Introduction
Acute myeloid leukemia (AML) is a heterogeneous
malignant disorder characterized by uncontrolled proliferation of
hematopoietic progenitor cells and differentiation arrest (1). AML is the most common form of acute
leukemia and the second most common type of leukemia diagnosed
worldwide (2). The incidence of AML
is directly proportional to an increase in age (3). Previous studies have reported that AML
results from mutations in different genes involved in the processes
of cell proliferation, survival and apoptosis (4). For instance, mutations in RAS,
tyrosine kinase signaling pathways (CBL, FLT3, JAK2, KIT,
PTPN11), chromatin modifiers (ASXL1/2, BCOR/L1, TET2)
and an additional 50 genes have been found to be associated with
occurrence of AML (5). According to
cytogenetic information, patients with AML can be divided into
three risk-based subgroups: Favorable, intermediate and poor
(6). Although great progress has
been made in chemotherapy-based regimens and hematopoietic stem
cell transplantation, the prognosis of AML remains unsatisfactory,
and the 5-year survival rate is <30% (7). Early diagnosis of AML can increase the
overall survival rate of patients with AML (3). Therefore, it is necessary to identify
novel and sensitive biomarkers for the diagnosis and prognosis of
patients with AML.
Long non-coding RNAs (lncRNAs) are a group of
non-coding RNAs that are between 200 bp and 100 kb in size
(8). Due to the absence of open
reading frames, lncRNAs lack the ability to encode proteins
(9). Mounting evidence has
suggested that lncRNAs serve essential roles in the regulation of
various cellular biological processes, such as proliferation,
differentiation, metabolism and programmed cell death (10). Aberrant expression of non-coding
RNAs has been observed in multiple diseases, including cancer
(11). To date, numerous lncRNAs,
such as nuclear paraspeckle assembly transcript 1 (NEAT1), HOX
transcript antisense RNA (HOTAIR) and metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1), have been identified as
important factors in the regulation of AML (12). lncRNA 00460 (LINC00460) is a newly
discovered lncRNA located on human chromosome 13q33.2 with a
transcript size of 935 bp (13).
Previous studies have revealed that LINC00460 functions as an
oncogene in various malignancies, such as lung cancer, colorectal
cancer, gastric cancer, ovarian cancer and osteosarcoma (14–18).
However, the status of LINC00460 expression and its prognostic
significance in AML remain elusive. To address this question, the
present study aimed to investigate the diagnostic and prognostic
value of serum LINC00460 in patients with AML.
MicroRNAs (miRNAs/miRs) are another type of
non-coding RNA, with a size of ~20 nucleotides, that can inhibit
target mRNAs at the post-transcriptional level (19). Previous studies have reported that
miRNAs serve essential roles in the initiation and progression of
various cancer types, including AML (20). Among the various miRNAs, miR-320b
has been implicated in the tumorigenesis of different cancer types,
such as glioma, colorectal cancer, lung cancer and prostate cancer
(21–24). However, knowledge regarding the role
of miR-320b in AML is currently lacking.
PBX homeobox 3 (PBX3) is a homeodomain-containing
transcription factor that belongs to the pre-B cell leukemia family
(25). The role of PBX3 has been
extensively studied in AML. For instance, enhanced expression of
PBX3 with its cofactor Meis homeobox 1 (MEIS1) can transform normal
hematopoietic stem cells into AML in mice (26). Furthermore, silencing PBX3 can
increase the sensitivity of AML cells to chemotherapy agents
(27). However, to the best of the
authors' knowledge, there are no reports investigating the
association between LINC00460, miR-320b and PBX3.
The present study examined the prognostic and
biological roles of LINC00460 in AML. The in vivo expression
of LINC00460 was measured by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR). The effects of LINCO00460 on
the viability, cell cycle distribution and apoptosis were assayed
in vitro. Furthermore, the mechanisms underlying the
functions of LINCO00460 were also investigated. The findings
suggested that LINC00460 might be a prognostic biomarker and
therapeutic target for AML.
Materials and methods
Patient samples
A total of 80 diagnosed AML cases were recruited
between December 2018 and January 2010 at the First Affiliated
Hospital of Wenzhou Medical University, which were classified as
favorable-risk cytogenetic (20 subjects), intermediate-risk
cytogenetic (42 subjects) and poor-risk cytogenetic (18 subjects).
The patients consisted of 38 females and 42 males, aged 37–87
years. The median value of expression of LINC00460 was 2.36 and was
used as a cutoff value. All patients were divided into two groups:
High LINC00460 expression group (>2.36; n=36) and low LINC00460
expression group (≤2.36; n=44). Of the 80 patients with AML, 45
cases were classified as cytogenetically normal-AML (CN-AML). The
diagnosis of patients with AML was made according to the
French-American-British (FAB) and World Health Organization
criteria combined with immunophenotyping and cytogenetic analysis
(28). Additionally, 67 healthy
volunteers without any type of malignancy or other benign disease
were enrolled as controls.
Firstly, the serum (0.5 ml) was collected from all
patients before any treatment. After receiving different treatments
[chemotherapy and (n=41)/or targeted therapy (n=1)], 42 patients
achieved complete remission (CR), the serum (0.5 ml) of these
patients was collected again.
This research was approved by the Ethics Committee
of Wenzhou Medical University. Written informed consent was
obtained from all patients prior to participation in the study.
Cell culture and transfection
Human bone marrow stromal cells (HS-5) and human AML
cells (THP-1, KG-1, K562 and HL-60) were purchased from the Cell
Bank of Shanghai Institute of Biological Sciences, Chinese Academy
of Sciences. All cells were cultured in RPMI-1640 medium (HyClone;
Cytiva) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Thermo
Fisher Scientific, Inc.) at 37°C with 5% CO2.
For transfection, small interfering RNA (siRNA)
against LINC00460 (si-LINC00460), negative control siRNA (si-NC),
pcDNA3.1 vector, LINC00460 overexpression vector (pcDNA-LINC00460),
miR-320b mimic, miR-NC, miR-320b inhibitor, miR-NC inhibitor and
PBX3 overexpression vector (pcDNA-PBX3) were synthesized by Suzhou
GenePharma Co., Ltd. The target sequences were as follows:
si-LINC00460, 5′-CACACUUCTCGGCUAAG-3′; si-NC,
5′-AACAGGCAUCCUACGACGCCA-3′; miR-320b mimics,
5′-AAAGCUGGGUUGAGAGGGCAA-3′; miR-NC, 5′-AAUUCUCCGAACGUGUCACTT-3′;
miR-320b inhibitor, 5′-UUGCCCUCUCAACCCAGCUUU-3′; and miR-NC
inhibitor, 5′-AAACACGGUUAUAUCACCAUCGCAUUA-3′. All transfections
were conducted using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Cells were seeded into 6-well plate at the density of
3×105 cells/well in 2 ml complete medium. After 24 h of
incubation at 37°C, the medium was changed to 300 µl Opti-MEM
(Gibco; Thermo Fisher Scientific, Inc.) containing 2 µg pcDNA-PBX3
or 2 µg pcDNA-LINC00460 with 50 mM siRNA. To modulate the
expression of miR-320b, 20 mM miRNA mimics or 20 mM miRNA inhibitor
in 200 µl Opti-MEM containing 5 µl Lipofectamine®
2000was added into each well. A total of 4 h after incubation, 1.5
ml complete medium was added into each well and the cells were
continually incubated at 37°C for 48 h for following experiments.
Experiments were repeated ≥3 times.
Cell viability assay
Cell viability was measured using the MTT assay as
described previously (29). Cells
were seeded in 6-well plates at a density of 1×105
cells/well. After transfection for the indicated time, 20 µl 0.5
mg/ml MTT (Beyotime Institute of Biotechnology) was added to the
medium and incubated for another 4 h at 37°C. Before measurement,
150 µl DMSO was added, and a microplate reader (SpectraMax 190;
Molecular Devices, LLC) was used to measure the optical density at
450 nm. Experiments were repeated ≥3 times.
Cell cycle measurement
Cell cycle status was detected via flow cytometry
(FACScan; Becton, Dickinson and Company) and analyzed with FlowJo
v10.4 software (FlowJo LLC). Following the different transfections
as described above, the cells were harvested and cell pellets were
fixed in cold ethanol (75%) overnight at −20°C. The fixed cells
were then resuspended in 1 mg/ml RNase A (Sigma-Aldrich; Merck
KGaA) in PBS and incubated for 1 h at 37°C. The cells were stained
with 50 µg/ml PI (Sigma-Aldrich; Merck KGaA) for 0.5 h at room
temperature in the dark. The cells were then analyzed, and the
results are presented as the mean values from three independent
measurements. Experiments were repeated ≥3 times.
Cellular apoptosis measurement
The apoptosis of cells was measured using the Cell
Death Detection ELISAplus kit (Roche Diagnostics GmbH)
according to the manufacturer's instructions. The amount of
histone-coupled DNA was quantified by measuring the absorbance at
405 nm by the BioTek synergy multimode microplate reader (BioTek
Instruments, Inc.). The results were analyzed by the Gen5 v1.0
(BioTek Instruments, Inc.). Experiments were repeated ≥3 times.
Caspase-3 activity assay
The activity of caspase-3 was measured using the
caspase-3 Activity Assay kit (Fluorometric; Abcam) according to the
manufacturer's instructions. Experiments were repeated ≥3
times.
