Introduction
Erythropoiesis is a multistep process that produces
erythroid cells, and it is regulated by transcription factors and
oxygen concentration (1). The body
oxygen level is one of the most critical factors affecting
erythropoiesis. For example, low oxygen levels trigger
erythropoiesis via erythropoietin release (2). Moreover, the oxygen level in the bone
marrow microenvironment mediates the interaction between erythroid
progenitors and stromal cells, further promoting erythroid
differentiation (2). Changes in the
oxygen level also alter the expression levels of globin genes, such
as γ-globin (3–5).
GATA binding protein 1 (GATA-1), a member of the
GATA transcription factor family, is expressed in hematopoietic
progenitor cells, megakaryocytes, eosinophil granulocyte cells,
testicular mast cells and testicular cells (6). GATA-1 is essential for normal
erythroid cell proliferation and development (7,8). Under
normoxia, GATA-1 modulates erythroid cells by inducing the
expression of various erythroid differentiation- and
maturation-related genes, such as zinc finger protein, FOG family
member 1 and erythroid Krüppel-like factor (9,10).
Accumulating evidence has indicated that microRNA (miRNA/miR) is a
downstream target of GATA-1. For instance, GATA-1 activates
miR-451a and completes a regulatory circuit that modulates
erythroid maturation (11).
However, the mechanism underlying GATA-1 regulation of erythroid
differentiation under hypoxia remains unknown.
miRNAs are members of a large class of non-coding
RNAs that control gene expression and regulate a wide array of
biological processes. miRNAs target mRNAs and induce translational
repression or mRNA degradation (12). Several miRNAs, including miR-451a
and miR-210-3p, profoundly alter the erythroid phenotype by
regulating early cell maturation and proliferation, the expression
level of fetal γ-globin genes and enucleation (13). miR-210-3p, known as ‘hypoxamiR,’
contributes to the cellular adaptation to hypoxia (14). Moreover, miR-210-3p is associated
with an elevated expression level of fetal γ-globin in
mithramycin-induced K562 cells (13). miR-210-3p expression is also
reportedly elevated during murine fetal liver erythroid cell
differentiation in vitro (15). Thus, miR-210-3p may mediate
erythropoiesis during hypoxia.
K562 cells, a myelogenous leukemia cell line derived
from the highly undifferentiated progenitor of the erythrocytic and
megakaryocytic lineages (16), have
the potential for megakaryocyte and erythroid cell differentiation,
thereby providing an excellent model system for investigating
cellular differentiation-related mechanisms (17).
The present study used a K562 cellular erythroid
differentiation model under hypoxia to investigate the expression
level of GATA-1 and the mechanism underlying its effects on
erythroid cell differentiation and miR-210-3p expression
regulation, which ultimately affects SMAD2 expression.
Materials and methods
Cell lines and lentivirus vectors
K562 cells were purchased from The Cell Bank of Type
Culture Collection of Chinese Academy of Sciences and were
maintained in RPMI-1640 medium (Hyclone; Cytiva) supplemented with
10% FBS (Zhejiang Tianhang Biotechnology Co., Ltd.) and 1%
penicillin/streptomycin solution (Beijing Solarbio Science &
Technology Co., Ltd.), at 37°C and 5% CO2. Lentivirus
vectors (LV-GATA1, cat. no. 26211-1; LV-GATA1-RNAi, cat. no.
18817-1; LV-hsa-mir-210, cat. no. 41113-2; LV-hsa-
miR-210-3p-inhibition, cat. no. 5212-1; LV-SMAD2-RNAi, cat. no.
15901-1) and transfection reagent (HitransG A&P, cat. no.
REVG003-1) were purchased from Shanghai GeneChem Co., Ltd.
Experimental grouping
K562 cells were divided into a normoxic group (21%
O2, 5% CO2, 37°C, saturation humidity) and a
hypoxic group (1% O2, 5% CO2, 94%
N2, 37°C, saturation humidity). Hemin was added (40
mM/l; 37°C; Beijing Solarbio Science & Technology Co., Ltd.),
and incubation was performed for 96 h.
Western blot analysis
K562 cells were lysed with mammalian cell lysis
buffer (Nanjing KeyGen Biotech Co., Ltd.) containing protease and
phosphatase (both Nanjing KeyGen Biotech Co., Ltd.) inhibitors. The
BCA protein determination method was used to detect the amount of
protein. The total protein content was extracted with a 10%
Tris-HCl gradient gel (Bio-Rad Laboratories, Inc.) and transferred
onto PVDF membranes, which was blocked using 5% non-fat milk in
TBS/Tween-20 (0.1%) for 2 h at room temperature. The membrane was
then probed with antibodies for GATA-1 (monoclonal rabbit
anti-human; 1:10,000; cat. no. ab181544; Abcam), SMAD2 (monoclonal
rabbit anti-human; 1:10,000; cat. no. ab40855; Abcam) and α-tubulin
(monoclonal mouse anti-human; 1:10,000; cat. no. ab7291; Abcam) and
incubated overnight at 4°C. The following day, the PVDF membrane
was taken out of the refrigerator, reheated at room temperature for
1 h and washed in TBS/Tween-20 (0.1%). The secondary antibodies
(Goat anti-mouse IgG, HRP conjugate, cat. no. SA00001-1; Goat
Anti-Rabbit IgG, HRP conjugate, cat. no. SA00001-2; ProteinTech
Group, Inc.) were added and incubated at room temperature for 1 h,
followed by washing with TBS/Tween-20 (0.1%). The visualization
reagent (cat. no. 34094; Thermo Fisher Scientific, Inc.) was
prepared according to the instructions of the kit. Quantity One
software (4.6.2; Bio-Rad Laboratories, Inc.) was used for density
analysis of protein bands.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the cells harvested
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
RNA was quantified using an Ultra-microspectrophotometer (Nanodrop
2000; Thermo Fisher Scientific, Inc.) at 260 nm. cDNA was
synthesized using a reverse transcriptase kit (60 min at 42°C and
then 5 min at 70°C; cat. no. K1691; Thermo Fisher Scientific, Inc.)
from 1 µg total RNA. For mRNAs, RT-qPCR was performed using the ABI
7500 real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and the QuantiNova SYBR Green PCR kit (Qiagen
Sciences, Inc.), according to the manufacturer's instructions
(95°C, 2 min for pre-degeneration; followed by 40 cycles at 95°C
for 5 sec and 60°C for 30 sec). The relative quantification of the
transcripts was performed using the 2−ΔΔCq method
(18). The primer sequences used
were as follows: γ-globin forward, 5′-GCAGCTTGTCACAGTGCAGTTC-3′ and
reverse, 5′-TGGCAAGAAGGTGCTGACTTC-3′; and β-actin forward,
5′-CCTGGCACCCAGCACAAT-3′ and reverse,
5′-GCTGATCCACATCTGCTGGAA-3′.
miRNA isolation and RT-qPCR
miRNA was isolated using a miRcute miRNA isolation
kit (Tiangen Biotech Co., Ltd.) and transcribed into cDNA,
according to the manufacturer's instructions (42°C for 60 min, 95°C
for 3 min; cat. no. KR211; Tiangen Biotech Co., Ltd.). RT-qPCR was
performed using ABI 7500 real-time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and a miRcute Plus miRNA qPCR
Detection kit (FP411-02, Tiangen Biotech Co., Ltd.), according to
the manufacturer's instructions (initial denaturation at 95°C for
15 min; followed by 40 cycles at 94°C for 20 sec and 60°C for 34
sec). The relative quantification of the transcripts was performed
using the 2−ΔΔCq method (18). Primers for internal reference U6
(cat. no. CD201-0145) and miR-210-3p (cat. no. CD201-0293) were
purchased from Tiangen Biotech Co., Ltd.
Benzidine staining
A total of 1×106 cells were collected
into a 1.5-ml EP tube with 500 µl PBS, following which 15 µl 0.4%
benzidine solution and 10 µl 3% H2O2 were
added and the suspension was incubated for 3 min at room
temperature. Then, 1 µl 5% sodium nitroprusside solution was added.
Then, 10 min later at room temperature, 10 µl suspension was
collected for observation (magnification, ×200) using an inverted
microscope (Olympus Corporation). The positive cells were
blue-black in color, and 200 cells were counted to calculate the
positive cell rate.
Wright's-Giemsa staining
A 10-µl cell suspension was coated on the slide.
After drying at room temperature, the staining solution A
(Wright's-Giemsa Staining Solution) was added for 1 min and the
staining solution B (PBS) was added and mixed for 8–10 min at room
temperature. After rinsing the excess dye with running water, the
cells were observed under an inverted microscope (magnification,
×1,000; Olympus Corporation). In total, 200 cells were counted in
each preparation, and the proportion of cells with increased
volume, nuclear migration and nuclear shrinkage was determined.
Lentivirus transfection
For the GATA-1 overexpression assay, K562 cells were
transfected with either the GV358 vector with a full-length GATA-1
[multiplicity of infection (MOI)=30] or with an empty vector, and a
transfection reagent was added into medium. For the GATA-1
knockdown assay, K562 cells were transfected with either a GV248
vector with the sequence 5′-ACAGAGCATGGCCTCCAGA-3′ (MOI=80) or with
an empty vector, and a transfection reagent was added into medium.
