Introduction
Preeclampsia is a common serious disorder of
obstetrics and is a hypertensive complication that occurs after the
20th week of gestation, characterized by new-onset hypertension
(1). The primary cause of maternal
mortality in preeclampsia has not yet been elucidated. At present,
it is hypothesized that placental maternal surface spiral arterial
remodeling disorder and insufficient extravillous trophoblast
invasion are key factors leading to preeclampsia (2,3).
MicroRNAs (miRNAs/miRs) are a type of small
non-coding RNAs that have been reported to be post-transcriptional
gene regulators and tumor suppressors (4,5).
Previous studies have investigated the role of miR-524-5p in
numerous types of cancer. For example, Chen et al (6) demonstrated that high/low miR-524-5p
expression is associated with improved/worse survival rate and
pathological grade of patients with glioma, and Liu et al
(7) detected lower levels of
miR-524-5p expression in human melanoma, whereas overexpression of
miR-524-5p effectively inhibited melanoma cell proliferation and
migration. Furthermore, Liu et al (7) demonstrated that tumors overexpressing
miR-524-5p were significantly smaller than those in negative
control (NC) mice. Additionally, a previous study has demonstrated
that miR-524-5p expression is downregulated in preeclampsia
(8). However, the role of
miR-524-5p in preeclampsia has not been fully elucidated.
NUMB endocytic adaptor protein (NUMB), a cell fate
determinant, serves an important role in asymmetric cell division
(9). NUMB is a key negative
regulator of the Notch signaling pathway (10) and is involved in numerous
physiological processes, such as differentiation/proliferation
balance, apoptosis regulation, cell migration and tissue
regeneration (11–13). To the best of our knowledge, NUMB
expression in placental tissues with extremely active growth and
differentiation has not yet been identified. Therefore, the present
study investigated the potential roles of miR-524-5p and NUMB in
trophoblast proliferation and invasion.
Materials and methods
Patients
Patients at Hainan Provincial People's Hospital
(Haikou, China) were enrolled in the present study between
September 2017 and January 2019. A total of 40 patients with
preeclampsia and 40 healthy pregnant women were admitted in the
present study. The risk factors for the development of
pre-eclampsia were recorded according to a previous study (14). According to the American College of
Obstetricians and Gynecologists practice bulletin for diagnosis and
management of preeclampsia and eclampsia, severe preeclampsia was
defined as either sustained systolic blood pressure ≥160 mmHg
and/or diastolic blood pressure ≥110 mmHg (measured twice, ≥6 h
apart) or severe proteinuria (>5 g/24 h specimen or ≥3 g/l in ≥2
random samples collected 4 h apart) (15). Normal systolic blood pressure is
<140 mmHg and diastolic blood pressure is <110 mmHg. Age, BMI
and smoking history were also recorded but were not used as
exclusion criteria. Patients with non-severe preeclampsia according
to the criterion listed in the International Society for the Study
of Hypertension in Pregnancy were excluded. Clinical information
was recorded up to the expected date of delivery and the placenta
was obtained for follow-up study. The present study was approved by
the Ethics Committee of Hainan General Hospital and all patients
provided written informed consent.
Cell culture
The human HTR-8/SVneo trophoblast cell line was
purchased from Shanghai Enzyme Research Biotechnology Co., Ltd.
HTR-8/SVneo cells were seeded in a 10-cm cell culture dish with
DMEM medium (Gibco; Thermo Fisher Scientific, Inc.) containing 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin at 37°C in a humidified atmosphere with 5%
CO2.
miR inhibitor and mimic
transfection
miR-524-5p inhibitor, mimic and negative control
(NC) oligonucleotides (20 µM; Guangzhou RiboBio Co., Ltd.) were
transfected into cells using 5 µl Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) in 6-well plates at
37°C for 24 h. The cells were divided into four groups: Scrambled
Inhibitor NC (5′-GAGAAAGUGCUUCGGUUUUUUG-3′), miR-524-5p inhibitor
(5′-GAGAAAGUGCUUCCCUUUGUAG-3′), scrambled mimic NC
(5′-CAAAAAACCGAAGCACUUUCUC-3′) and miR-524-5p mimic
(5′-CUACAAAGGGAAGCACUUUCUC-3′).
Cell counting Kit-8 (CCK-8) assay
Cells were seeded into 6-well plates at a density of
1×104 cells/ml for 0, 24, 48 or 72 h. Cells were
incubated with 10 µl CCK-8 solution (Beijing Solarbio Science &
Technology Co., Ltd.) at 37°C for 2 h, and cell viability was
measured. Absorbance values were measured using a microplate reader
at a wavelength of 450 nm.
Bioinformatics analysis
The target genes of miR-524-5p were predicted using
TargetScan (http://www.targetscan.org/mamm_31/) and microRNA.org
databases (http://www.mirbase.org/) (16,17).
Binding sites between miR-524-5p and the target genes were searched
to identify potential interactions between them.
Luciferase reporter assay
A luciferase reporter assay was conducted to explore
the association between miR-524-5p and NUMB, as described
previously (18). HTR-/SVneo cells
were divided into two groups, wild-type and mutant NUMB with
3′-untranslated region (UTR) reporters. HTR-8/SVneo cells at 80%
confluence were co-transfected at 37°C for 24 h with wild-type or
mutant NUMB 3′-UTR reporters (16 µg/ml; Generay Biotech Co., Ltd.)
together with miR-524-5p mimic (50 pmol/ml) or NC using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Subsequently, Dual-Luciferase Reporter Assay
System (Promega Corporation) was performed according to the
manufacturer's protocol. Cells were harvested and lysed for the
assay 24 h after transfection. Renilla luciferase was used
to normalize the data.
Transwell invasion assay
Transwell assays were carried out as described
previously (19). Cells
(2×104 cells/well) were cultured at 37°C for 24 h in
24-well plates with Transwell inserts precoated with
Matrigel® (37°C for 30 min; BD Pharmingen; BD
Biosciences). DMEM medium was added to the upper chamber, and DMEM
containing 10% FBS in the lower chamber. Subsequently, cells were
stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) at
20°C for 10 min and the number of cells that had invaded through
the Matrigel membrane was counted using a light microscope
(magnification, ×100; Leica Microsystems GmbH).