RNA purification and RT-qPCR
Whole blood samples (5 ml) were obtained from the
healthy controls and patients with AML before receiving any
therapy. All serum specimens were centrifuged at 500 × g for 5 min
and then centrifuged at 3,000 × g for 5 min, both at 4°C. Serum
samples were then stored at −80°C until further analysis.
Total RNA was isolated using TRIzol®
reagent (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The quality and concentration of RNA
were measured using a NanoDrop ND-1000 (Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using the
PrimerScript™ RT reagent kit (Takara Biotechnology Co., Ltd.). The
reverse transcription was conducted at 65°C for 10 min. qPCR was
performed on an Applied Biosystems 7500 Detection System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) using SYBR Premix ExTaq
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The thermocycling conditions were: 95°C for 10 min,
followed by 40 cycles of 95°C for 5 sec and 55°C for 5 sec and 72°C
for 5 sec, followed by 73°C for 30 sec as a final extension. The
relative expression of LINC00460 was assessed using the comparative
2−ΔΔCq method (30). The
expression levels of LINC00460 and miR-320b were normalized to the
expression levels of GAPDH and U6, respectively. The following
primers were used: LINC00460 forward, 5′-GGATGAACCACCATTGCC-3′ and
reverse, 5′-CCCACGCTCAGTCTTTCT-3′; miR-320b forward,
5′-TCCGAAACGGGAGAGTTGG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′,
GAPDH forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse,
5′-AGGGGCCATCCACAGTCTTC-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. Experiments were repeated ≥3
times.
Western blot analysis
Total cell lysates were collected using RIPA buffer
(Beyotime Institute of Biotechnology). The concentration of
proteins was measured using a BCA kit (Beyotime Institute of
Biotechnology). Equal amounts of protein (20 µg) were subjected to
12% SDS-PAGE and then transferred to PVDF membranes (Beyotime
Institute of Biotechnology). The membranes were blocked with 10%
skimmed milk for 1 h at room temperature and then incubated with
the primary antibodies at 4°C overnight. The following primary
antibodies were used: Bcl-2 (cat. no. 1507; 1:1,000), Bcl-xl (cat.
no. 2762; 1:1,000), caspase-3 (cat. no. 14220; 1:1,000), PBX3 (cat.
no. ab183849; 1:1,000; Abcam, USA), GAPDH (cat. no. 5174; 1:5,000).
The membranes were washed with PBS and then incubated with
secondary antibody at room temperature for 1 h. The following
secondary antibodies were used: HRP linked anti-mouse antibody
(cat. no. 7076; 1:5,000), HRP linked anti-rabbit antibody (cat. no.
7074; 1:5,000). All antibodies were purchased from Cell Signaling
Technology, Inc., and diluted at the ratio recommended by the
manufacturer. Finally, the protein bands were visualized using the
ECL reagent (Beyotime Institute of Biotechnology). Images were
analyzed using Chemidoc Touch Imaging System v1.2 (Bio-Rad
Laboratories, Inc.). The experiments were repeated ≥3 times.
Dual-luciferase assays
The wild-type (WT) and mutant (MUT) fragments of
LINC00460 and the 3′-untranslated region (UTR) of PBX3 containing
the miR-320b targeting sequences were synthesized and inserted into
the pGL3 promoter vector (Promega Corporation). Cells were seeded
into 6-well plate at the density of 3×105 cells/well in
2 ml complete medium. After 24 h of incubation at 37°C, the medium
was changed to 500 µl Opti-MEM (Gibco; Thermo Fisher Scientific,
Inc.) containing 1 µg plasmid, 20 mM miRNA mimics and 5 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were then cultured at 37°C. Finally, 48 h
after transfection, cells were collected and the Dual-Luciferase
Reporter Assay system (Promega Corporation) was used to measure
luminescence. Renilla luciferase activity was used to
normalize the firefly luciferase activity. Experiments were
repeated ≥3 times.
Bioinformatic analysis
StarBase Ver 3.0 (http://starbase.sysu.edu.cn) and TargetScan Ver 3.1
(http://www.targetscan.org/mamm_31/)
were used to predict the potential binding miRNAs of LINC00460 and
targets of miR-320b.
Statistical analysis
Statistical analyses were performed using SPSS 18.0
(SPSS, Inc.) or GraphPad 8.0 (GraphPad Software, Inc.) software.
Data were presented as mean ± standard deviation. The difference in
the relative serum LINC00460 expression between groups was
determined using the Mann-Whitney U test or Kruskal-Wallis test
followed by Dunn's post hoc test. The median value of LINC00460
expression was used as the cut-off value to designate the patients
with AML into a high LINC00460 group and a low LINC00460 group. A
receiver operating characteristic (ROC) curve and the area under
the curve (AUC) were used to assess the diagnostic value of serum
LINC00460 expression in patients with AML and healthy controls. The
Pearson χ2 test was used to assay intergroup
differences. The Cox proportional hazards regression model was
applied for univariate and multivariate analyses to estimate the
prognostic factors for survival prediction. Survival curves were
constructed via Kaplan-Meier survival analysis with the log-rank
test. Paired Student's t-test and One-way ANOVA followed by the
post hoc Tukey test were applied for comparing the difference
between two groups or multiple groups, respectively. P<0.05 was
considered to indicate a statistically significant difference.
Results
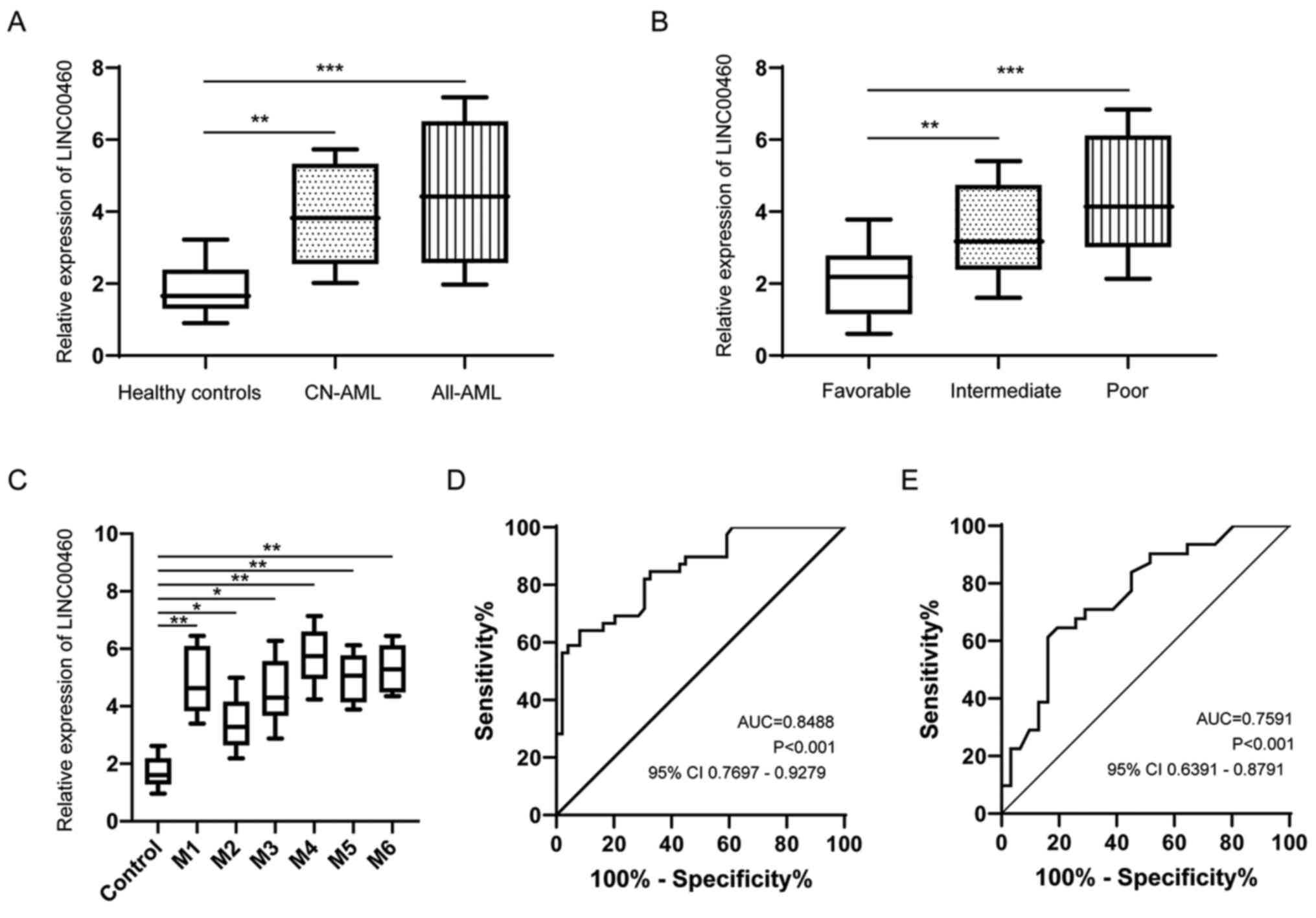
Expression of LINC00460 is upregulated
in patients with AML
First, the expression of LINC00460 in serum from 80
patients with AML and 67 healthy controls was measured via RT-qPCR.