FLAG was inserted into the lentivirus vector of GATA-1
overexpression, and the high-molecular-weight bands in western
blotting results were FLAG protein bands. After adding lentivirus
reagent in the cell medium (operates at room temperature), the
culture was maintained in the incubator at 37°C with 5%
CO2. After 96 h, the cells were observed using an
inverted fluorescence microscope (magnification, ×100; Olympus
Corporation). Transfection efficiency was determined using western
blotting.
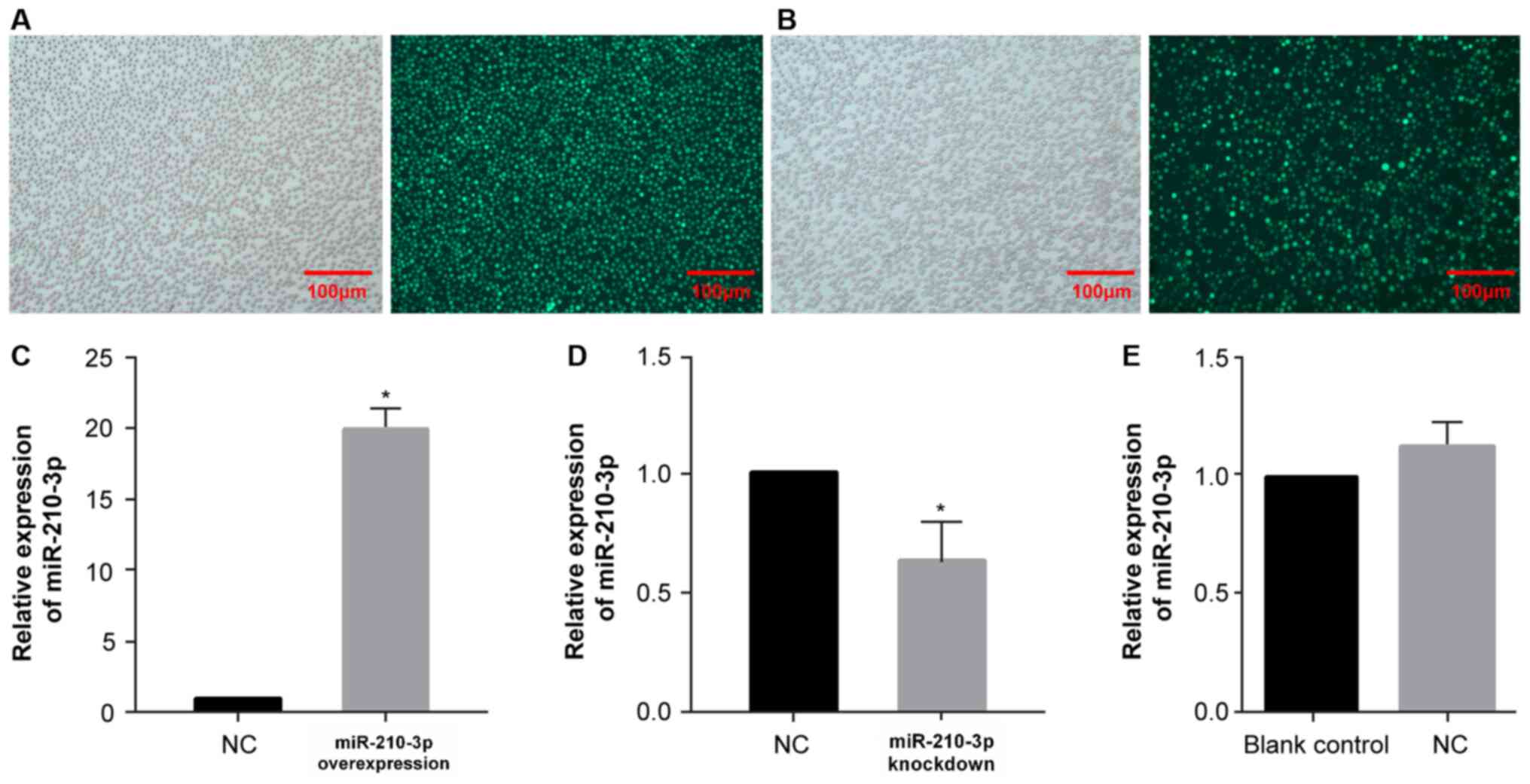
For the miR-210-3p overexpression and knockdown
assay, K562 cells were transfected with either a GV309 vector with
a full-length cDNA of miR-210-3p (MOI=30) or a GV280 vector with a
reverse complementary sequence (5′-TCAGCCGCTGTCACACGCACAG-3′;
MOI=50), and a transfection reagent was added into medium. K562
cells transfected with lentiviruses with the sequence,
5′-TTCTCCGAACGTGTCACGT-3′ (MOI=30), were used as negative controls
(NC) for both reactions. After adding lentivirus reagent in the
cell medium (operates at room temperature), the culture was
continued in the incubator at 37°C with 5% CO2. After 96
h, the cells were observed using an inverted fluorescence
microscope (magnification, ×100; Olympus Corporation). Transfection
efficiency was confirmed via RT-qPCR.
For the SMAD2 inhibition assay, K562 cells were
transfected with either a GV248 vector with the sequence
5′-CGATTAGATGAGCTTGAGAAA-3′ (MOI=80) or with an empty vector, and a
transfection reagent was added into medium. After adding lentivirus
reagent in the cell medium (operates at room temperature), the
culture was continued in the incubator at 37°C with 5%
CO2. After 96 h, the cells were observed using an
inverted fluorescence microscope (magnification, ×100; Olympus
Corporation). Transfection efficiency was confirmed using western
blotting.
Flow cytometry analysis of CD235a
K562 cells were harvested at the indicated time
points (induced by hemin for 0 and 96 h) and washed twice at 4°C
with PBS. Cells were incubated with APC-Cy7-conjugated anti-CD235a
antibodies for 15 min (cat. no. 349116; BioLegend, Inc.). Flow
cytometry was conducted in K562 cells using the
fluorescence-activated cell sorter Beckman CytoFLEX flow cytometer
(Beckman Coulter, Inc.) to analyze the primary erythroid cell
surface marker CD235a.
Luciferase reporter assay
Cells in the logarithmic growth stage were
resuspended, counted, inoculated in a 24-well culture plate
(~105 cells, depending on the size of the cells) and
cultured at 37°C in a 5% CO2 incubator until the degree
of cell fusion reached ~60%. PGL3 plasmid (Shanghai GeneChem Co.,
Ltd.) containing firefly and adrenal luciferase reporter genes was
transfected into cells at room temperature using the X-tremeGENE HP
transfection reagent (cat. no. 06366236001; Roche Diagnostics) for
24–48 h to observe the fluorescent marker gene expression on the
plasmid and to determine the transfection efficiency, or cells were
transfected for 48 h to detect luciferase activity using a
Dual-Luciferase® Reporter Assay system (cat. no. E2910;
Promega Corporation), according to the manufacturer's instructions
(19–21).
miRNA target prediction
The downstream target genes of miR-210-3p were
predicted by using databases such as miRBase (miRbase, http://www.mirbase.org/) and TargetScan
(TargetScanHuman 7.2, http://www.targetscan.org/vert_72/). Finally, from a
literature review, the target genes related to erythroid
development were screened and sequence matching analysis was
carried out.
Statistical analysis
SPSS 19.0 software (IBM, Corp.) was used for data
processing. Normally distributed data are presented as mean ± SD
from three independent experiments. One-way ANOVA was used to
compare multiple groups. When the variances were homogeneous, the
Tukey's test was used for post hoc analysis. When the variances
were uneven, Tamhane's T2 test was used for post hoc analysis. An
unpaired Student's t-test was used to compare two groups. P<0.05
was considered to indicate a statistically significant difference.
Experiments was repeated for three times.