Reverse transcription-quantitative
(RT-q)PCR analysis
Total RNA was extracted from cells using
TRIzol® reagent and quantified using NanoDrop 2000c
(both Thermo Fisher Scientific, Inc). For miR-524-5p detection, RNA
was reverse transcribed to cDNA using the RevertAid First Strand
cDNA kit (Thermo Fisher Scientific, Inc), according to the
manufacturer's protocol. qPCR was performed in 96-well plates in
the ABI Step-One plus Real-Time PCR system (Thermo Fisher
Scientific, Inc.) using SYBR Green Master Mix (Takara Biotechnology
Co., Ltd.). Expression levels of miR-524-5p were normalized to
those of small nuclear RNA U6, while GAPDH was used as an
endogenous control for NUMB. Primers were obtained from Invitrogen
(Thermo Fisher Scientific, Inc.). The primer sequences were as
follows: miR-524-5p forward, 5′-CTACAAAGGGAAGCACTTTTCTCAA-3′ and
reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward,
5′-CGCTTCGGCAGCACATATAC-3′ and reverse, 5′-CAGGGGCCATGCTAATCTT-3′;
NUMB forward, 5′-AAGGCTTCTTTGGAAAAACTGG-3′ and reverse,
5′-CATGGCTCAACCTTTCACCT-3′; and GAPDH forward,
5′-TGTTCGTCATGGGTGTGAAC-3′ and reverse, 5′-ATGGCATGGACTGTGGTCAT-3′.
Each well contained 1 µl template, 10 µl Master Mix, 0.5 µl forward
primer (10 µM), 0.5 µl reverse primer (10 µM) and 8 µl diethyl
pyrocarbonate H2O in a 20-µl reaction system. qPCR
thermocycling conditions were as follows: Initial denaturation at
95°C for 2 min, followed by 40 cycles of denaturation at 95°C for
20 sec, annealing at 60°C for 45 sec and extension at 72°C for 30
sec. Finally, gene expression levels were calculated using the
2−ΔΔCq method (20).
Western blotting
Proteins were extracted from cells using RIPA buffer
(EMD Millipore) and quantified using a BCA kit (Beijing Solarbio
Science & Technology Co., Ltd.). Target proteins (30 µg) were
separated according to their mass via 10% SDS-PAGE, then
transferred to a PVDF membrane. Membranes were incubated with
primary antibodies against proliferating cell nuclear antigen
(PCNA; 1:1,000; cat. no. ab92552), Ki67 (1:2,000; cat. no.
ab92742), NUMB (1:1,000; cat. no. ab220362), Bcl-2 (1:500; cat. no.
ab196495), Notch1 (1:1,000; cat. no. ab52627), cyclin D1 (1:500;
cat. no. ab40754), CDK6 (1:2,000; cat. no. ab151247) and GAPDH
(1:10,000; cat. no. ab181602) (all Abcam) at 4°C overnight, then
incubated with horseradish peroxidase-conjugated goat anti-rabbit
IgG secondary antibody (1:5,000; cat. no. A0208; Beyotime Institute
of Biotechnology) at 25°C for 1 h. Bands were visualized using the
ECL reagent (Merck KGaA) and density was measured via ImageJ
software (v 2.1.4.7).
Statistical analysis
Data are presented as the mean ± standard deviation
of three independent experiemnts. Statistical comparisons were
performed using SPSS 17.0 software (SPSS, Inc.) Differences between
two groups were compared using an unpaired Student's t-test.
One-way ANOVA followed by Tukey's multiple comparison test was used
for comparisons among three groups. Pearson's correlation analysis
was used to examine the correlation between NUMB and miR-524-5p
expression. The χ2 test was used to analyze the number
of volunteers who smoked. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-524-5p is downregulated in
preeclamptic placenta compared with in normal placenta
A total of 40 patients with preeclampsia and 40
normal pregnant females (controls) were enrolled in the present
study. Clinical characteristics of patients and normal pregnant
females were recorded, including age, BMI, smoking history and
systolic/diastolic blood pressure (Table I). Maternal age, BMI and smoking
history exhibited no significant difference between the two groups.