LINC00460 expression was significantly upregulated in patients with
AML or CN-AML compared with those in healthy controls (Fig. 1A). Next, the patients with AML were
divided into favorable, intermediate and poor groups according to
their cytogenetic features (31).
It was identified that LINC00460 expression in patients with AML
with intermediate or poor cytogenetic risk subtypes was greatly
increased compared with that in patients with favorable cytogenetic
risk subtypes (Fig. 1B). Moreover,
the expression of LINC00460 was compared among different FAB
subtypes, but there was no significant difference among the various
subtypes (Fig. 1C). ROC curve
analysis demonstrated that serum LINC00460 could differentiate
patients with AML from healthy controls with an AUC value of 0.8488
(Fig. 1D). In addition, serum
LINC00460 could serve as a reliable biomarker for differentiating
patients with CN-AML from healthy controls with an AUC value of
0.7591 (Fig. 1E).
Association between the expression of
LINC00460 and clinicopathologic features of AML
The correlation between the expression of serum
LINC00460 and the clinicopathological characteristics of AML was
analyzed. The 80 patients with AML were grouped into a high serum
LINC00460 group (n=36) and a low serum LINC00460 group (n=44)
according to the median serum LINC00460 expression. As presented in
Tables I and II, serum LINC00460 levels were
significantly associated with clinicopathological features, such as
FAB classification and cytogenetics. However, there was no
significant correlation of LINC00460 expression with other clinical
features, including sex, age, white blood cell count, BM blasts,
extramedullary disease and CR (Table
I).
 | Table I.Relationship between serum LINC00460
expression and clinicopathologic features in acute myeloid
leukemia. |
Table I.
Relationship between serum LINC00460
expression and clinicopathologic features in acute myeloid
leukemia.
|
|
| Serum Linc0062
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | No. | High | Low | P-value |
|---|
| Sex |
|
|
|
|
|
Male | 42 | 18 | 24 | 0.685 |
|
Female | 38 | 18 | 20 |
|
| Age, years |
|
|
|
|
|
<60 | 47 | 21 | 26 | 0.945 |
|
≥60 | 33 | 15 | 18 |
|
| BM blasts, % |
|
|
|
|
|
<50 | 41 | 15 | 26 | 0.079 |
|
≥50 | 39 | 21 | 18 |
|
| PLT counts,
×109/l |
|
|
|
|
|
<50 | 34 | 18 | 16 | 0.220 |
|
≥50 | 46 | 18 | 28 |
|
| WBC counts,
×109/l |
|
|
|
|
|
<10 | 45 | 16 | 29 | 0.054 |
|
≥10 | 35 | 20 | 15 |
|
| Extramedullary
disease |
|
|
|
|
| No | 52 | 22 | 30 | 0.509 |
|
Yes | 28 | 14 | 14 |
|
| Complete
remission |
|
|
|
|
|
Yes | 51 | 24 | 27 | 0.624 |
| No | 29 | 12 | 17 |
|
| Cytogenetics |
|
|
|
|
|
Favorable | 20 | 5 | 15 | 0.018 |
|
Intermediate | 42 | 20 | 22 |
|
|
Unfavorable | 18 | 11 | 7 |
|
 | Table II.Univariate and multivariate analyses
of prognostic factors in acute myeloid leukemia. |
Table II.
Univariate and multivariate analyses
of prognostic factors in acute myeloid leukemia.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Variables | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Sex | 1.332 | 0.691–2.412 | 0.372 | – | – | – |
| Male
vs. female |
|
|
|
|
|
|
| Age, years | 1.448 | 0.732–2.578 | 0.228 | – | – | – |
| <60
vs. ≥60 |
|
|
|
|
|
|
| WBC counts
×109/l | 1.541 | 0.811–2.635 | 0.175 | – | – | – |
| <10
vs. ≥10 |
|
|
|
|
|
|
| Blast in BM, % | 1.493 | 0.824–2.674 | 0.213 | – | – | – |
| <50
vs. ≥50 |
|
|
|
|
|
|
| Extramedullary
disease | 1.712 | 0.631–2.215 | 0.302 | – | – | – |
| No vs.
Yes |
|
|
|
|
|
|
| Complete
remission | 1.515 | 0.703–2.113 | 0.117 | – | – | – |
| Yes vs.
No |
|
|
|
|
|
|
| FAB
classification | 3.126 | 1.643–5.189 | 0.004 | 2.916 | 1.352–4.715 | 0.009 |
| M1-M5
vs. M6-M7 |
|
|
|
|
|
|
| Cytogenetics | 3.372 | 1.769–5.376 | 0.003 | 2.747 | 1.116–4.032 | 0.017 |
|
Unfavorable vs.
favorable/intermediate |
|
|
|
|
|
|
| Serum LINC00899
expression | 3.762 | 1.818–6.177 | 0.001 | 3.015 | 1.462–5.018 | 0.004 |
| High
vs. low |
|
|
|
|
|
|
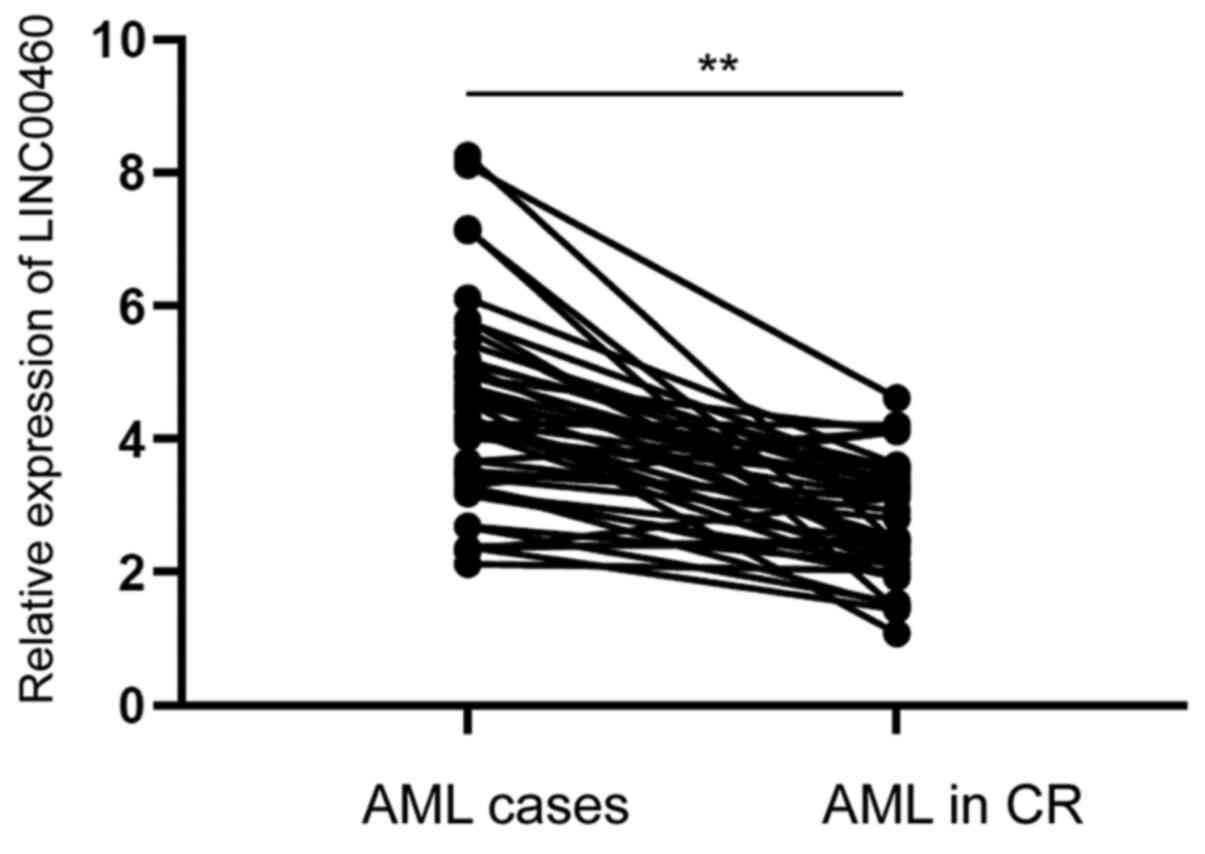
Association between serum LINC00460
expression and treatment response
After receiving different treatments (chemotherapy
and/or targeted therapy), 42 patients achieved CR. To further
investigate the effect of serum LINC00460 on the progression of
AML, the expression of serum LINC00460 in patients with AML was
compared before and after achieving CR. It was demonstrated that
serum LINC00460 expression was markedly decreased in patients with
AML after treatment, suggesting that serum LINC00460 expression was
closely associated with treatment response (Fig. 2).