Results
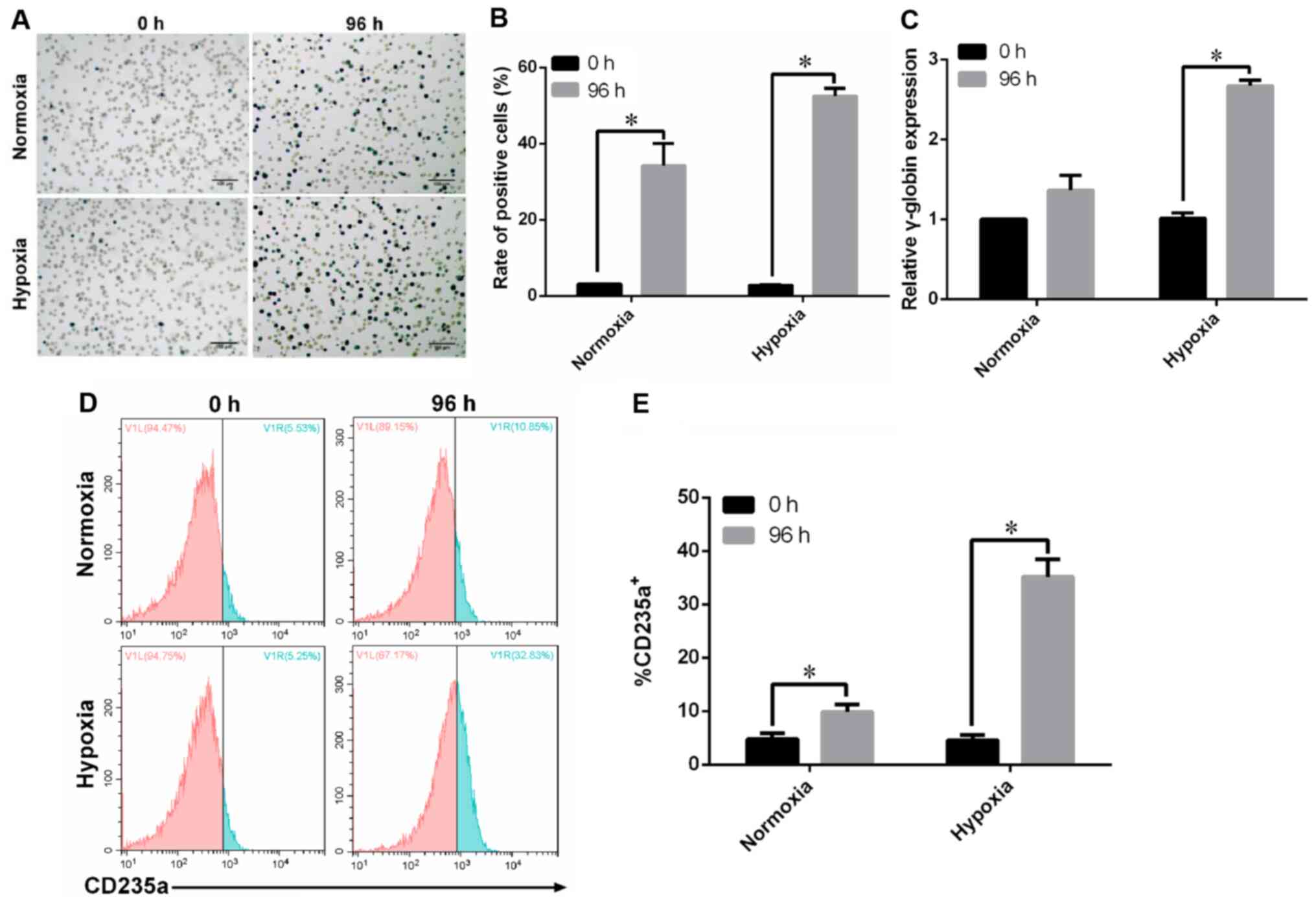
K562 cells successfully differentiate
into erythroid cells under hypoxia
An erythroid cell differentiation model under
hypoxia was established using hemin-induced K562 cells. Benzidine
staining was performed to identify hemoglobin-containing cells as
an indicator of erythrocyte differentiation. The results
demonstrated that hemin treatment significantly increased the
proportion of benzidine-positive K562 cells under hypoxia (Fig. 1A and B). Moreover, under hypoxia, a
2.6-fold elevation of γ-globin expression was observed at 96 h
compared with the level at 0 h (Fig.
1C). The results also indicated that the 96-h hemin treatment
increased the proportion of CD235a+ K562 cells compared
with the 0-h treatment group (Fig. 1D
and E). Thus, it was found that K562 cells successfully
differentiated into erythroid cells under hypoxia.
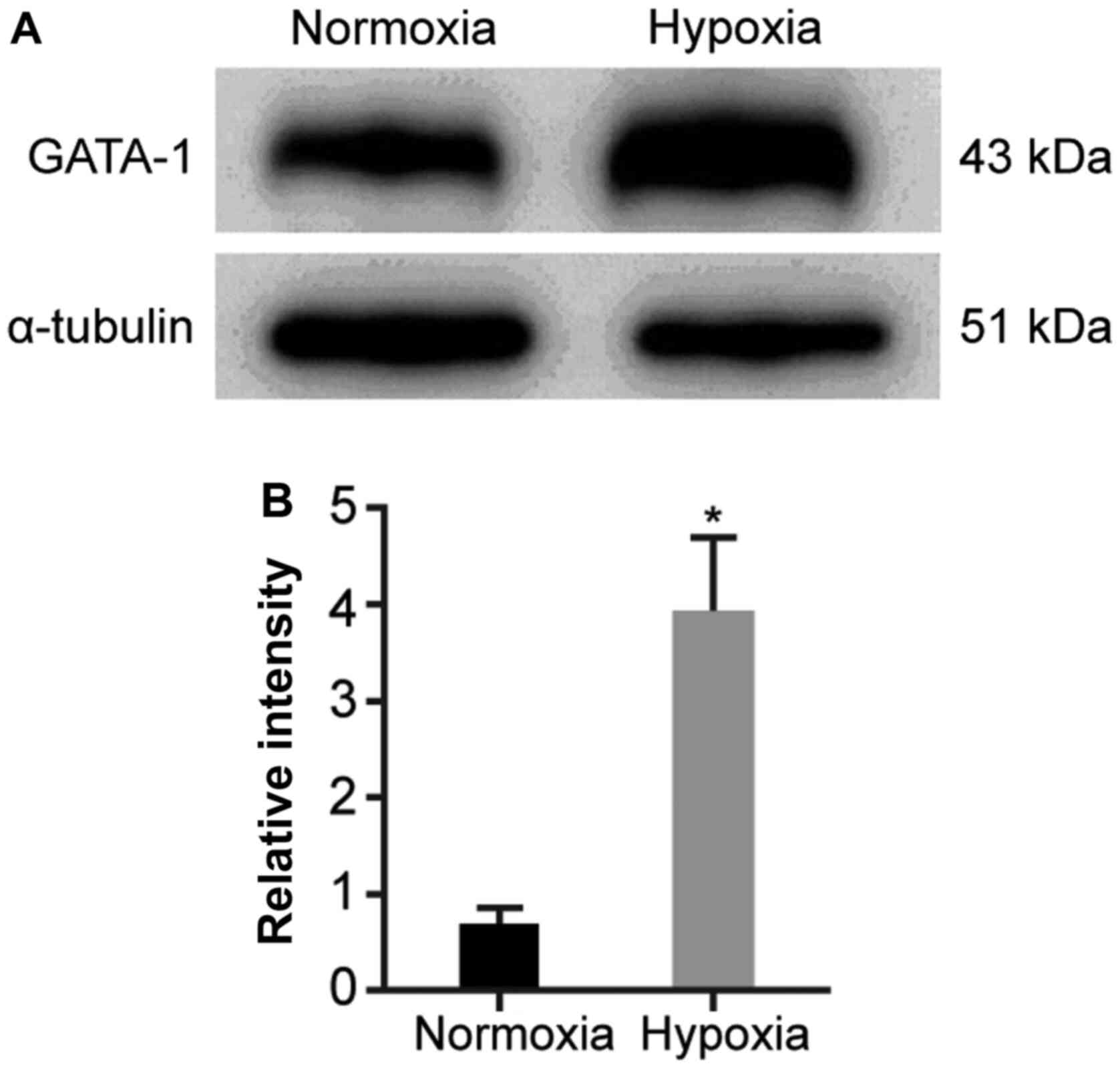
GATA-1 expression is upregulated in
hemin-induced K562 cells under hypoxia
To assess the influence of hypoxia on GATA-1
expression during erythroid cell differentiation, GATA-1 expression
levels were determined in hemin-treated K562 cells using western
blotting. The results demonstrated that the expression level of
GATA-1 was significantly increased in the hypoxia group compared
with that in the normoxia group (Fig.
2).