Patients with preeclampsia had a significantly shorter gestational
age and lower fetal birth weight, while systolic and diastolic
blood pressure in patients with preeclampsia were significantly
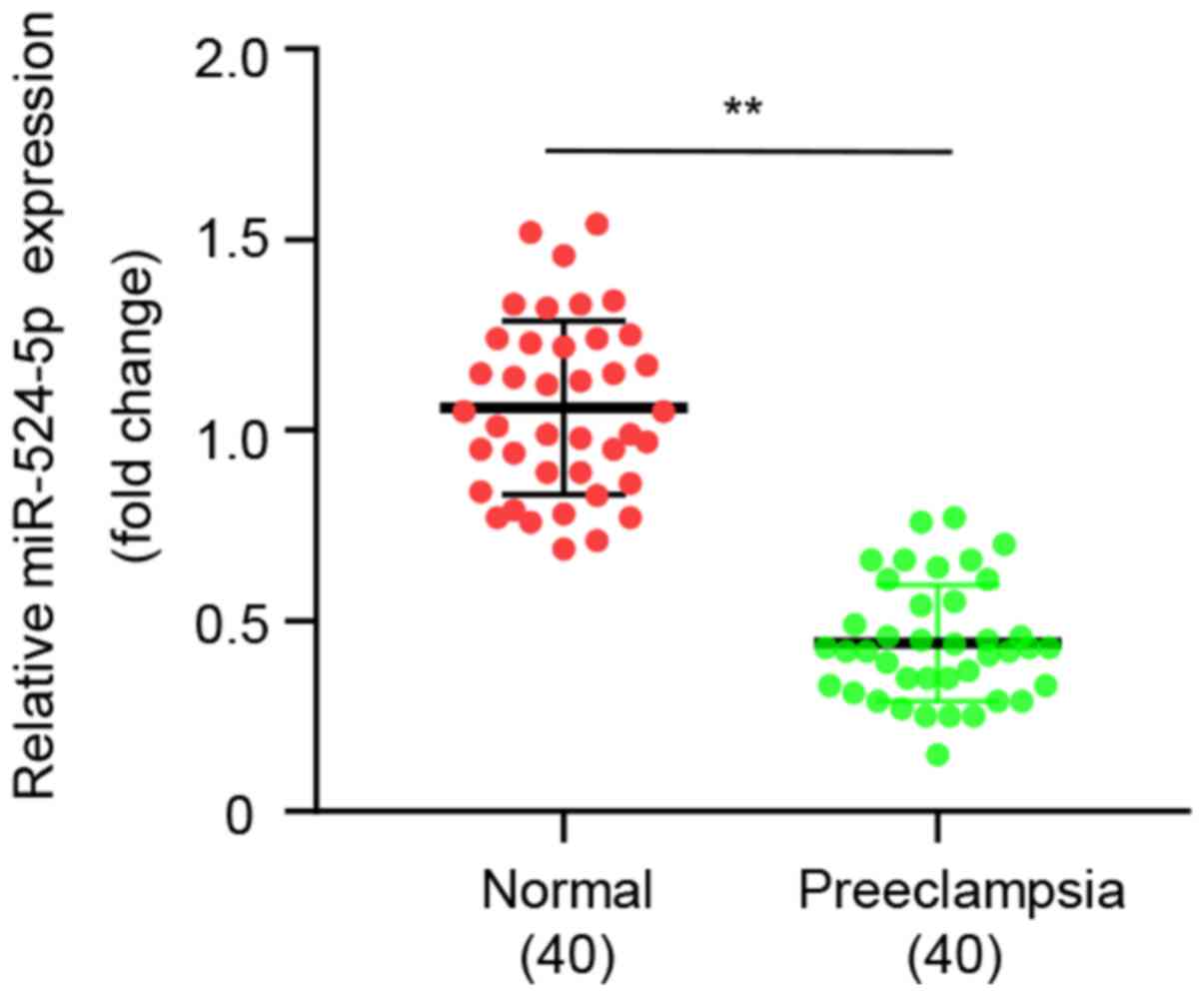
higher than in normal controls. qPCR demonstrated that expression
levels of miR-524-5p were significantly lower in patients with
preeclampsia than in normal controls (Fig. 1), which indicated that
downregulation of miR-524-5p expression may be a biomarker of
preeclampsia.
 | Table I.Clinical information of patients with
PE and normal pregnant females. |
Table I.
Clinical information of patients with
PE and normal pregnant females.
| Parameter | Control (n=40) | PE (n=40) | P-value |
|---|
| Maternal age at
delivery, years |
29.53±2.90 |
30.43±3.15 | 0.1878 |
| Gestational age,
weeks |
38.18±2.24 |
36.70±3.15 | <0.05 |
| Proteinuria, g/24
h | – |
4.57±1.40 | <0.001 |
| Systolic blood
pressure, mmHg |
105.81±13.41 | 168.43±5.43 | <0.001 |
| Diastolic blood
pressure, mmHg |
77.38±4.33 | 116.06±3.75 | <0.001 |
| Fetal birth weight,
g |
3,556.88±309.88 |
2,689.85±428.90 | <0.001 |
| BMI | 23.42±3.28 |
23.01±3.66 | 0.594 |
| Number of
individuals who smoked, n | 3.00 | 4.00 | 0.694 |
miR-524-5p regulates proliferation and
invasion of human HTR-8/SVneo trophoblasts
In order to investigate the effect of miR-524-5p on
the proliferative and invasive abilities of human HTR-8/SVneo
trophoblasts, miR-524-5p mimic and inhibitor were transfected into
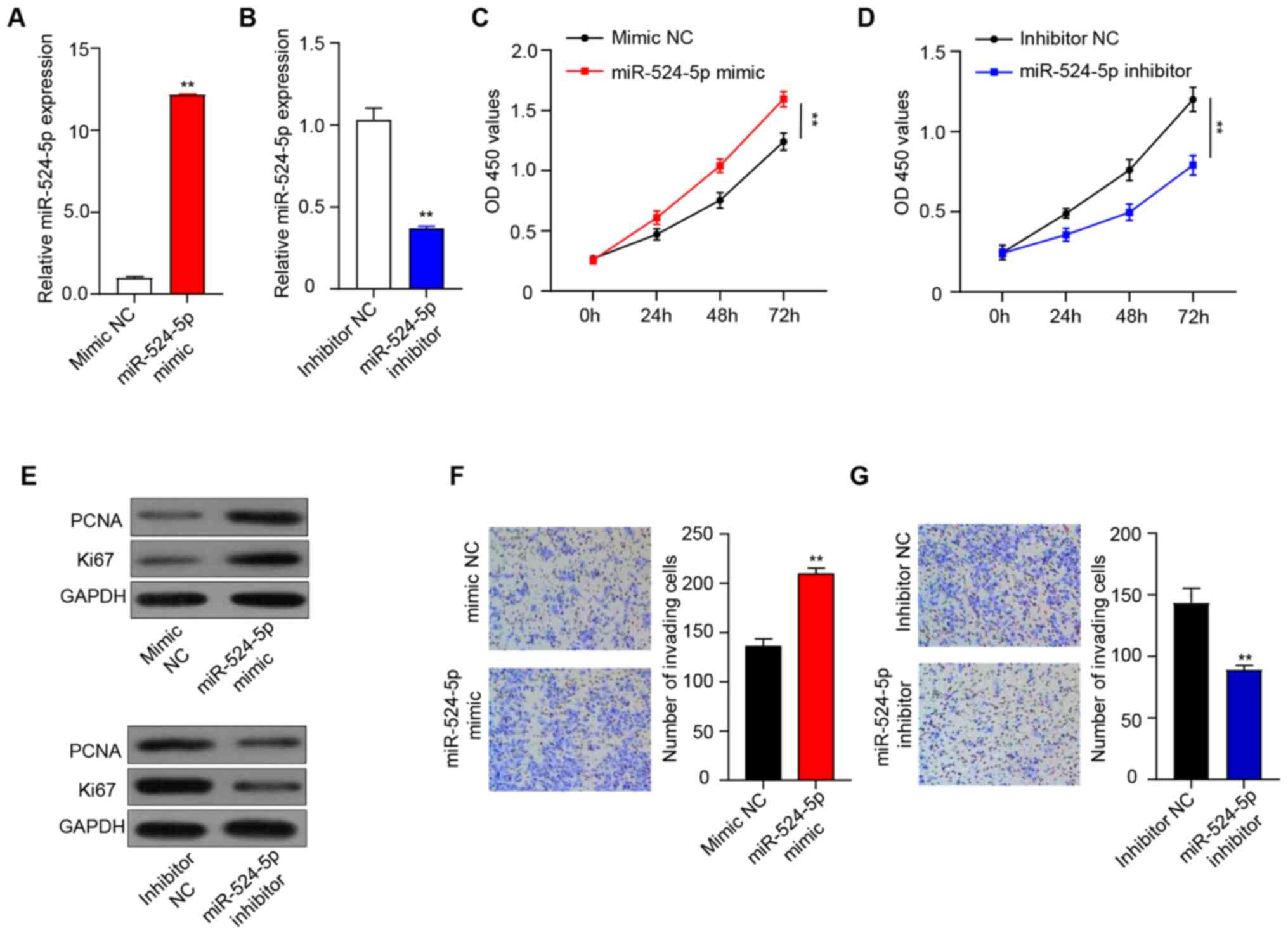
cells. Expression levels of miR-524-5p were significantly higher in
the miR-524-5p mimic group than in the mimic NC group (Fig. 2A) and significantly lower in the
miR-524-5p inhibitor group than in the inhibitor NC group (Fig. 2B), which indicated that mimic and
inhibitor had been transfected successfully. The CCK-8 assay
demonstrated that cell viability of the miR-524-5p mimic group was
significantly higher than that of the mimic NC group at 72 h
(Fig. 2C), while cell viability of
the miR-524-5p inhibitor group was significantly lower compared
with the inhibitor NC group (Fig.
2D). Expression levels of PCNA and Ki67 were determined by
western blotting to investigate the underlying mechanism of cell