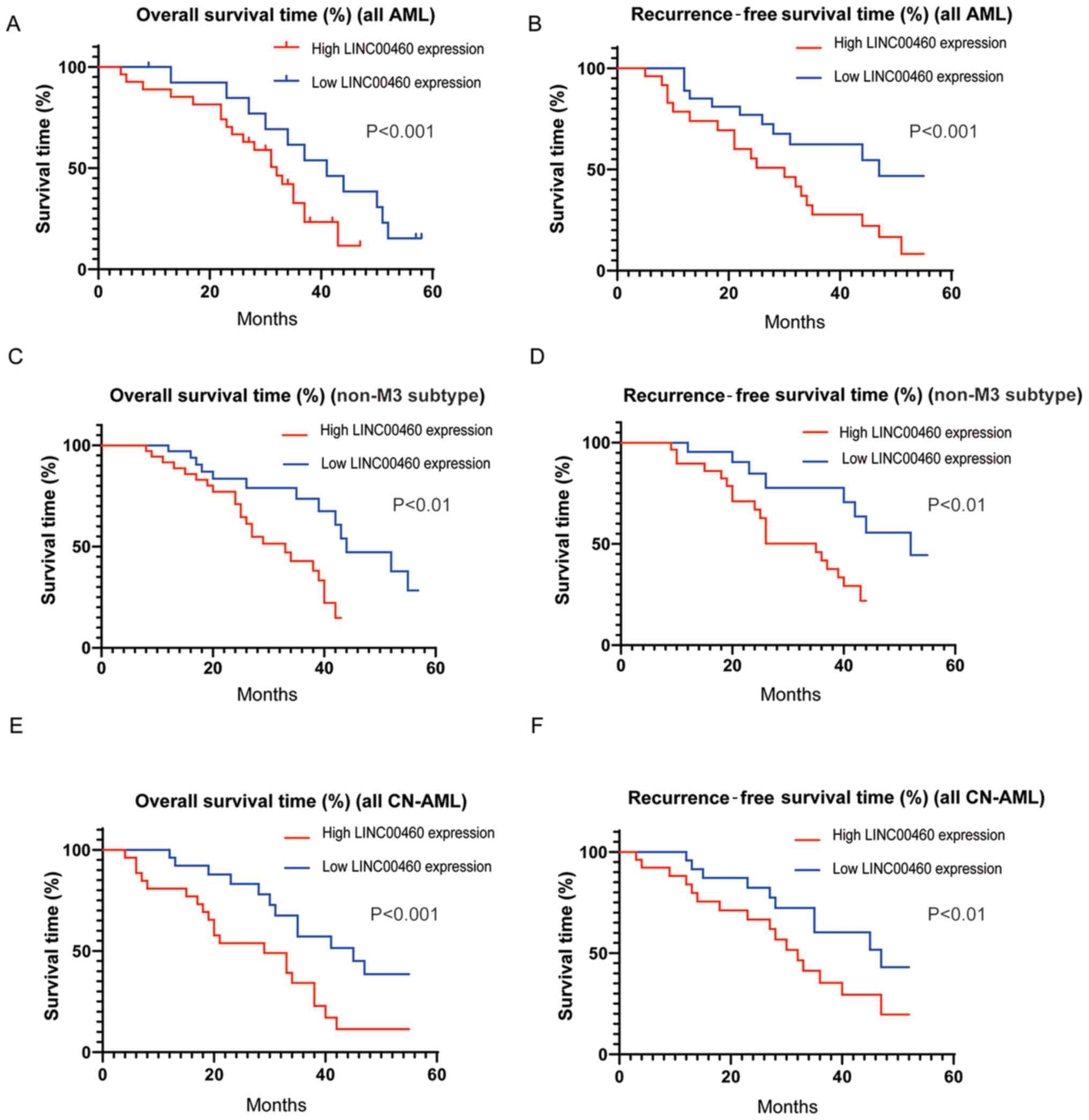
Prognostic evaluation of LINC00460
expression in patients with AML
Clinical follow-up was conduct to determine the
prognostic value of serum LINC00460 expression in patients with
AML. The log-rank test and Kaplan-Meier analysis were performed.
The results demonstrated that the patients with AML in the low
LINC00460 expression group had a significantly longer 5-year
overall survival time (Fig. 3A) and
recurrence-free survival (Fig. 3B)
compared with those in the high expression group. Similar results
were obtained in non-M3 subtype cases (non-acute promyelocytic
leukemia; Fig. 3C and D). In
addition, among the 45 patients with CN-AML, higher expression of
LINC00460 was associated with shorter 5-year overall survival
(Fig. 3E) and recurrence-free
survival (Fig. 3F) compared with
the lower expression group.
Univariate analysis identified that FAB
classification, cytogenetics and serum LINC00460 expression were
significantly correlated with poor overall survival of patients
with AML (P<0.05; Table II).
Further multivariate analysis demonstrated that serum LINC00460
expression, FAB classification and cytogenetics were independent
prognostic indicators of the overall survival of patients with AML
(Table II). These data suggested
that LINC00460 was a potential prognostic biomarker for patients
with AML.
Knockdown of LINC00460 inhibits
viability, as well as induces cell cycle arrest and apoptosis in
AML cells
Next, the function of LINC00460 were investigated
in vitro. The expression of LINC00460 was measured in a
normal human bone marrow stromal cell line (HS-5) and AML cells
(THP-1, KG-1, ME-1 and HL-60) via RT-qPCR. It was found that the
expression of LINC00460 was significantly upregulated in AML cells
(Fig. 4A).
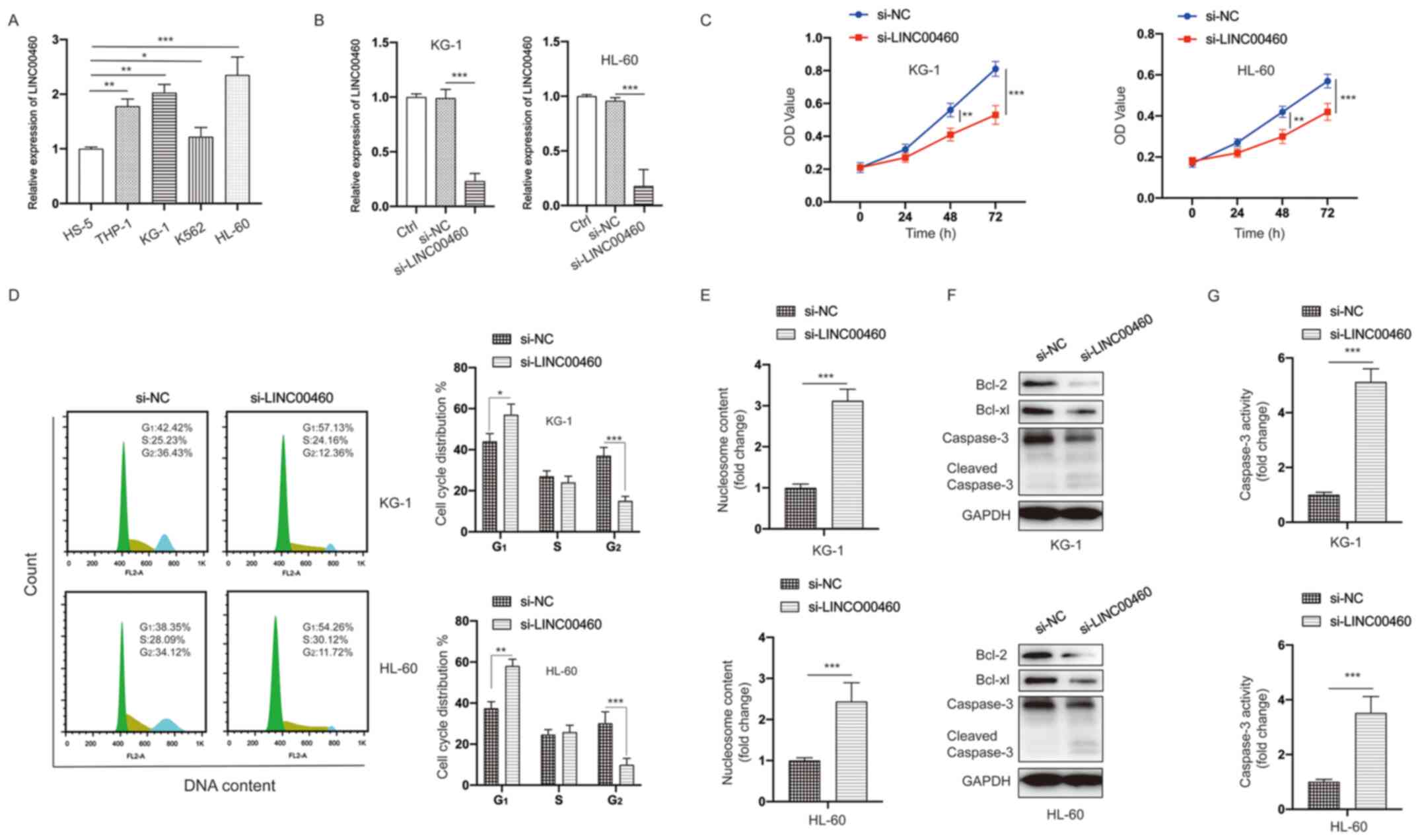 | Figure 4.LINC00460 functions as an oncogene in
AML cells. (A) Expression of LINC00460 was measured in bone marrow
stromal cells (HS-5) and AML cells (THP-1, KG-1, K562 and HL-60).