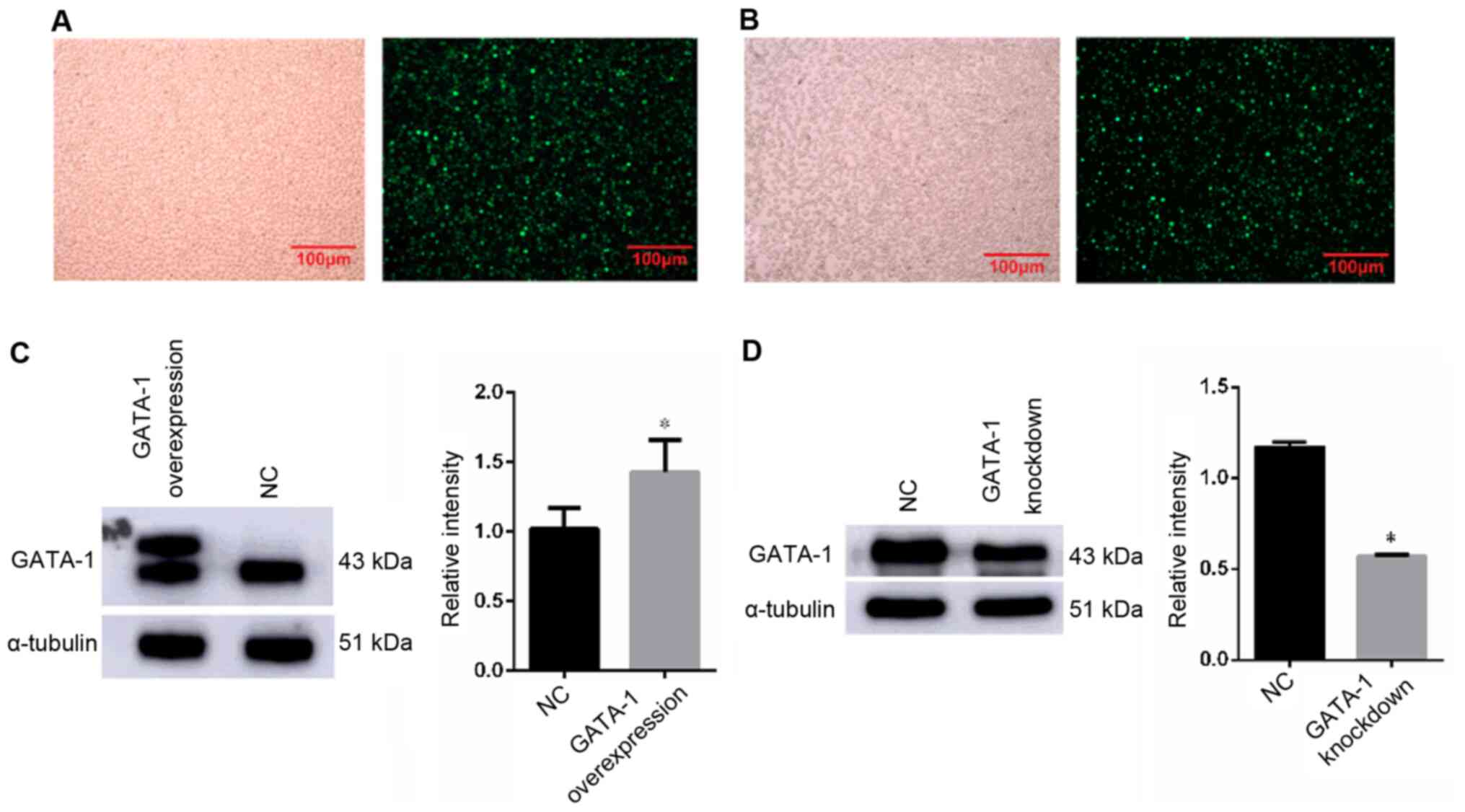
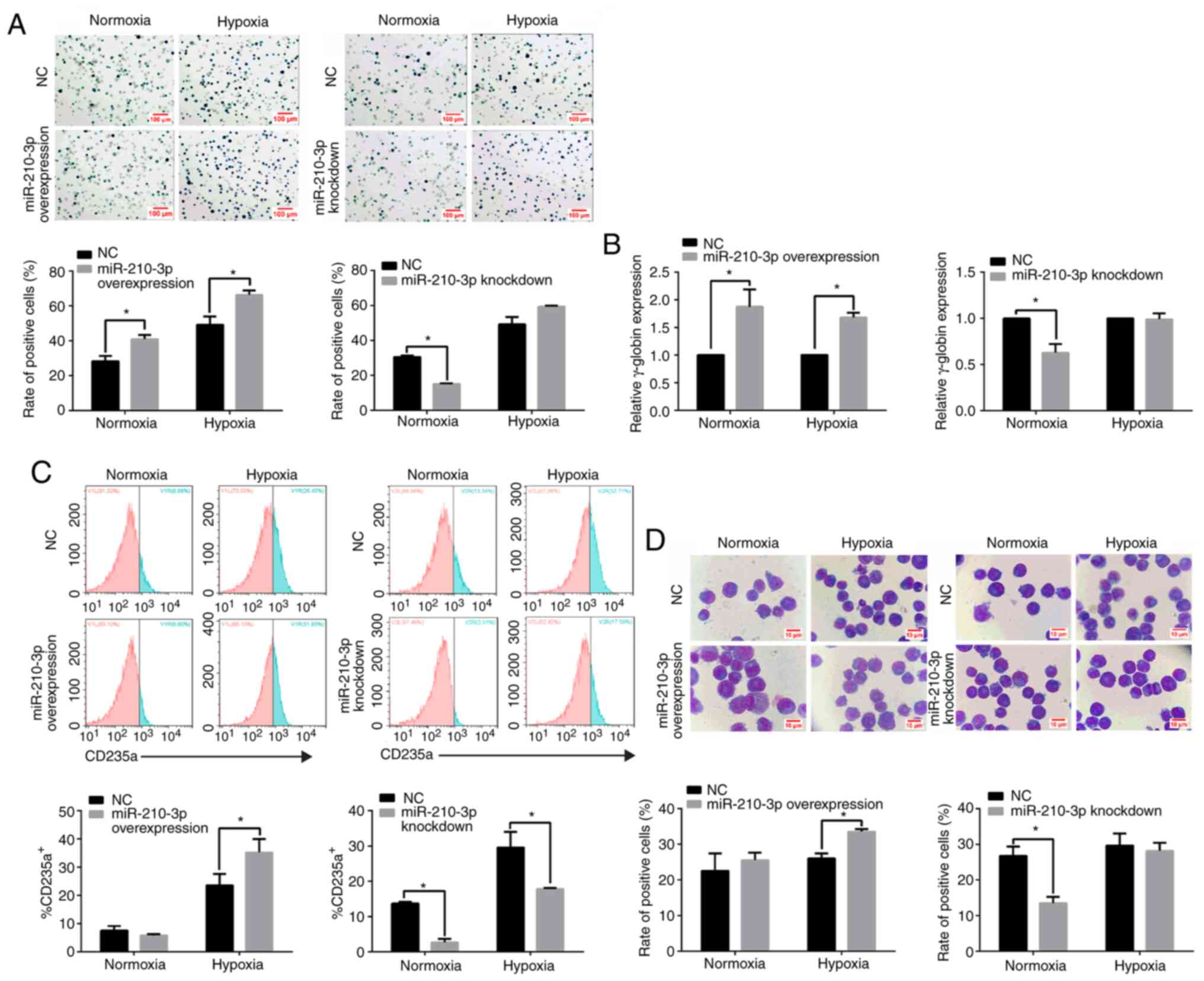
GATA-1 promotes the erythroid
differentiation of K562 cells under hypoxia
To further validate the role of GATA-1 in erythroid
differentiation under hypoxia, the corresponding erythroid
differentiation index was detected after GATA-1 overexpression or
knockdown. Under fluorescence microscope, the cells showed green
fluorescence, indicating successful lentivirus transfection
(Fig. 3A and B). The results of
western blot analysis of GATA-1 protein expression showed that its
expression level was higher in the overexpression group compared
with that of the NC group (Fig.
3C); In addition, the results of western blot analysis of
GATA-1 protein expression indicated that the relative expression
level in the knockdown group was significantly lower compared with
that of the NC group (Fig. 3D).