proliferation. Overexpression of miR-524-5p increased PCNA and Ki67
protein expression, while PCNA and Ki67 expression in the
miR-524-5p inhibitor group was lower than in the inhibitor NC
group, which demonstrated that inhibition of miR-524-5p inhibited
cell proliferation (Fig. 2E).
In order to investigate the influence of miR-524-5p
on the invasive ability of human HTR-8/SVneo trophoblasts, the
degree of cell invasion was determined by Transwell invasion assay.
Overexpression and inhibition of miR-524-5p in HTR-8/SVneo cells
regulated cell invasion. After 24 h, the invasive ability of cells
in the miR-524-5p mimic group was significantly increased compared
with that in the mimic NC group (Fig.
2F), while the invasive ability of cells in the miR-524-5p
inhibitor group was significantly decreased compared with that in
the inhibitor NC group (Fig.
2G).
miR-524-5p regulates the expression
levels of NUMB via binding to the 3′-UTR of NUMB mRNA
In order to determine the target genes of
miR-524-5p, TargetScan and microRNA.org databases were used.
Results from the databases indicated that NUMB was a candidate
target gene regulated by miR-524-5p, and microRNA.org database was
used to predict the binding sites (Fig.
3A).
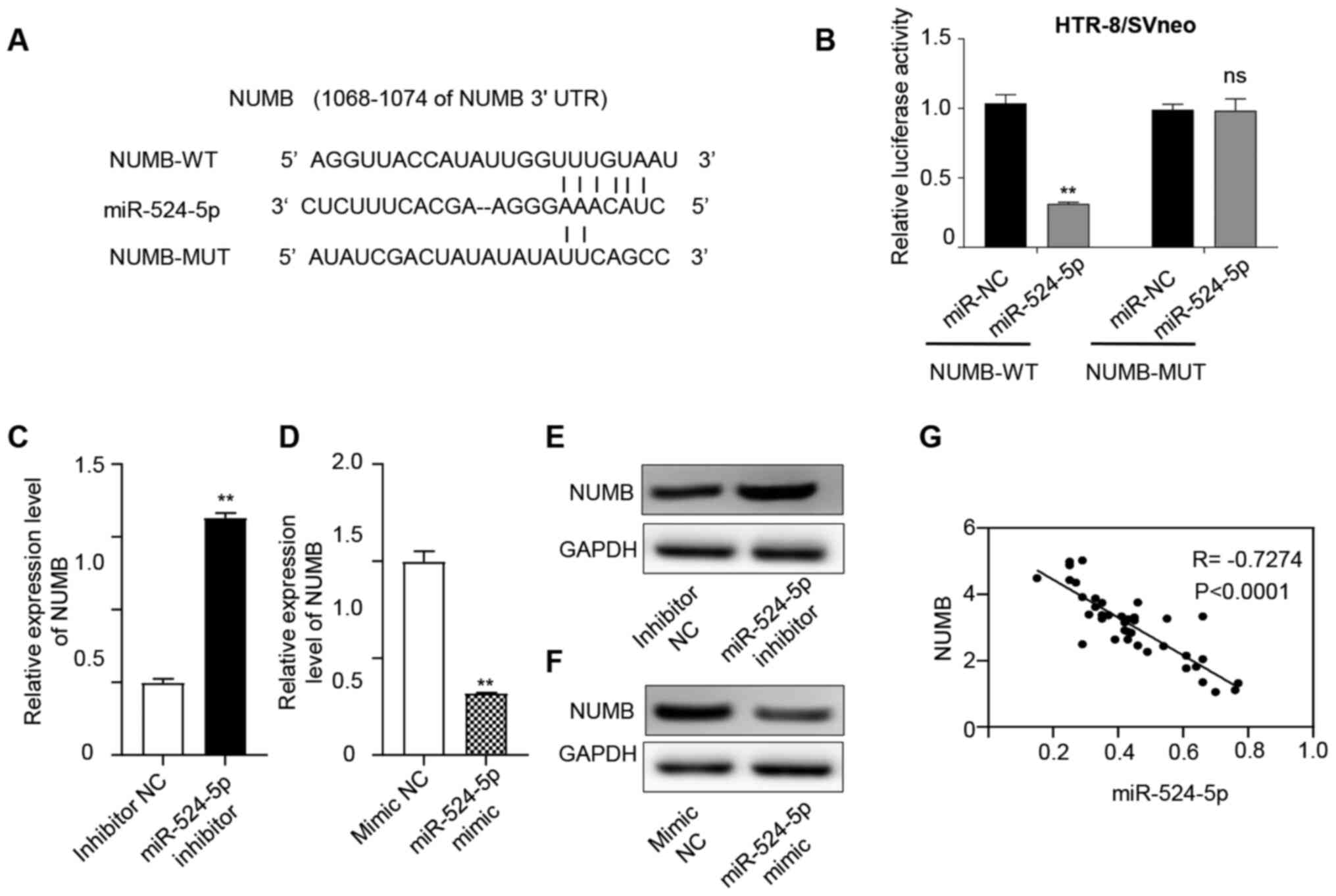 | Figure 3.miR-524-5p regulates the expression
levels of NUMB. (A) microRNA.org database was used to predict
binding sites between miR-524-5p and NUMB. (B) Fluorescence
intensity was measured using a dual luciferase assay. Overexpressed
miR-524-5p bound to NUMB-WT and fluorescence intensity was
weakened. NUMB-MUT exhibited no significant difference in
fluorescence intensity. RT-qPCR was used to detect NUMB expression
in the (C) inhibitor and (D) mimic groups. Compared with the
inhibitor NC group, NUMB expression was significantly upregulated
in the miR-524-5p inhibitor group, while compared with the mimic NC
group, NUMB expression in the miR-524-5p mimic group was
significantly downregulated. Western blotting was used to detect
NUMB expression in the (E) inhibitor and (F) mimic groups. Compared
with the inhibitor NC group, NUMB expression was significantly
upregulated in the miR-524-5p inhibitor group, while it was
significantly downregulated in the miR-524-5p mimic group compared