(B) AML cells were transfected as indicated, and the expression of
LINC00460 was measured. (C) AML cells were treated as indicated,
and cell viability was measured at different time points. (D) AML
cells were treated as indicated, and cell cycle distribution was
assayed. (E) AML cells were treated as indicated, and cell
apoptosis was assayed. (F) AML cells were treated as indicated, and
total cell lysates were subjected to western blotting with the
specified antibodies. (G) AML cells were treated as indicated, and
Caspase-3 activity was assayed. All experiments were conducted ≥3
times. Data are presented as the mean ± SD. *P<0.05;
**P<0.01; ***P<0.001. NC, negative control; siRNA, small
interfering RNA; AML, acute myeloid leukemia; LINC00460, long
non-coding RNA 00460; OD, optical density. |
To further address the biological functions of
LINC00460 in AML cells, siRNA-mediated knockdown of LINC00460 was
performed in AML cells. The si-LINC00460-transfected cells
demonstrated a significant decrease in LINC00460 expression
compared with cells transfected with si-NC (Fig. 4B). The MTT assay indicated that the
viability of AML cells was significantly inhibited after silencing
LINC00460 compared with the control group (Fig. 4C). Cell cycle distribution analysis
identified that knockdown of LINC00460 caused cell cycle arrest at
the G2 phase in AML cells (Fig. 4D). Moreover, a significant increase
in apoptotic AML cells was observed after silencing LINC00460
(Fig. 4E). Western blotting and
caspase-3 activity results also suggested that knockdown LINC00460
induced apoptosis in AML cells (Fig. 4F
and G). Taken together, these data indicated that knockdown
LINC00460 inhibited viability, as well as induced cell cycle arrest
and apoptosis in AML cells.
LINC00460 acts as a sponge of
miR-320b
Considering that lncRNAs can act as sponges to bind
miRNAs, two bioinformatic tools (TargetScan and StarBase 2.0) were
applied for predicting putative miRNAs. The putative binding sites
between LINC00460 and miR-320b are presented in Fig. 5A. It was found that the expression
of miR-320b was significantly lower in AML cells compared with in
HS-5 cells (Fig. 5B). The
transfection efficiency of the miR-320b mimic is presented in
Fig. 5C. The interaction between
miR-320b and LINC00460 was then examined via measuring luciferase
activity. The results demonstrated that overexpression of miR-320b
significantly inhibited the luciferase activity of AML cells
transfected with LINC00460-WT, while its efficacy was absent with
respect to the LINC00460-MUT group (Fig. 5D). The expression of LINC00460 was
significantly upregulated following transfection with
pcDNA.LINC00460 (Fig. 5E). The
expression of miR-320b was significantly decreased by LINC00460
overexpression, but was increased by LINC00460 knockdown in AML
cells (Fig. 5E). Further analysis
identified that the viability of AML cells was inhibited by
transfection with the miR-320b mimic (Fig. 5F). Similar to the knockdown of
LINC00460, overexpression of miR-320b caused cell cycle arrest at
the G2 phase and increased apoptosis of AML cells
(Fig. 5G and H). Thus, these data
indicated that LINC00460 acted as a sponge of miR-320b.
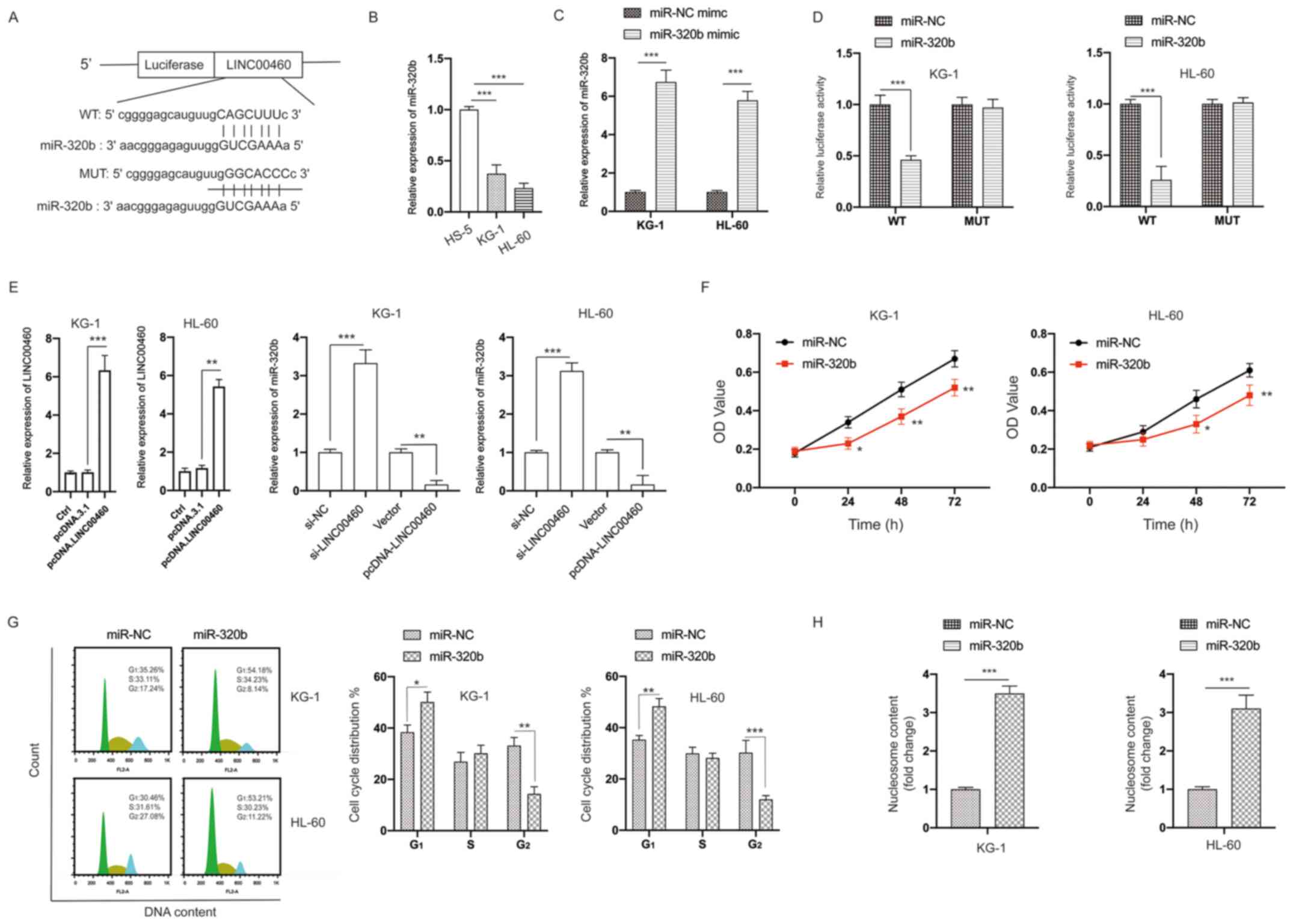 | Figure 5.LINC00460 acts as a sponge of
miR-320b. (A) Constructed luciferase reporter plasmids containing
the predicted WT or MUT miR-320b binding sites on LINC00460. (B)
Expression of miR-320b in HS-5, KG-1 and HL-60 cells was measured.
(C) KG-1 and HL-60 cells were transfected with miR-NC mimic or
miR-320b mimic, the expression of miR-320b was measured via reverse
transcription-quantitative PCR. (D) A luciferase reporter assay was
performed to measure the luciferase activity in AML cells after
co-transfection of miRNA mimics and LINC00460 WT or MUT. (E) AML
cells were transfected as indicated, and the expression of miR-320b
was measured. (F) AML cells were transfected with miR-320b mimics
or miR-NC mimics for the indicated times, and cell viability was
measured. (G) AML cells were transfected as indicated, and the cell
cycle distribution was assayed. (H) AML cells were transfected as
indicated, and cell apoptosis was measured. All experiments were
conducted ≥3 times. Data are presented as the mean ± SD.
*P<0.05; **P<0.01; ***P<0.001. NC, negative control; miR,
microRNA; AML, acute myeloid leukemia; LINC00460, long non-coding
RNA 00460; OD, optical density; Ctrl, control; WT, wild-type; MUT,
mutant. |
Inhibition of miR-320b partially
blocks the effects of LINC00460 knockdown on AML cells
To further analyze the correlation between miR-320b
and LINC00460, transfection of the miR-320b inhibitor was
conducted, which successfully decreased the expression of miR-320b
(Fig. 6A). It was found that
knockdown of miR-320b partially reversed the effects of silencing
LINC00460 on the viability of AML cells (Fig. 6B). Moreover, the cell cycle arrest
at the G2 phase (Figs.
S1 and 6C) and apoptosis
(Fig. 6D) induced by silencing
LINC00460 could be partially rescued by knockdown of miR-320b.
Therefore, these data further demonstrated the relationship between
LINC00460 and miR-320b.