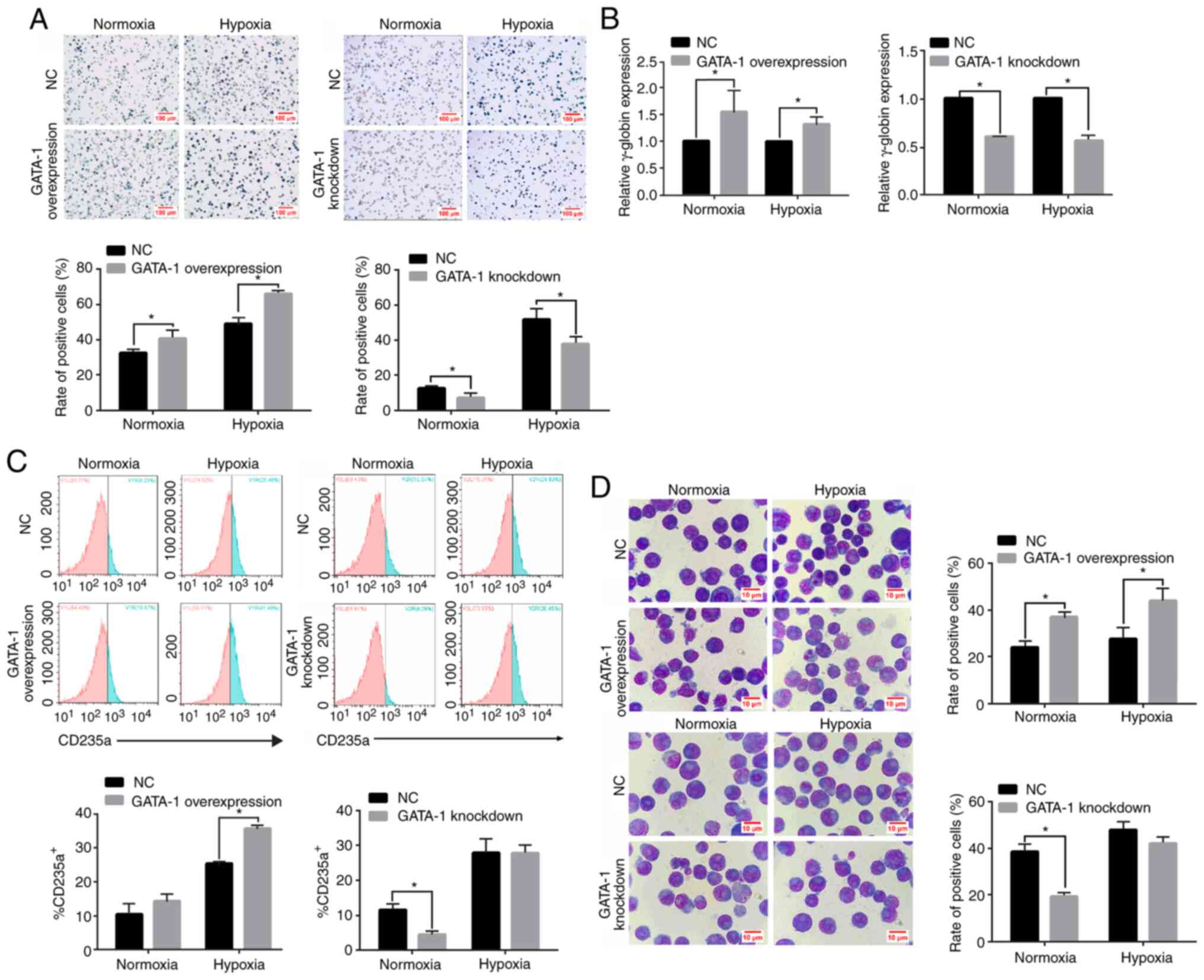
An elevation in the percentage of benzidine-positive
cells (Fig. 4A; left panel) and
γ-globin expression (Fig. 4B; left
panel) was observed at 96 h in GATA-1-overexpressing K562 cells
compared with the NC group. Additionally, the percentage of
CD235a+ (Fig. 4C; left
panel) in GATA-1-overexpressing K562 cells showed an expected
increase at 96 h under hypoxia. Wright's-Giemsa staining also
demonstrated that the proportion of cells with a larger cell volume
and a smaller nucleus accumulated at one side was significantly
higher in the GATA-1-overexpressing group compared with that in the
NC group (Fig. 4D; upper panel).
Conversely, transfection with the GV248 vector (knockdown group)
inhibited the expression of GATA-1 in K562 cells, as indicated by
the decrease in benzidine-positive cells (Fig. 4A; right panel) and repressed
γ-globin accumulation (Fig. 4B;
right panel) compared with the phenotype observed in the NC group.
Concurrently, the reduced percentage of CD235a+ cells
(Fig. 4C; right panel), as well as
the proportion of cells with an increased volume and a lopsided
nucleus were significantly lower in the GATA-1-knockdown group
compared with in the NC group; however, no significant difference
was observed with hypoxia (Fig. 4D;
bottom panel). These results indicated that GATA-1 promoted
erythroid differentiation in K562 cells under hypoxia.
miR-210-3p is a direct target gene of
GATA-1 in erythroid differentiation
qPCR was conducted to validate the expression level
of miR-210-3p in K562 cells during erythroid differentiation under
hypoxia. The expression level of miR-210-3p in the hypoxia group
was higher compared with that in the normoxia group (Fig. 5A). To further determine the direct
relevance of miR-210-3p upregulation in response to GATA-1
activation during erythroid cell differentiation, qPCR was
conducted to analyze miR-210-3p expression in K562 cells after
GATA-1 overexpression or knockdown. The results demonstrated that
miR-210-3p showed a significant increase in GATA-1-overexpressing
K562 cells under hypoxia (Fig. 5B).
Moreover, miR-210-3p was downregulated after the GATA-1 knockdown
(Fig. 5C), which revealed the
mechanism via which GATA-1 could activate miR-210-3p during
erythroid cell differentiation under hypoxia.
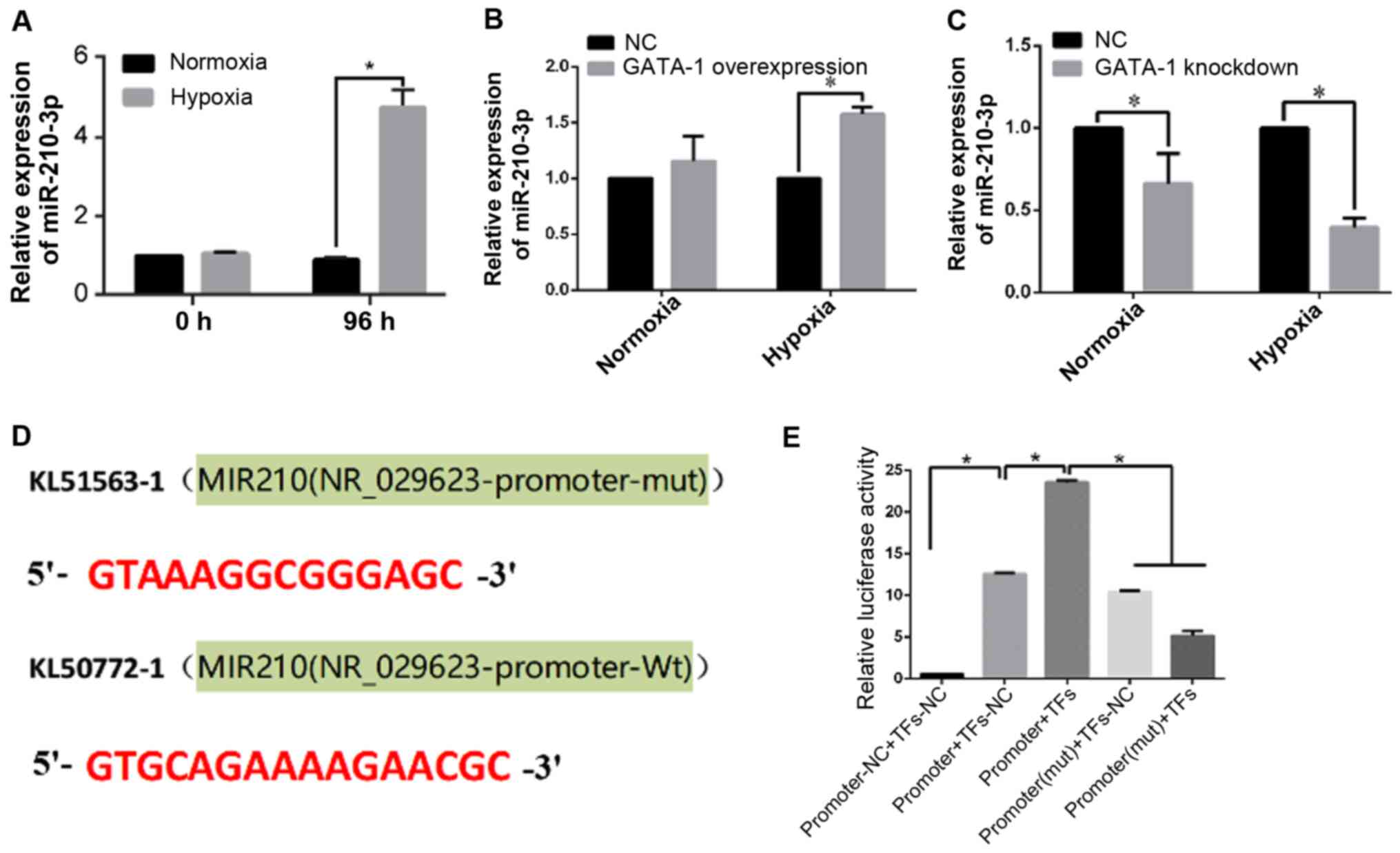 | Figure 5.miR-210-3p is a direct target of
GATA-1 in erythroid differentiation. (A) Reverse
transcription-quantitative PCR analysis of miR-210-3p expression in
K562 cells after hemin treatment for 0 and 96 h under hypoxia and
normoxia. *P<0.05, two-tailed Student's t-test, n=3. Reverse
transcription-quantitative PCR analysis of miR-210-3p expression in
hemin-induced K562 cells with (B) GATA-1 overexpression or (C)
GATA-1 knockdown. Data are presented as the mean ± SD from three
independent experiments. *P<0.05, two-tailed Student's t-test,
n=3. (D) Prediction of binding sites between the miR-210-3p
promoter region and GATA-1. (E) Relative luciferase activity of the
indicated reporter constructs. Firefly luciferase activity was
normalized to the activity of co-expressed Renilla
luciferase. Data are presented as the mean ± SD from three
independent experiments. *P<0.05, univariate ANOVA, n=3. GATA-1,
GATA binding protein 1; NC, negative control; miR, microRNA; mut,
mutant; Wt, wild-type; TFs, transcription factors. |
To confirm these findings, a dual-luciferase
reporter assay was performed to determine the binding site of
miR-210-3p and GATA-1. The binding site prediction of the
miR-210-3p promoter region is shown in Fig. 5D. The reporter assays revealed a
GATA-1-dependent activation of the miR-210-3p promoter. Notably,
mutations of the GATA-1-binding site abolished this upregulation,
as evidenced by the luciferase activity assay (Fig. 5E).
miR-210-3p promotes the erythroid
differentiation of K562 cells under hypoxia
To examine the role of miR-210-3p in erythroid cell
differentiation under hypoxia, a miR-210-3p-overexpressing and
knockdown lentivirus was transfected into K562 cells. Under
fluorescence microscope, the cells showed green fluorescence,
indicating successful lentivirus transfection (Fig. 6A and B), and RT-qPCR was performed
to assess the transfection efficiency (Fig. 6C and D). The effect of NC lentivirus
on the expression level of miR-210-3p was also detected via
RT-qPCR. The results demonstrated that, compared with the blank
control group, the NC lentivirus did not affect the expression
level of miR-210-3p itself, which was convenient for conducting
subsequent experiments (Fig.