with the mimic NC group. (G) RT-qPCR was used to detect the
expression levels of NUMB in 40 preeclampsia tissues and Pearson's
correlation coefficient was used to analyze the correlation between
NUMB and miR-524-5p expression. There was a significant negative
association between NUMB and miR-524-5p expression. **P<0.01.
miR, microRNA; NUMB, NUMB endocytic adaptor protein; WT, wild-type;
MUT, mutant; RT-q, reverse transcription-quantitative; NC, negative
control; UTR, untranslated region; ns, not significant. |
In order to determine whether inhibition of NUMB by
miR-524-5p occurred via these predicted miR-524-5p binding sites,
the binding site was mutated. The luciferase reporter assay
indicated that transfection with the NUMB mutant 3′-UTR did not
lead to a reduction in the relative luciferase activity in the
presence of miR-524-5p, compared with miR NC (Fig. 3B). Subsequently, the expression
levels of NUMB were determined following miR-524-5p mimic or
inhibitor treatment. Inhibition of miR-524-5p increased the
expression levels of NUMB mRNA and protein (Fig. 3C and E); conversely, when miR-524-5p
was overexpressed, NUMB mRNA and protein expression levels were
decreased (Fig. 3D and F).
Additionally, Pearson's correlation analysis showed that NUMB
exhibited a negative correlation with miR-524-5p (Fig. 3G), which suggested that the highly
conserved sequence of the NUMB 3′-UTR may be the primary binding
site of miR-524-5p, and further confirmed that miR-524-5p directly
inhibited NUMB.
miR-524-5p regulates proliferation and
invasion of HTR-8/SVneo cells by targeting NUMB to regulate the
Notch signaling pathway
pcDNA3.1-NUMB and miR-524-5p mimic were transfected
into human HTR-8/SVneo trophoblasts. Cells were divided into 4
groups, including mimic NC+ pcDNA 3.1, miR-524-5p mimic + pcDNA
3.1, mimic NC + pcDNA 3.1-NUMB and miR-524-5p mimic + pcDNA
3.1-NUMB. Cell viability was determined by CCK-8 assay, and the
results revealed that cell viability was highest in the miR-524-5p
mimic + pcDNA 3.1 group and lowest in mimic NC + pcDNA 3.1-NUMB,
which indicated that activation of miR-524-5p may inhibit the
expression levels of NUMB and cell viability was increased due to
elevated miR-524-5p expression at 72 h (Fig. 4A).