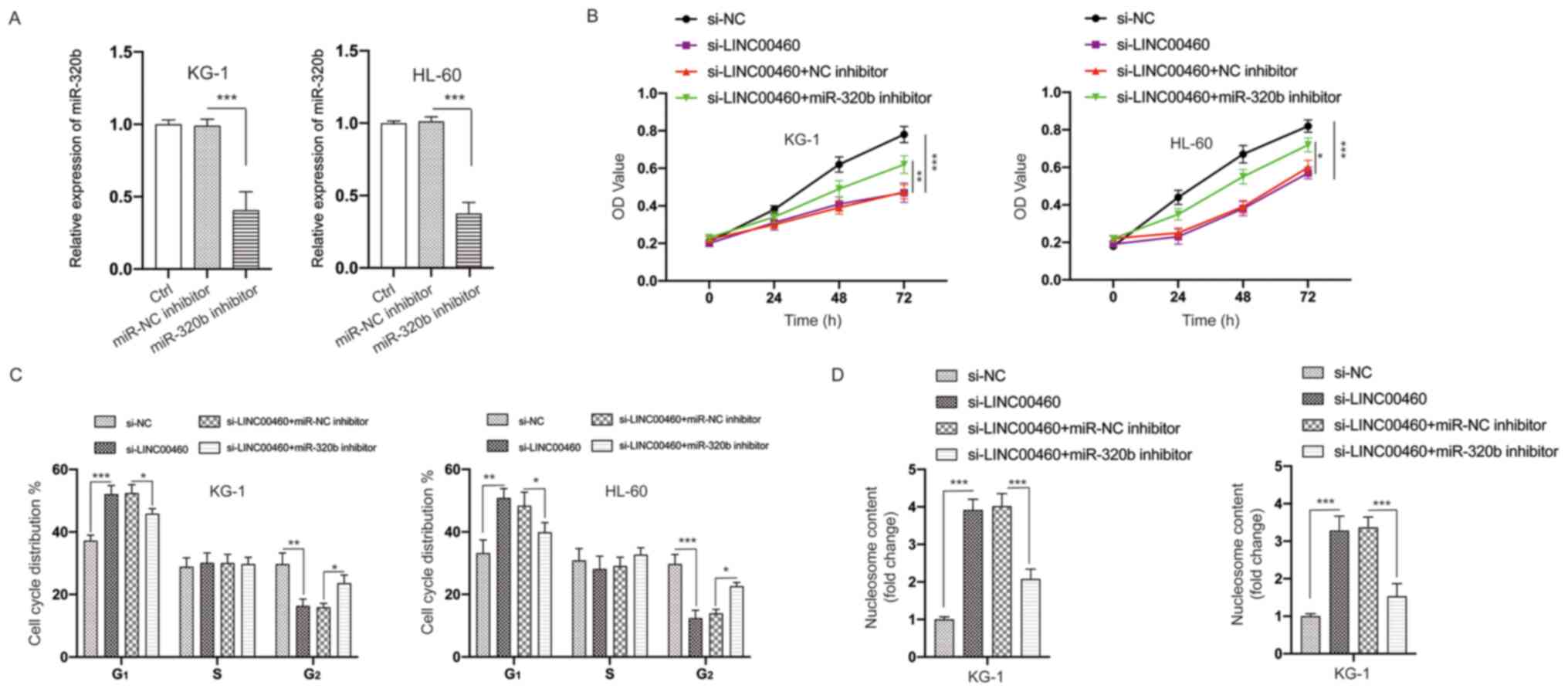 | Figure 6.Knockdown of miR-320b suppresses the
effects of silencing LINC00460 on AML cells. (A) AML cells were
transfected as indicated, and the expression of miR-320b was
measured. (B) AML cells were transfected as indicated, and the
viability of cells was measured using the MTT assay. (C) AML cells
were transfected as indicated, and the cell cycle distribution was
detected. (D) AML cells were transfected as indicated, and cell
apoptosis was evaluated. All experiments were conducted ≥3 times.
Data are presented as the mean ± SD. *P<0.05; **P<0.01;
***P<0.001. NC, negative control; miR, microRNA; AML, acute
myeloid leukemia; LINC00460, long non-coding RNA 00460; OD, optical
density; Ctrl, control; siRNA, small interfering RNA. |
PBX3 is a direct target of
miR-320b
Next, the possible direct targets of miR-320b were
investigated. Using bioinformatics analysis, the 3′-UTR of the PBX3
gene was predicted to bind with miR-320b (Fig. 7A). The interaction between miR-320b
and PBX3 was further validated via the dual-luciferase reporter
assay, which indicated significantly decreased luciferase activity
caused by miR-320b mimics in AML cells expressing the WT PBX3
3′-UTR sequence, while there was little change in luciferase
activity in cells expressing the MUT PBX3 3′-UTR sequence (Fig. 7B). The transfection of miR-320b
mimics decreased the protein expression level of PBX3 in AML cells
(Fig. 7C). Moreover, knockdown and
overexpression of LINC00460 led to decreased and increase of PBX3
expression in AML cells, respectively (Fig. 7D). This finding suggested that
LINC00460 may exert its function via regulation of the
miR-320b/PBX3 axis.
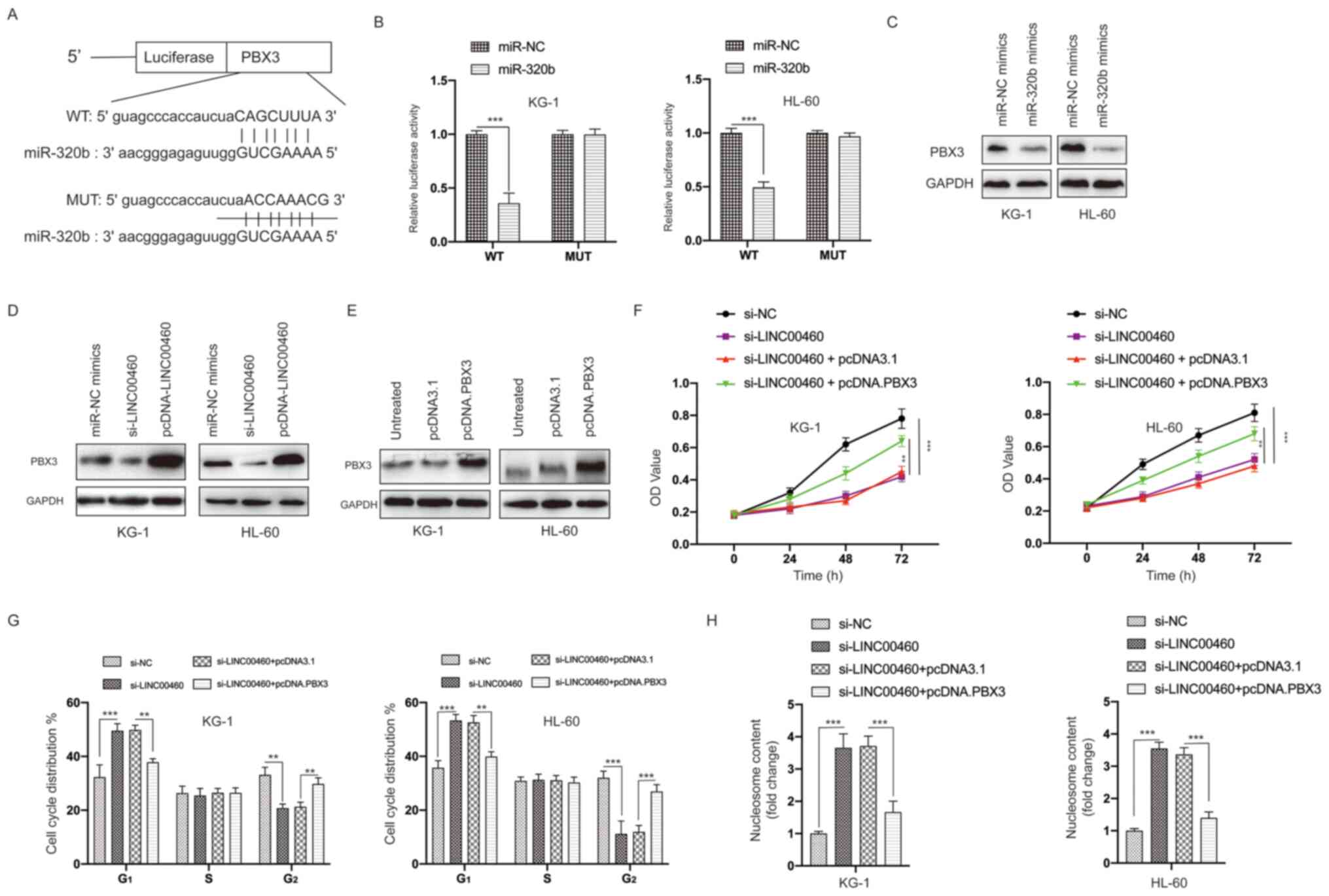 | Figure 7.PBX3 is a direct target of miR-320b.
(A) Constructed luciferase reporter plasmids containing the
predicted WT or MUT miR-320b binding sites on PBX3. (B) A
luciferase reporter assay was performed to measure the luciferase
activity in AML cells after co-transfection of miRNA mimics and
PBX3 WT or MUT. (C) AML cells were transfected with miR-NC mimics
or miR-320b mimics and the protein levels of PBX3 measured by
western blotting. (D) AML cells were transfected with miR-NC
mimics, si-LINC00460 or pcDNA-LINC00460 and the protein levels of
PBX3 measured by western blotting. (E) AML cells were transfected
with pcDNA3.1 or pcDNA.PBX3 and the protein expression levels of
PBX3 measured via western blotting. (F) AML cells were transfected
as indicated, and cell viability was measured using the MTT assay.
(G) AML cells were transfected as indicated, and the cell cycle
distribution was detected. (H) AML cells were transfected as
indicated, and cell apoptosis was examined. All experiments were
conducted ≥3 times. Data are presented as the mean ± SD.