6E).
The benzidine staining results demonstrated that
miR-210-3p overexpression increased the proportion of
benzidine-positive cells after 96 h of hemin treatment in K562
cells under hypoxia (Fig. 7A; left
panel). Furthermore, a 1.7-fold increase in γ-globin expression was
observed at 96 h in miR-210-3p-overexpressing K562 cells compared
with that in the NC group under hypoxia (Fig. 7B; left panel). It was found that
miR-210-3p overexpression increased the percentage of
CD235a+ cells compared with that in the NC group under
hypoxia (Fig. 7C; left panel).
Wright's-Giemsa staining also showed that miR-210-3p served a vital
role in erythroid cell differentiation by increasing the cell
volume, nuclear shrinkage and incidence of a lopsided nucleus under
hypoxic conditions (Fig. 7D; left
panel). In agreement with the gain-of-function data, a significant
decrease was observed in the results of the erythroid cell markers
γ-globin and CD235a, combined with the benzidine staining, under
normoxia, which demonstrated that loss of miR-210-3p function
impaired erythroid cell maturation (Fig. 7; right panels of A-D). Thus, it was
suggested miR-210-3p promoted erythroid cell differentiation.
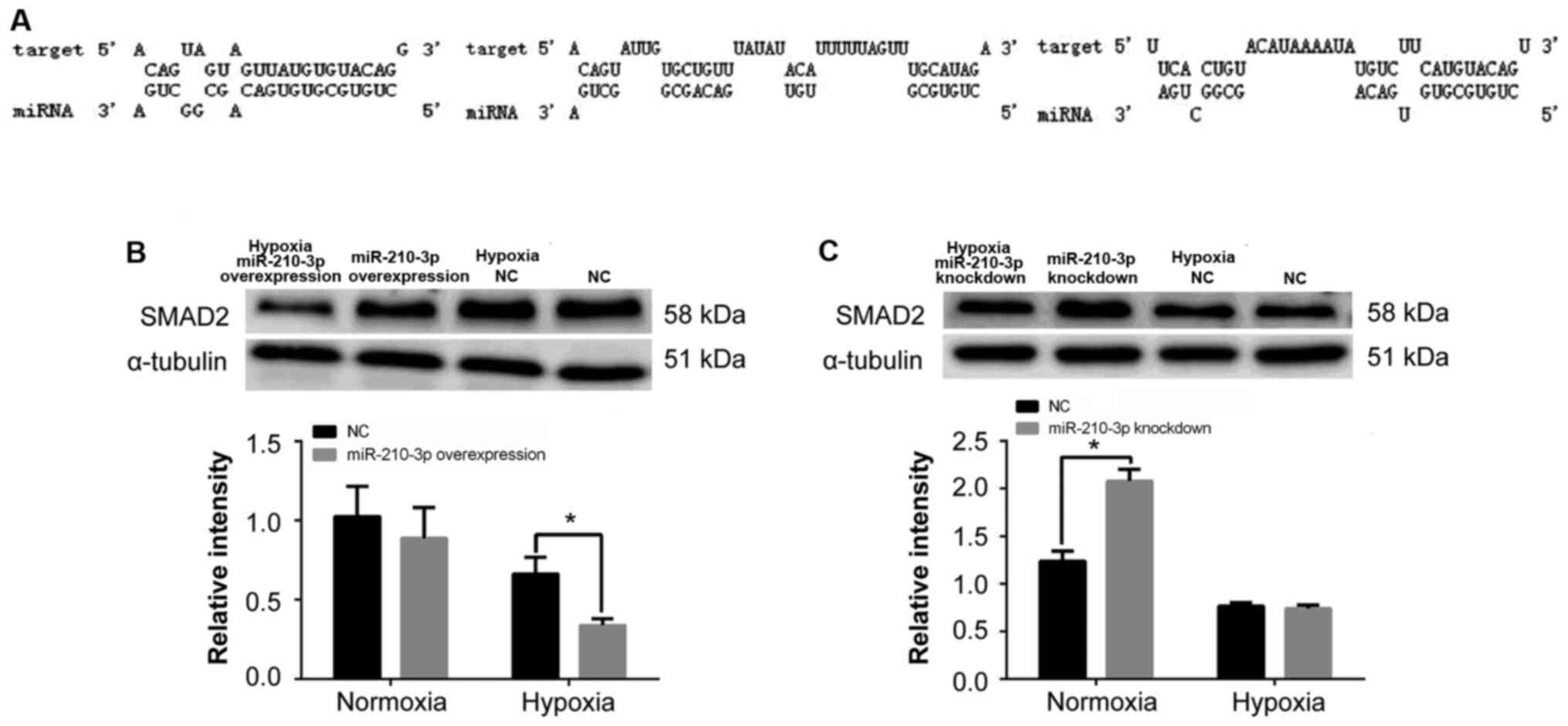
SMAD2 is a potential downstream target
gene for miR-210-3p
As miRNAs function by translationally repressing
their targets (22), the current
study aimed to examine the targets of miR-210-3p. To this end,
TargetScan was used to predict the downstream target genes of
miR-210-3p (Fig. 8A). When further
establishing the interaction between miR-210-3p and SMAD2, a
significant decrease in SMAD2 expression was observed in the
presence of lentivirus-mediated miR-210-3p overexpression in K562
cells under hypoxia compared with the NC group, in which no effect
was observed (Fig. 8B). Conversely,
SMAD2 expression was increased after the endogenous knockdown of
miR-210-3p by the inhibition lentivirus under normoxia (Fig. 8C).
SMAD2 acts as a negative regulator of
erythroid cell differentiation under hypoxia
To date, the function of SMAD2 in erythropoiesis
remains unknown. Thus, to investigate its biological role in
erythroid cell differentiation under hypoxia, a loss-of-function
experiment was conducted using lentivirus-mediated SMAD2-inhibition
in K562 cells. Under fluorescence microscope, the cells showed
green fluorescence, indicating successful lentivirus transfection,
western blotting was performed to assess transfection efficiency
(Fig. 9A). The percentage of
CD235a+ cells (Fig. 9B)
and benzidine-positive cells (Fig.
9C) was increased in SMAD2 inhibition lentivirus-transfected
K562 cells under normoxic and hypoxic conditions. Furthermore,
Wright's-Giemsa staining revealed that, under normoxic and hypoxic
conditions, the percentage of cells with larger volumes, lopsided
nucleus and nuclear shrinkage was higher compared with that in the
NC group (Fig. 9D). As expected,
SMAD2 knockdown increased the expression level of γ-globin in K562
cells at 96 h under normoxic and hypoxia (Fig. 9E). The aforementioned results
suggested that the increase was more significant under hypoxia.
Therefore, it was indicated SMAD2 negatively regulated erythroid
cell differentiation under hypoxia.