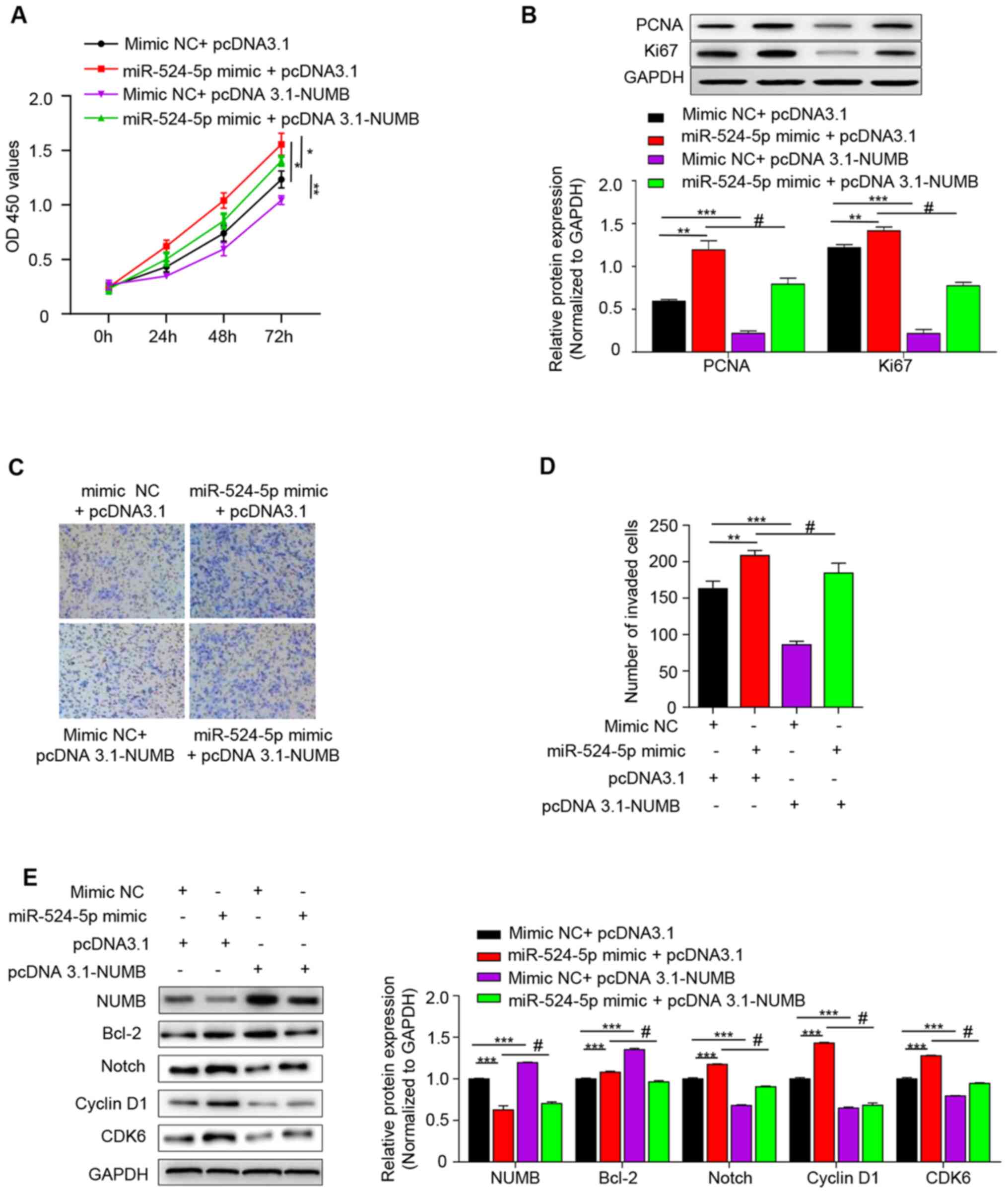 | Figure 4.miR-524-5p regulates the Notch
signaling pathway via NUMB. (A) Cell Counting Kit-8 was used to
detect the proliferation ability of each group for 0, 24, 48 and 72
h. (B) Western blot analysis of PCNA and Ki67 protein expression.
(C) Invasive ability was detected using Transwell assay (Scale bar,
100 µm) and (D) quantified. (E) Western blot was used to detect the
expression levels of NUMB, Notch1, Bcl-2, Cyclin D1 and CDK6 in the
Notch signaling pathway. *P<0.05; **P<0.01; ***P<0.001 vs.
mimic NC+ pcDNA3.1 group; #P<0.001 vs. miR-524-5p
mimic + pcDNA3.1 group. miR, microRNA; NUMB, NUMB endocytic adaptor
protein; PCNA, proliferating cell nuclear antigen; NC, negative
control; OD, optical density; EV, empty vector. |
Apoptosis rate is significantly increased in the
syncytiotrophoblast in preeclampsia (21). The effect of miR-524-5p and NUMB on
cell proliferation was investigated. PCND and Ki67 are common
proliferation markers in apoptosis (22), and therefore western blot analysis
was performed to detect the expression levels of these two
proteins. Expression levels of PCNA and Ki67 were highest in the
miR-524-5p mimic + pcDNA 3.1 group and lowest in the mimic NC +
pcDNA 3.1-NUMB group, which indicated that overexpression of
miR-524-5p decreased NUMB and increased the cell proliferative
ability (Fig. 4B). Additionally,
the effect of miR-524-5p mimic on cell invasion was investigated.
The results revealed that the miR-524-5p mimic + pcDNA3.1 group had
the highest number of invading cells and the mimic NC +
pcDNA3.1-NUMB group exhibited the lowest number. The number of
invading cells in the miR-524-5p mimic + pcDNA3.1-NUMB was between
the two aforementioned groups (Fig. 4C
and D).
The present results suggested that miR-524-5p mimic
inhibited NUMB expression and that high expression levels of NUMB
inhibited cell proliferation and invasion. As the mechanism remains
unknown, the present study investigated the specific mechanism by
which miR-524-5p stimulates proliferation and invasion of
HTR-8/SVneo cells by detecting the expression levels of signaling
molecules associated with the Notch signaling pathway. Protein
expression levels of Bcl-2, Notch, cyclin D1 and CDK6 were
determined by western blotting. Protein expression level trends of
Bcl-2, Notch, cyclin D1 and CDK6 in each group were downregulated
when pcDNA 3.1-NUMB were transformed simultaneously with NC or
mimic (Fig. 4E and S1). This indicated that miR-524-5p
stimulated proliferation and invasion of HTR-8/SVneo cells by
targeting NUMB to regulate the Notch signaling pathway.