**P<0.01; ***P<0.001. NC, negative control; miR, microRNA;
AML, acute myeloid leukemia; LINC00460, long non-coding RNA 00460;
OD, optical density; siRNA, small interfering RNA; PBX3, PBX
homeobox 3; WT, wild-type; MUT, mutant. |
To further verify the role of PBX3 in the function
of LINC00460, the expression of PBX3 was successfully overexpressed
in AML cells via transfection of an expression vector (Fig. 7E). The MTT assays results
demonstrated that overexpression of PBX3 partially blocked the
inhibitory effects of LINC00460 knockdown on the viability of AML
cells (Fig. 7F). Furthermore,
overexpression of PBX3 reversed the effects of silencing LINC00460
on the cell cycle distribution (Figs.
S2 and 7G) and apoptosis
(Fig. 7H) of AML cells.
Collectively, these data suggested that PBX3 was a direct target of
miR-320b and that LINC00460 exerts its effects, at least, partially
via regulation of the miR-320b/PBX3 axis.
Discussion
AML, characterized by a subpopulation of long-term
proliferative progenitor cells, is a common type of hematological
malignant tumor that threatens human life (32). Its poor prognosis means it is
necessary to identify novel therapeutic agents and sensitive
biomarkers of AML (33). In recent
years, accumulating evidence has revealed that serum lncRNAs could
be applied as potential biomarkers for the diagnosis and prognosis
of different malignancies, including AML. For instance, both
LINC00265 and LINC00899 are found to be upregulated in the serum of
patients with AML and could be used as a potential biomarker for
AML (34,35). In the current study, it was
identified that serum LINC00460 expression in patients with AML was
significantly upregulated compared with that in healthy controls.
Moreover, serum LINC00460 expression was significantly increased in
AML subjects with poor-risk cytogenetics. ROC analysis demonstrated
that serum LINC00460 expression could effectively differentiate
patients with AML from healthy controls. Moreover, downregulation
of serum LINC00460 was detected in patients with AML with a CR. The
present data also indicated that patients with high serum LINC00460
expression had a significantly poorer overall survival time and
recurrence-free survival time compared with those with low serum
LINC00460 expression. Additionally, serum LINC00460 could serve as
an independent prognostic indicator for patients with AML. To the
best of our knowledge, the present study was the first report on
the clinical significance of serum LINC00460 in patients with
AML.
The current findings are consistent with other
studies showing that LINC00460 functioned as an oncogene in various
types of cancer. For example, Feng et al (36) reported that LINC00460 was
significantly upregulated in glioma tissues and cell lines compared
with non-tumor healthy tissues. In epithelial ovarian cancer,
LINC00460 expression is markedly increased in both cancer tissues
and cell lines (17). Moreover,
upregulated LINC00460 expression promotes tumorigenicity by binding
miR-338-3p in vitro (17).
LINC00460 was also observed to be upregulated in both head and neck
squamous cell carcinoma (HNSCC) tissues and cell lines, as well as
predicted a poor prognosis in patients with HNSCC (37). In breast cancer, upregulation of
LINC00460 expression was observed in breast cancer tissues and was
found to be associated with aggressive clinical characteristics
(38). Furthermore, knockdown of
LINC00460 results in the inhibition of viability, migration and
invasion of breast cancer cells (38). In a recent study, LINC00460 was
found to be notably upregulated in gastric cancer tissues compared
with non-tumor tissues, and LINC00460 could be used as an
independent prognostic marker in gastric cancer (39).
To date, accumulating evidence suggests that lncRNAs
can be released into various body fluids, such as urine, saliva and
serum, as a result of cancer cell excretion (40). The easy accessibility of serum and
the stability of lncRNA in serum makes lncRNAs ideal biomarkers for
the diagnosis and prognosis of cancer. There are several
explanations regarding the stability of lncRNAs in serum: i)
lncRNAs may be selectively enclosed into membrane-covered vesicles,
which can provide protection against degradation; ii) lncRNAs may
bind with proteins to avoid degradation; and iii) lncRNAs may fold
into stable complex secondary and tertiary structures (41). Although the present study identified
the oncogenic role of LINC00460 and its underlying mechanisms in
AML, there remain some limitations. First, the clinical sample size
was relatively small. Second, most participants were recruited from
the same hospital. Therefore, further multicenter cohort studies
should be performed in the future to further confirm the current
findings. Moreover, it would be interesting to investigate
LINC00460 in serum samples of patients with other malignant
diseases.
The present study investigated the biological
functions of LINC00460 in vitro. It was identified that
knockdown of LINC00460 inhibited the viability, as well as induced
cell cycle arrest and apoptosis of AML cells. These findings are in
line with previous studies, which also reported that LINC00460
acted as an oncogene in lung, colorectal and ovarian cancer cells
(22,42,43).
Furthermore, Lian et al (15) revealed that inhibition of LINC00460
induced cell cycle arrest at the G1 phase in colorectal
cells, while the present study identified that silencing LINC00460
caused cell cycle arrest at the G2 phase. This
discrepancy may be caused by the use of different cell lines, and
additional investigations are required to further elucidate the
role of LINC00460 in the control of cell cycle progression.
It is well known that miRNAs participate widely in
the progression of various human cancer types (44). In addition, lncRNAs can act as
competing endogenous RNAs to bind and inhibit the functions of
miRNAs (45). The present study
demonstrated that LINC00460 acted as a sponge of miR-320b, which
has been identified as a tumor suppressor in different human cancer
types, such as colorectal cancer, glioma, lung cancer and prostate
cancer (21–23). Previous studies have reported that
the expression and functions of miR-320b in tumorigenesis could be
regulated by lncRNAs. For instance, lncRNA X inactive specific
transcript can regulate the progression of osteosarcoma by
targeting and suppressing miR-320b expression (46). In addition, miR-320b is targeted and
repressed by the lncRNA NR2F2-antisense 1 in modulating the
tumorigenesis of lung cancer (23).
Consistent with previous studies, the present results suggested
that miR-320b could be a downstream target of LINC00460, which
exerts its function at least partly via regulation of miR-320b.
PBX3 acts as a transcription factor and has been
intensively studied in the pathogenesis of AML (26,27).
More importantly, inactivation of PBX3 could suppress the stemness
and survival of leukemia cells (47). PBX3 was also found to serve
essential roles in the tumorigenesis of various solid cancer types,
such as colorectal cancer, prostate cancer, gastric cancer,
cervical cancer and pancreatic cancer (48). Therefore, targeting PBX3 may be a
potential strategy for the treatment of cancer, including AML. The
present study identified novel mechanistic events involved in the
regulation of PBX3, which may be applied in the field of treatment
of various cancer types.
In conclusion, the present study provided evidence
that serum LINC00460 expression was significantly upregulated in
patients with AML, and was closely associated with poor clinical
outcome and unfavorable clinical variables. To the best of our
knowledge, the present study identified the role of LINC00460 in
AML for the first time. Furthermore, it was suggested that PBX3,
which may also be a potential target for AML, could be negatively
regulated by miR-320b. The in vitro studies demonstrated
that LINC00460 exerted its functions via the miR-320b/PBX3 axis in
AML cells. Therefore, LINC00460 may be used as a potential
indicator and target for the prognosis and treatment of AML.
Supplementary Material
Supporting Data
Acknowledgements
The authors thank Dr Wenming Wang (Laboratory of
Biochemistry, Shanghai University of Science and Technology) for
helpful suggestions during the present study.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ performed the experiments and drafted the
manuscript, ZJ performed the experiments, XZ conducted the
statistical analysis, TJ repeated some of the experiments and LX
designed the study and drafted the manuscript. All authors reviewed
and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Wenzhou Medical University. Written informed consent was obtained
from all patients prior to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Courville EL, Wu Y, Kourda J, Roth CG,
Brockmann J, Muzikansky A, Fathi AT, de Leval L, Orazi A and
Hasserjian RP: Clinicopathologic analysis of acute myeloid leukemia
arising from chronic myelomonocytic leukemia. Mod Pathol.
26:751–761. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Appelbaum FR, Gundacker H, Head DR, Slovak
ML, Willman CL, Godwin JE, Anderson JE and Petersdorf SH: Age and
acute myeloid leukemia. Blood. 107:3481–3485. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Döhner H, Weisdorf DJ and Bloomfield CD:
Acute myeloid leukemia. N Engl J Med. 373:1136–1152. 2015.
View Article : Google Scholar
|
|
5
|
Christen F, Hoyer K, Yoshida K, Hou HA,
Waldhueter N, Heuser M, Hills RK, Chan W, Hablesreiter R, Blau O,
et al: Genomic landscape and clonal evolution of acute myeloid
leukemia with t(8;21): An international study on 331 patients.
Blood. 133:1140–1151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gregory TK, Wald D, Chen Y, Vermaat JM,
Xiong Y and Tse W: Molecular prognostic markers for adult acute
myeloid leukemia with normal cytogenetics. J Hematol Oncol.
2:232009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kurosawa S, Miyawaki S, Yamaguchi T,
Kanamori H, Sakura T, Moriuchi Y, Sano F, Kobayashi T, Yasumoto A,
Hatanaka K, et al: Prognosis of patients with core binding factor
acute myeloid leukemia after first relapse. Haematologica.