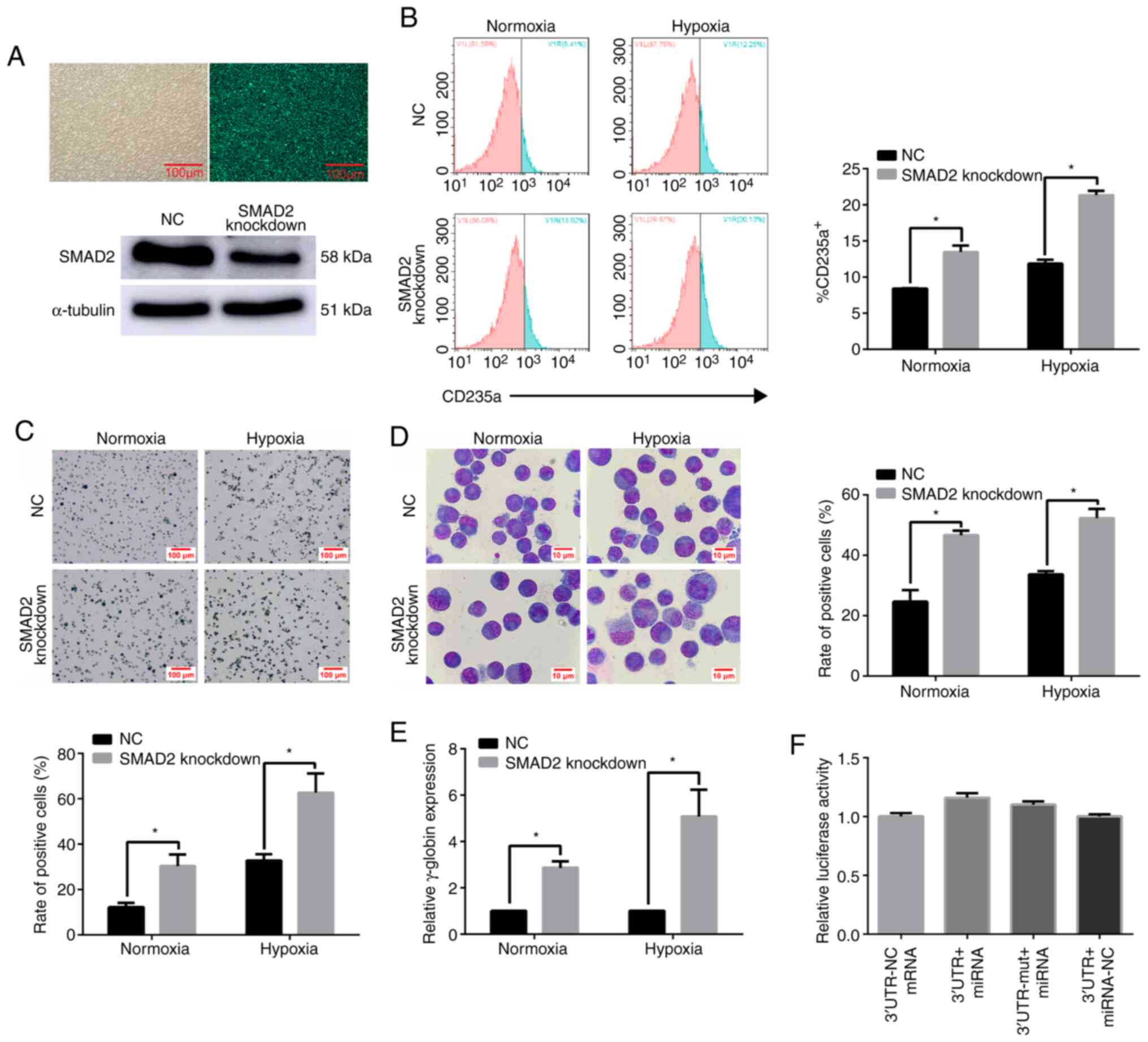 | Figure 9.SMAD2 acts as a negative regulator of
erythroid differentiation under hypoxia. (A) The fluorescence
results of SMAD2 knockdown lentivirus transfection efficiency were
detected. The field of view under an optical microscope (left). The
field of view under a fluorescence microscope (right). Scale bar,
100 µm. Western blot analysis of SMAD2 expression in K562 cells at
96 h after treatment with SMAD2-knockdown lentivirus. (B) FACS
analysis of K562 cells after SMAD2 knockdown and hemin induction
for 96 h. (C) Benzidine staining of K562 cells after SMAD2
knockdown and hemin treatment for 96 h. Hemoglobinized cells were
stained dark blue/black. Scale, bar 100 µm. (D) Representative
Wright's-Giemsa staining of K562 cells with SMAD2 knockdown and
hemin induction for 96 h. Scale bar, 10 µm. (E) Reverse
transcription-quantitative PCR analysis of γ-globin expression in
K562 cells with SMAD2 knockdown, following hemin induction.
γ-globin expression was significantly increased in SMAD2 knockdown
K562 cells compared with the NC under hypoxia. *P<0.05,
two-tailed Student's t-test, n=3. (F) Relative luciferase activity
of the indicated reporter constructs. Firefly luciferase activity
was normalized to the activity of co-expressed Renilla
luciferase, univariate ANOVA, n=3. NC, negative control; miRNA,
microRNA; UTR, untranslated region; UTR-mut, UTR-mutant; Wt, wild
type. |
SMAD2 does not bind to miR-210-3p
directly
To further determine the regulatory relationship
between miR-210-3p and SMAD2, direct binding sites between
miR-210-3p and SMAD2 were detected using a dual-luciferase assay.
The results demonstrated that the relative luciferase activity of
miR-210-3p-overexpressing vectors combined with SMAD2 wild-type
vectors was not significantly different compared with that of SMAD2
mutant vectors. Moreover, miR-210-3p could not bind directly to the
3′ untranslated region (UTR) of SMAD2 (Fig. 9F).
Discussion
Erythropoiesis, the process of erythroid cell
production, is controlled by several factors, including oxygen
levels (23). Hypoxia occurs at
high-altitude areas and in several physiological and pathological
processes, such as rapid tissue growth and acute and chronic
ischemia (24). Recently, studies
have been performed from the perspective that various geographical
and specific environmental differences may influence erythroid cell
differentiation. In fact, it has been reported that ‘special
environments’ (e.g., high-altitude areas) significantly influence
various physiological functions of the human body, including
erythroid cell differentiation (25). The effect of high-altitude hypoxia
on erythroid cell development continues to gain increased attention
from researchers. Of note, hypoxia may affect the core regulatory
factors of healthy erythroid cell differentiation (26).
GATA-1 regulates numerous erythroid cell
differentiation-specific genes by binding to its target protein via
double zinc finger domains. By activating target genes, GATA-1
helps to establish and maintain erythroid phenotypes (27). Moreover, it has been reported that
the maturation of GATA-1-deficient erythroid progenitor cells was
inhibited, and apoptosis was induced. However, the erythroid cells
matured when GATA-1 activity was restored in GATA-1 knockout
erythroid lines (28). GATA-1 can
modulate erythroid cell differentiation by modulating critical
miRNAs, and the mechanism underlying gene regulation via these
post-transcriptional inhibitors is being gradually revealed by
experimental observations (11).
Previous studies have confirmed that miRNAs are essential
regulators of all stages of hematopoiesis and hematopoietic
disorders (29,30). Some miRNAs reportedly prevent the
differentiation of early-stage progenitor cells or regulate the
terminal stages of hematopoietic development (31).
In the present study, an erythroid differentiation
model of K562 cells under hypoxia was used to investigate the
effects and functional relationship of GATA-1 and miR-210-3p on
erythroid differentiation and elucidate the possible regulatory
mechanism of erythroid differentiation under hypoxia. The
expression level of GATA-1 protein in the K562 cell erythroid
differentiation model was significantly higher compared with that
in the normoxic group. Additionally, the current study evaluated
the GATA-1-mediated promotion of erythroid development in K562
cells during hypoxia through gain- and loss-of-function
experiments. The results demonstrated that GATA-1 promoted
erythroid cell differentiation under hypoxic conditions. Other
regulatory pathways under hypoxia, however, may also influence
erythroid cell differentiation (32). Hypoxia can upregulate the expression
level of GATA-1 and accelerate erythroid cell differentiation; this
may be why some indicators of erythroid differentiation after
inhibiting GATA-1 under hypoxia showed no significant difference
compared with the NC group.
The present study demonstrated that the expression
level of miR-210-3p was associated with the upregulation and
downregulation of GATA-1 during erythroid differentiation of K562
cells under hypoxia. A dual-luciferase assay was used to verify the
relationship between GATA-1 and miR-210-3p. The results
demonstrated that the transcription factor could bind to the
wild-type miR-210-3p vector, increasing the level of fluorescence
expression, thus suggesting the presence of a direct binding site
between GATA-1 and the miR-210-3p promoter.
miR-210-3p reportedly mediates hypoxia-induced K562
and erythroid progenitor cell differentiation (33). miR-210-3p also participates in the
regulation of erythrocytic maturation, proliferation (13) and γ-globin gene expression in early
erythrocytes. Previous research has reported that miR-210-3p
enhanced CD34+ erythroid progenitor cell differentiation
(34). Moreover, a notable increase
in miR-210-3p expression during erythroid differentiation of a
murine fetal liver cell culture has been observed (15), and the induction of erythropoiesis
after phenylhydrazine-induced hemolytic anemia increased miR-210-3p
levels (15). Based on these
results, it was suggested that miR-210-3p could affect erythroid
differentiation under hypoxia. Therefore, the current study aimed
to upregulate and to inhibit miR-210-3p expression to detect the
corresponding erythroid differentiation indexes. It was found that
miR-210-3p positively regulated erythroid cell differentiation
under hypoxia.
To determine the possible underlying mechanism, a
previous study investigated miR-210-3p using
miRBase/Targetscan/KEGG (Kyoto Encyclopedia of Genes and Genomes)
(35) pathway analyses using
bioinformatics software and identified SMAD2, which is involved in
the proliferation (36), apoptosis
(37) and differentiation (38) of several types of cells, as the
possible downstream target gene involved in the regulation of
erythroid differentiation. SMAD2, which exerts an inhibitory
influence under normal steady-state conditions, has emerged as an
important regulator of erythropoiesis (39–42).
Additionally, overactivation or dysregulation of SMAD2 signaling
has been implicated in diseases characterized by impaired erythroid
cell differentiation (43–46). Histological examination has shown
that SMAD2 was activated in hematopoietic progenitor cells and
participated in the regulation of TGF-β-mediated proliferation and
erythroid differentiation (43).