Discussion
In recent years, numerous studies have continued to
investigate preeclampsia (1,23), but
the exact cause has not yet been elucidated. There is still no
ideal animal model that replicates all pathological conditions of
preeclampsia that are produced. Preeclampsia is hypothesized to be
associated with insufficient cell invasion and endothelial cell
dysfunction (24). In the present
study, patients with severe PE symptoms exhibited higher systolic
and diastolic blood pressure and proteinuria levels, whereas in
normal controls, proteinuria was not detected.
miRNAs serve roles in a number of diseases, such as
sclerosis, lung cancer and neurodegenerative diseases like
Alzheimer's disease (25). The
discovery of dysregulated miRNAs and their gene-regulatory roles in
placental development has provided a novel approach for elucidating
the underlying mechanisms of pregnancy-specific diseases (26). In the present study, bioinformatics
analysis was used to predict the target genes of a specific miRNA,
revealing that miR-524-5p was expressed at lower levels in patients
with preeclampsia compared with normal controls. miR-524-5p
expression in patients with preeclampsia should continue to be
monitored in future studies. The predicted target gene of
miR-524-5p was NUMB (confirmed by luciferase reporter assay);
expression levels of NUMB, a negative regulator of the Notch
signaling pathway, were regulated by miR-524-5p (10). Notch is a type of receptor that
mediates transmembrane communication and cell fate (27). Bcl-2 is a factor that regulates
apoptosis in the intrinsic mitochondrial apoptosis pathway
(28). Cyclin D1 is one member of
the Cyclin-D family; it is commonly overexpressed and known to be a
direct target of Jagged1-mediated Notch signaling in breast cancer
(29). Inactive CDK6 kinase is
reported to disrupt Notch-dependent survival and proliferation by
altering the expression levels of Notch target gene (30).
NUMB facilitates the migration and invasion of
trophoblastic cells (31). A number
of mechanisms have been suggested to explain the dysregulation of
NUMB in trophoblastic cells. NUMB serves a key role in asymmetric
division (32), as well as in
regulating cell recognition, differentiation, tissue renewal and
stabilization of the differentiation environment (33). In addition, NUMB acts as an
important negative regulator of the Notch signaling pathway
(10). The present study
demonstrated that miR-524-5p negatively regulated both mRNA and
protein levels of NUMB. Viability of HTR-8/SVneo cells was
decreased when miR-524-5p expression was inhibited, while cell
viability was increased when miR-524-5p was activated using a miR
mimic. Subsequently, the effect of miR-524-5p on the proliferative
and invasive abilities of HTR-8/SVneo cells were investigated.
While low expression levels of miR-524-5p significantly inhibited
cell proliferation and migration compared with the control group,
activation of miR-524-5p promoted cell proliferation and migration.
Next, western blotting was performed to detected the association
between miR-524-5p and NUMB, revealing that inhibition of
miR-524-5p resulted in upregulation of NUMB. Finally, to
investigate the mechanism underlying the miR-524-5p-mediated
upregulation of cell proliferation and invasion, Notch signaling
pathway-associated proteins were detected via western blotting,
including Bcl-2, Notch, cyclin D1 and CDK6. Larger patient cohorts
and animal studies are required to validate the results, as these
were the two primary limitations of the present research.
In conclusion, the present results demonstrated that
miR-524-5p regulated proliferation and invasion of HTR-8/SVneo
cells by targeting NUMB to regulate the Notch signaling
pathway.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by Provincial Natural
Science Foundation of China Hainan (grant no. 819MS119).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LS conceptualized the study and gave final approval
of the version to be published. LZ and JS designed and performed
the experiments, and wrote and revised the manuscript. LW, RT, XC,
DW and HC performed the experiments, analyzed the data and prepared
figures and tables. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Hainan General Hospital. All patients provided written
informed consent prior to participation in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NUMB
|
NUMB endocytic adaptor protein
|
|
PCNA
|
proliferating cell nuclear antigen
|
References
|
1
|
Cheng D, Jiang S, Chen J, Li J, Ao L and
Zhang Y: Upregulated long noncoding RNA Linc00261 in pre-eclampsia
and its effect on trophoblast invasion and migration via regulating
miR-558/TIMP4 signaling pathway. J Cell Biochem. 120:13243–13253.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Veerbeek JH, Brouwers L, Koster MP, Koenen
SV, van Vliet EO, Nikkels PG, Franx A and van Rijn BB: Spiral
artery remodeling and maternal cardiovascular risk: The spiral
artery remodeling (SPAR) study. J Hypertens. 34:1570–1577. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shyu MK, Chen CW, Lin NY, Liao WC, Chen
CH, Lin CJ, Huang HC, Lee JJ, Huang MJ, Tseng GF, et al: MUC1
expression is elevated in severe preeclamptic placentas and
suppresses trophoblast cell invasion via β1-integrin signaling. J
Clin Endocrinol Metab. 96:3759–3767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
He J, Xiang D and Yin L: MicroRNA-708
inhibits the proliferation and invasion of osteosarcoma cells by
directly targeting ZEB1. Mol Med Rep. 19:3948–3954. 2019.PubMed/NCBI
|
|
5
|
Bhagirath D, Yang TL, Tabatabai ZL,
Shahryari V, Majid S, Dahiya R, Tanaka Y and Saini S: Role of a
novel race-related tumor suppressor microRNA located in frequently
deleted chromosomal locus 8p21 in prostate cancer progression.
Carcinogenesis. 40:633–642. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen L, Zhang W, Yan W, Han L, Zhang K,
Shi Z, Zhang J, Wang Y, Li Y, Yu S, et al: The putative tumor
suppressor miR-524-5p directly targets Jagged-1 and Hes-1 in
glioma. Carcinogenesis. 33:2276–2282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu SM, Lu J, Lee HC, Chung FH and Ma N:
miR-524-5p suppresses the growth of oncogenic BRAF melanoma by
targeting BRAF and ERK2. Oncotarget. 5:9444–9459. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hromadnikova I, Kotlabova K, Ondrackova M,
Pirkova P, Kestlerova A, Novotna V, Hympanova L and Krofta L:
Expression profile of C19MC microRNAs in placental tissue in
pregnancy-related complications. DNA Cell Biol. 34:437–457. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang XR, Sun J, Wang J and Lu YY: Advances
in research on cell fate determinant Numb regulating liver cancer.
Zhonghua Gan Zang Bing Za Zhi. 26:714–717. 2018.(In Chinese).
PubMed/NCBI
|
|
10
|
Zhang J, Shao X, Sun H, Liu K, Ding Z,
Chen J, Fang L, Su W, Hong Y and Li H and Li H: NUMB negatively
regulates the epithelial-mesenchymal transition of triple-negative
breast cancer by antagonizing Notch signaling. Oncotarget.