98:1525–1531. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang L, Froberg JE and Lee JT: Long
noncoding RNAs: Fresh perspectives into the RNA world. Trends
Biochem Sci. 39:35–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Böhmdorfer G and Wierzbicki AT: Control of
chromatin structure by long noncoding RNA. Trends Cell Biol.
25:623–632. 2015. View Article : Google Scholar
|
|
10
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu R, Yao J and Ren Y: A novel circRNA,
circNUP98, a potential biomarker, acted as an oncogene via the
miR-567/PRDX3 axis in renal cell carcinoma. J Cell Mol Med.
24:10177–10188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY,
Bin-Zhou, Wu JB, Tang LY and Gao SM: Long non-coding RNA HOTAIR
modulates c-KIT expression through sponging miR-193a in acute
myeloid leukemia. FEBS Lett. 589:1981–1987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang Y, Wu Y, Chen X, Zhang S, Wang K,
Guan X, Yang K, Li J and Bai Y: A novel long noncoding RNA
linc00460 up-regulated by CBP/P300 promotes carcinogenesis in
esophageal squamous cell carcinoma. Biosci Rep. 37:BSR201710192017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yue QY and Zhang Y: Effects of Linc00460
on cell migration and invasion through regulating
epithelial-mesenchymal transition (EMT) in non-small cell lung
cancer. Eur Rev Med Pharmacol Sci. 22:1003–1010. 2018.PubMed/NCBI
|
|
15
|
Lian Y, Yan C, Xu H, Yang J, Yu Y, Zhou J,
Shi Y, Ren J, Ji G and Wang K: A novel lncRNA, LINC00460, affects
cell proliferation and apoptosis by regulating KLF2 and CUL4A
expression in colorectal cancer. Mol Ther Nucleic Acids.
12:684–697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang F, Liang S, Liu X, Han L, Wang J and
Du Q: LINC00460 modulates KDM2A to promote cell proliferation and
migration by targeting miR-342-3p in gastric cancer. Onco Targets
Ther. 11:6383–6394. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu X, Wen J, Wang H and Wang Y: Long
non-coding RNA LINC00460 promotes epithelial ovarian cancer
progression by regulating microRNA-338-3p. Biomed Pharmacother.
108:1022–1028. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lian H, Xie P, Yin N, Zhang J, Zhang X, Li
J and Zhang C: Linc00460 promotes osteosarcoma progression via
miR-1224-5p/FADS1 axis. Life Sci. 233:1167572019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sayed D and Abdellatif M: MicroRNAs in
development and disease. Physiol Rev. 91:827–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao Q, Wang B, Li X and Jiang G: miRNAs
in acute myeloid leukemia. Oncotarget. 8:3666–3682. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lv QL, Du H, Liu YL, Huang YT, Wang GH,
Zhang X, Chen SH and Zhou HH: Low expression of microRNA-320b
correlates with tumorigenesis and unfavorable prognosis in glioma.
Oncol Rep. 38:959–966. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang H, Cao F, Li X, Miao H, e J, Xing J
and Fu CG: miR-320b suppresses cell proliferation by targeting
c-Myc in human colorectal cancer cells. BMC Cancer. 15:7482015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang S, Zhang X, Sun Q, Zhuang C, Li G,
Sun L and Wang H: LncRNA NR2F2-AS1 promotes tumourigenesis through
modulating BMI1 expression by targeting miR-320b in non-small cell
lung cancer. J Cell Mol Med. 23:2001–2011. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lieb V, Weigelt K, Scheinost L, Fischer K,
Greither T, Marcou M, Theil G, Klocker H, Holzhausen HJ, Lai X, et
al: Serum levels of miR-320 family members are associated with
clinical parameters and diagnosis in prostate cancer patients.
Oncotarget. 9:10402–10416. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li Z, Zhang Z, Li Y, Arnovitz S, Chen P,
Huang H, Jiang X, Hong GM, Kunjamma RB, Ren H, et al: PBX3 is an
important cofactor of HOXA9 in leukemogenesis. Blood.
121:1422–1431. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Chen P, Su R, Hu C, Li Y, Elkahloun
AG, Zuo Z, Gurbuxani S, Arnovitz S, Weng H, et al: PBX3 and MEIS1
cooperate in hematopoietic cells to drive acute myeloid leukemias
characterized by a core transcriptome of the MLL-rearranged
disease. Cancer Res. 76:619–629. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dickson GJ, Liberante FG, Kettyle LM,
O'Hagan KA, Finnegan DP, Bullinger L, Geerts D, McMullin MF, Lappin
TR, Mills KI, et al: HOXA/PBX3 knockdown impairs growth and
sensitizes cytogenetically normal acute myeloid leukemia cells to
chemotherapy. Haematologica. 98:1216–1225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Müller-Berndorff H, Haas PS, Kunzmann R,
Schulte-Mönting J and Lübbert M: Comparison of five prognostic
scoring systems, the French-American-British (FAB) and World Health
Organization (WHO) classifications in patients with myelodysplastic
syndromes: Results of a single-center analysis. Ann Hematol.
85:502–513. 2006. View Article : Google Scholar
|
|
29
|
Yu R, Yu BX, Chen JF, Lv XY, Yan ZJ, Cheng
Y and Ma Q: Anti-tumor effects of Atractylenolide I on bladder
cancer cells. J Exp Clin Cancer Res. 35:402016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang ML and Bailey NG: Acute myeloid
leukemia genetics: Risk stratification and implications for
therapy. Arch Pathol Lab Med. 139:1215–1223. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Carneiro BA, Altman JK, Kaplan JB,
Ossenkoppele G, Swords R, Platanias LC and Giles FJ: Targeted
therapy of acute myeloid leukemia. Expert Rev Anticancer Ther.
15:399–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Estey EH: Acute myeloid leukemia: 2013
update on risk-stratification and management. Am J Hematol.
88:318–327. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Li Y, Song HQ and Sun GW: Long
non-coding RNA LINC00899 as a novel serum biomarker for diagnosis
and prognosis prediction of acute myeloid leukemia. Eur Rev Med
Pharmacol Sci. 22:7364–7370. 2018.PubMed/NCBI
|
|
35
|
Ma L, Kuai WX, Sun XZ, Lu XC and Yuan YF:
Long noncoding RNA LINC00265 predicts the prognosis of acute
myeloid leukemia patients and functions as a promoter by activating
PI3K-AKT pathway. Eur Rev Med Pharmacol Sci. 22:7867–7876.
2018.PubMed/NCBI
|
|
36
|
Feng L, Rao M, Zhou Y, Zhang Y and Zhu Y:
Long noncoding RNA 00460 (LINC00460) promotes glioma progression by
negatively regulating miR-320a. J Cell Biochem. 120:9556–9563.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xie X, Xiong G, Wang Q, Ge Y and Cui X:
Long non-coding RNA LINC00460 promotes head and neck squamous cell
carcinoma cell progression by sponging miR-612 to up-regulate AKT2.
Am J Transl Res. 11:6326–6340. 2019.PubMed/NCBI
|
|
38
|
Zhu Y, Yang L, Chong QY, Yan H, Zhang W,
Qian W, Tan S, Wu Z, Lobie PE and Zhu T: Long noncoding RNA
Linc00460 promotes breast cancer progression by regulating the
miR-489-5p/FGF7/AKT axis. Cancer Manag Res. 11:5983–6001. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang J, Lian Y, Yang R, Lian Y, Wu J, Liu
J, Wang K and Xu H: Upregulation of lncRNA LINC00460 facilitates GC
progression through epigenetically silencing CCNG2 by EZH2/LSD1 and
indicates poor outcomes. Mol Ther Nucleic Acids. 19:1164–1175.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pardini B, Sabo AA, Birolo G and Calin GA:
Noncoding RNAs in extracellular fluids as cancer biomarkers: The
new frontier of liquid biopsies. Cancers (Basel). 11:11702019.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chiabotto G, Gai C, Deregibus MC and
Camussi G: Salivary extracellular vesicle-associated exRNA as
cancer biomarker. Cancers (Basel). 11:8912019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li K, Sun D, Gou Q, Ke X, Gong Y, Zuo Y,
Zhou JK, Guo C, Xia Z, Liu L, et al: Long non-coding RNA linc00460
promotes epithelial-mesenchymal transition and cell migration in
lung cancer cells. Cancer Lett. 420:80–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang X, Mo FM, Bo H, Xiao L, Chen GY, Zeng
PW, Huang YN, Lei Z, Yuan WJ and Chen ZH: Upregulated expression of
long non-coding RNA, LINC00460, suppresses proliferation of
colorectal cancer. J Cancer. 9:2834–2843. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu X, Dinglin X, Wang X, Luo W, Shen Q, Li
Y, Gu L, Zhou Q, Zhu H, Li Y, et al: Long noncoding RNA XIST
promotes malignancies of esophageal squamous cell carcinoma via
regulation of miR-101/EZH2. Oncotarget. 8:76015–76028. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang W, Zhao C, Zhao J, Zhu Y, Weng X,
Chen Q, Sun H, Mi JQ, Li J, Zhu J, et al: Inactivation of PBX3 and
HOXA9 by down-regulating H3K79 methylation represses NPM1-mutated
leukemic cell survival. Theranostics. 8:4359–4371. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Morgan R and Pandha HS: PBX3 in cancer.
Cancers (Basel). 12:4312020. View Article : Google Scholar : PubMed/NCBI
|