for the treatment of pancreatic
cancer (2016). Clin Transl Oncol. 18:1172–1178. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Xu T, Wu J, Han P, Zhao Z and Song X:
Circular RNA expression profiles and features in human tissues: A
study using RNA-seq data. BMC Genomics. 18 (Suppl 6):S6802017.
View Article : Google Scholar
|
|
97
|
Liu Y, Xia L, Dong L, Wang J, Xiao Q, Yu X
and Zhu H: circHIPK3 promotes gemcitabine (GEM) resistance in
pancreatic cancer cells by sponging miR-330-5p and targets RASSF1.
Cancer Manag Res. 12:921–929. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Rahman A, O'Connor DB, Gather F, Koscic S,
Gilgan J, Mockler D, Bashir Y, Memba R, Duggan SN and Conlon KC:
Clinical classification and severity scoring systems in chronic
pancreatitis: A systematic review. Dig Surg. 37:181–191. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Wang L, Luo T, Bao Z, Li Y and Bu W:
Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic
rats. Biochem Biophys Res Commun. 505:644–650. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Wang J, Li X, Liu Y, Peng C, Zhu H, Tu G,
Yu X and Li Z: circHIPK3 promotes pyroptosis in acinar cells
through regulation of the miR-193a-5p/GSDMD axis. Front Med
(Lausanne). 7:882020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Holmström L, Haukilahti A, Vähätalo J,
Kenttä T, Appel H, Kiviniemi A, Pakanen L, Huikuri HV, Myerburg RJ
and Junttila J: Electrocardiographic associations with myocardial
fibrosis among sudden cardiac death victims. Heart. 106:1001–1006.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y,
Shen G and Wang F: Inhibition of circHIPK3 prevents angiotensin
II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol.
292:188–196. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Gotua M, Gamkrelidze A, Rukhadze M,
Abramidze T, Bochorishvili E, Shengelidze G, Dolidze N,
Chkhartishvili E, Bachert C, Pfaar O, et al: 2020 Aria care
pathways for allergic rhinitis-georgia. Georgian Med News. 108–117.
2019.PubMed/NCBI
|
|
104
|
Zhu X, Wang X, Wang Y and Zhao Y: The
regulatory network among circHIPK3, LncGAS5, and miR-495 promotes
Th2 differentiation in allergic rhinitis. Cell Death Dis.
11:2162020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Taneja K and Williamson SR: Updates in
pathologic staging and histologic grading of renal cell carcinoma.
Surg Pathol Clin. 11:797–812. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Lai J, Xin J, Fu C and Zhang W: circHIPK3
promotes proliferation and metastasis and inhibits apoptosis of
renal cancer cells by inhibiting MiR-485-3p. Cancer Cell Int.
20:2482020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li H, Heng B, Ouyang P, Xie X, Zhang T,
Chen G, Chen Z, Cheang K and Lai C: Comprehensive profiling of
circRNAs and the tumor suppressor function of circHIPK3 in clear
cell renal carcinoma. J Mol Histol. 51:317–327. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Han B, Shaolong E, Luan L, Li N and Liu X:
circHIPK3 promotes clear cell renal cell carcinoma (ccRCC) cells
proliferation and metastasis via altering of miR-508-3p/CXCL13
signal. Onco Targets Ther. 13:6051–6062. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Qian W, Huang T and Feng W: Circular RNA
HIPK3 promotes EMT of cervical cancer through sponging miR-338-3p
to up-regulate HIF-1α. Cancer Manag Res. 12:177–187. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Chen S, Fan X, Gu H, Zhang L and Zhao W:
Competing endogenous RNA regulatory network in papillary thyroid
carcinoma. Mol Med Rep. 18:695–704. 2018.PubMed/NCBI
|
|
112
|
Shu T, Yang L, Sun L, Lu J and Zhan X:
circHIPK3 promotes thyroid cancer tumorigenesis and invasion
through the Mirna-338-3p/RAB23 axis. Med Princ Pract. Oct
26–2020.(Epub ahead of print). doi: 10.1159/000512548. View Article : Google Scholar
|
|
113
|
Ji F, Yang CQ, Li XL, Zhang LL, Yang M, Li
JQ, Gao HF, Zhu T, Cheng MY, Li WP, et al: Risk of breast
cancer-related death in women with a prior cancer. Aging (Albany
NY). 12:5894–5906. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Howell A, Anderson AS, Clarke RB, Duffy
SW, Evans DG, Garcia-Closas M, Gescher AJ, Key TJ, Saxton JM and
Harvie MN: Risk determination and prevention of breast cancer.
Breast Cancer Res. 16:4462014. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chen ZG, Zhao HJ, Lin L, Liu JB, Bai JZ
and Wang GS: Circular RNA CirCHIPK3 promotes cell proliferation and
invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis.
Thorac Cancer. 11:2660–2671. 2020. View Article : Google Scholar : PubMed/NCBI
|