7:61036–61053. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao C, Guo H, Li J, Myint T, Pittman W,
Yang L, Zhong W, Schwartz RJ, Schwarz JJ, Singer HA, et al: Numb
family proteins are essential for cardiac morphogenesis and
progenitor differentiation. Development. 141:281–295. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng H, Wang L, Su Q, Yi K, Du J and Wang
Z: MiR-31-5p promotes the cell growth, migration and invasion of
colorectal cancer cells by targeting NUMB. Biomed Pharmacother.
109:208–216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
George RM, Biressi S, Beres BJ, Rogers E,
Mulia AK, Allen RE, Rawls A, Rando TA and Wilson-Rawls J:
Numb-deficient satellite cells have regeneration and proliferation
defects. Proc Natl Acad Sci USA. 110:18549–18554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rebahi H, Elizabeth Still M, Faouzi Y and
Rhassane El Adib A: Risk factors for eclampsia in pregnant women
with preeclampsia and positive neurosensory signs. Turk J Obstet
Gynecol. 15:227–234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
ACOG Committee on Obstetric Practice, .
ACOG practice bulletin. Diagnosis and management of preeclampsia
and eclampsia. Number 33, January 2002. American college of
obstetricians and gynecologists. Int J Gynaecol Obstet. 77:67–5.
2002.PubMed/NCBI
|
|
16
|
Zhao X, Ren Y, Cui N, Wang X and Cui Y:
Identification of key microRNAs and their targets in exosomes of
pancreatic cancer using bioinformatics analysis. Medicine
(Baltimore). 97:e126322018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen K, Chen H, Zhang K, Sun S, Mo J, Lu
J, Qian Z and Yang H: MicroRNA profiling and bioinformatics
analyses reveal the potential roles of microRNAs in chordoma. Oncol
Lett. 14:5533–5539. 2017.PubMed/NCBI
|
|
18
|
Tan J, Yang L, Liu C and Yan Z:
MicroRNA-26a targets MAPK6 to inhibit smooth muscle cell
proliferation and vein graft neointimal hyperplasia. Sci Rep.
7:466022017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song X, Li C, Li J, Liu L, Meng L, Ding H
and Long W: The long noncoding RNA uc.294 is upregulated in
early-onset pre-eclampsia and inhibits proliferation, invasion of
trophoblast cells (HTR-8/SVneo). J Cell Physiol. 234:11001–11008.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ishihara N, Matsuo H, Murakoshi H,
Laoag-Fernandez JB, Samoto T and Maruo T: Increased apoptosis in
the syncytiotrophoblast in human term placentas complicated by
either preeclampsia or intrauterine growth retardation. Am J Obstet
Gynecol. 186:158–166. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iatropoulos MJ and Williams GM:
Proliferation markers. Exp Toxicol Pathol. 48:175–181. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Benkő Z, Chaveeva P, de Paco Matallana C,
Zingler E, Wright A, Wright D and Nicolaides KH: Validation of
competing-risks model in screening for pre-eclampsia in twin
pregnancy by maternal factors. Ultrasound Obstet Gynecol.
53:649–654. 2019. View Article : Google Scholar
|
|
24
|
Haram K, Bjørge L and Guttu K:
Pathophysiology and clinical manifestations in pre-eclampsia.
Tidsskr Nor Laegeforen. 120:1426–1431. 2000.(In Norwegian).
PubMed/NCBI
|
|
25
|
Hewel C, Kaiser J, Wierczeiko A, Linke J,
Reinhardt C, Endres K and Gerber S: Common miRNA patterns of
Alzheimer's disease and parkinson's disease and their putative
impact on commensal gut microbiota. Front Neurosci. 13:1132019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang W, Song WY, Xie Y, Hu LL, Hou XM,
Wang R, Gao Y, Zhang JN, Zhang L, Li WW, et al: miR-181a-5p
suppresses invasion and migration of HTR-8/SVneo cells by directly
targeting IGF2BP2. Cell Death Dis. 9:162018. View Article : Google Scholar
|
|
27
|
Qi S, Lei Y, Zhao L, Mu YL, Li M, Zhao X,
Chen ZJ and Zhang H: Melatonin inhibits 17β-estradiol-induced
migration, invasion and epithelial-mesenchymal transition in normal
and endometriotic endometrial epithelial cells. Reprod Biol
Endocrinol. 16:622018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Frenzel A, Grespi F, Chmelewskij W and
Villunger A: Bcl2 family proteins in carcinogenesis and the
treatment of cancer. Apoptosis. 14:584–596. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Reedijk M, Reedijk M, Reedijk M, Cohen B,
Shimizu M, Ng N, Bukhman Y, Pan J and Dering J: Cyclin D1 is a
direct target of JAG-mediated Notch signaling in breast cancer.
Cancer Res. 69 (Suppl 24):S21502009.
|
|
30
|
Hu MG, Deshpande A, Schlichting N, Hinds
EA, Mao C, Dose M, Hu GF, Van Etten RA, Gounari F and Hinds PW:
CDK6 kinase activity is required for thymocyte development. Blood.
117:6120–6131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang J, Zhou T, Sun Z, Ye T, Zhou S, Li J,
Liu Y, Kong L, Tang J, Liu D and Xing HR: Zeb1 regulates the
symmetric division of mouse lewis lung carcinoma stem cells through
Numb mediated by miR-31. Int J Biol Sci. 14:1399–1410. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu YC, Lee KS, Song Y, Gehrke S and Lu B:
The bantam microRNA acts through Numb to exert cell growth control
and feedback regulation of Notch in tumor-forming stem cells in the
Drosophila brain. PLoS Genet. 13:e10067852017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tosoni D, Zecchini S, Coazzoli M, Colaluca
I, Mazzarol G, Rubio A, Caccia M, Villa E, Zilian O, Di Fiore PP
and Pece S: The Numb/p53 circuitry couples replicative self-renewal
and tumor suppression in mammary epithelial cells. J Cell Biol.
211:845–862. 2015. View Article : Google Scholar : PubMed/NCBI
|