Accumulating evidence has also suggested that luspatercept-mediated
inhibition of SMAD2 signaling promotes erythroid differentiation
(47). Moreover, inhibition of
SMAD2 increased the level of hepatocyte growth factor, an effective
angiogenic factor, in patients with squamous cell carcinoma
(48,49). Taken together, these results
indicate that SMAD2 could regulate the development of erythrocytes.
It has also been reported that miR-210-3p has a direct binding site
for members of the SMAD family, which regulates its expression
(50). Therefore, we hypothesized
that miR-210-3p may serve a role in promoting erythroid cell
differentiation by inhibiting the gene expression of SMAD2.
To verify whether SMAD2 was directly regulated by
miR-210-3p, a dual-luciferase assay was conducted. However, it was
found that miR-210-3p did not bind directly to the 3′UTR of SMAD2,
and it was identified that it negatively affected SMAD2 expression,
as determined detecting SMAD2 protein expression after the
overexpression and knockdown of miR-210-3p expression. Further
analysis demonstrated that SMAD2 knockdown enhanced erythroid cell
differentiation. A previous study revealed that miR-210-3p could
inhibit the activity of the TGF signaling pathway during osteoblast
differentiation (51). However,
SMAD-1/5/8 competitive inhibition may interfere with SMAD2/3
binding to Co-SMAD (52), thereby
accelerating osteoblast differentiation (53). Therefore, we hypothesized the
existence of other sequences in the 3′UTR of SMAD2 that could
directly bind to miR-210-3p. Alternatively, miR-210-3p may not bind
directly to the 3′UTR of SMAD2 but may indirectly inhibit SMAD2 by
suppressing the activity of the TGF signaling pathway, thus
facilitating erythroid differentiation.
In conclusion, the present data suggested that,
under hypoxia, GATA-1 overexpression significantly promoted
erythroid differentiation, possibly by modulating miR-210-3p
expression. Furthermore, the expression level of miR-210-3p
increased with the degree of differentiation as it regulated
several erythroid differentiation-related genes. Thus, with
increasing information regarding miRNA profiles and transcription
factor regulation, integrating these data may improve the current
understanding of the molecular mechanisms underlying human
adaptation and pathophysiology under hypoxic conditions.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant no. 81760333), the Basic
Research (Application) Project of the QingHai Science and
Technology Department (grant no. 2017-ZJ-722) and the Scientific
Research Fund for Young Scientist (grant no. 2019-kty-3). The
funding sources helped in the design of the study. First, when
applying for funding for a project, members of the foundation
reviewed the feasibility of the project and proposed suggestions
for modification. Second, the foundation conducted regular
inspections as the project progressed.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CH and LF conceptualized and designed this study. CH
and LF are responsible for confirming the authenticity of the raw
data. CY, CF, YY and JD acquired the data. CH, CF, JD, TL and SW
analyzed and interpreted the data. CH drafted the manuscript, and
CH, YY and LF edited the original draft. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNAs/miRs
|
microRNAs
|
|
NC
|
negative control
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
GATA-1
|
GATA binding protein 1
|
References
|
1
|
Kerenyi MA and Orkin SH: Networking
erythropoiesis. J Exp Med. 207:2537–2541. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Haase VH: Hypoxic regulation of
erythropoiesis and iron metabolism. Am J Physiol Ren Physiol.
299:F1–F13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Narayan AD, Ersek A, Campbell TA, Colón
DM, Pixley JS and Zanjani ED: The effect of hypoxia and stem cell
source on haemoglobin switching. Br J Haematol. 128:562–570. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rogers HM, Yu X, Wen J, Smith R, Fibach E
and Noguchi CT: Hypoxia alters progression of the erythroid
program. Exp Hematol. 36:17–27. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vlaski M, Lafarge X, Chevaleyre J, Duchez
P, Boiron JM and Ivanovic Z: Low oxygen concentration as a general
physiologic regulator of erythropoiesis beyond the EPO-related
downstream tuning and a tool for the optimization of red blood cell
production ex vivo. Exp Hematol. 37:573–584. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ferreira R, Ohneda K, Yamamoto M and
Philipsen S: GATA1 Function, a paradigm for transcription factors
in hematopoiesis. Mol Cell Biol. 25:1215–1227. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu M, Riva L, Xie H, Schindler Y, Moran
TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, et al: Insights
into GATA-1-mediated gene activation versus repression via
genome-wide chromatin occupancy analysis. Mol Cell. 36:682–695.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Scherzer CR, Grass JA, Liao Z, Pepivani I,
Zheng B, Eklund AC, Ney PA, Ng J, McGoldrick M, Mollenhauer B, et
al: GATA transcription factors directly regulate the Parkinson's
disease-linked gene alpha-synuclein. Proc Natl Acad Sci USA.
105:10907–10912. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crispino JD: GATA1 in normal and malignant
hematopoiesis. Semin Cell Dev Biol. 16:137–147. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cantor AB and Orkin SH: Transcriptional
regulation of erythropoiesis: an affair involving multiple
partners. Oncogene. 21:3368–3376. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dore LC, Amigo JD, Dos Santos CO, Zhang Z,
Gai X, Tobias JW, Yu D, Klein AM, Dorman C, Wu W, et al: A
GATA-1-regulated microRNA locus essential for erythropoiesis. Proc
Natl Acad Sci USA. 105:3333–3338. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nilsen TW: Mechanisms of microRNA-mediated
gene regulation in animal cells. Trends Genet. 5:243–249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bianchi N, Zuccato C, Finotti A, Lampronti
I, Borgatti M and Gambari R: Involvement of miRNA in erythroid
differentiation. Epigenomics. 4:51–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang X, Le QT and Giaccia AJ:
MiR-210-micromanager of the hypoxia pathway. Trends Mol Med.
16:230–237. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kosaka N, Sugiura K, Yamamoto Y, Yoshioka
Y, Miyazaki H, Komatsu N, Ochiya T and Kato T: Identification of
erythropoietin-induced microRNAs in haematopoietic cells during
erythroid differentiation. Br J Haematol. 142:293–300. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Rowley PT, Ohlsson-Wilhelm BM, Farley BA
and LaBella S: Inducers of erythroid differentiation in K562 human
leukemia cells. Exp Hematol. 9:32–37. 1981.PubMed/NCBI
|
|
17
|
Gahmberg CG and Andersson LC: K562-a human
leukemia cell line with erythroid features. Semin Hematol.
18:72–77. 1981.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mazan-Mamczarz K and Gartenhaus RB: Role
of microRNA deregulation in the pathogenesis of diffuse large
B-cell lymphoma (DLBCL). Leuk Res. 37:1420–1428. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fish JE, Santoro MM, Morton SU, Yu S, Yeh
RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY and Srivastava D:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abend JR, Uldrick T and Ziegelbauer JM:
Regulation of tumor necrosis factor-like weak inducer of apoptosis
receptor protein (TWEAKR) expression by kaposi's sarcoma-associated
herpesvirus MicroRNA prevents tweak-induced apoptosis and
inflammatory cytokine expression. J Virol. 84:12139–12151. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shivdasani RA: MicroRNAs: Regulators of
gene expression and cell differentiation. Blood. 108:3646–3653.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bapat A, Schippel N, Shi X, Jasbi P, Gu H,
Kala M, Sertil A and Sharma S: Hypoxia promotes erythroid
differentiation through the development of progenitors and
proerythroblasts. Exp Hematol. 97:32–46.e35. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaelin WG Jr and Ratcliffe PJ: Oxygen
sensing by metazoans: The central role of the HIF hydroxylase
pathway. Mol Cell. 30:393–402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Windsor JS and Rodway GW: Heights and
haematology: The story of haemoglobin at altitude. Postgrad Med J.
83:148–151. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Noguchi CT and Rogers H: Hypoxia alters
progression of the erythroid program. Exp Hematol. 2:163. 2007.
|
|
27
|
Welch JJ, Watts JA, Vakoc CR, Yao Y, Wang
H, Hardison RC, Blobel GA, Chodosh LA and Weiss MJ: Global
regulation of erythroid gene expression by transcription factor
GATA-1. Blood. 10:3136–3147. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu J, Zhou LQ, Yu W, Zhao ZG, Xie XM, Wang
WT, Xiong J, Li M, Xue Z, Wang X, et al: PML4 facilitates erythroid
differentiation by enhancing the transcriptional activity of
GATA-1. Blood. 123:261–270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Doss JF, Corcoran DL, Jima D, Telen MJ,
Dave SS and Chi JT: A comprehensive joint analysis of the long and
short RNA transcriptomes of human erythrocytes. BMC Genomics.
16:952. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rasmussen KD, Simmini S, Abreugoodger C,
Bartonicek N, Di Giacomo M, Bilbao-Cortes D, Horos R, Von Lindern
M, Enright AJ and O'Carroll D: The miR-144/451 locus is required
for erythroid homeostasis. J Exp Med. 207:1351–1358. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Undi RB, Kandi R and Gutti RK: MicroRNAs
as haematopoiesis regulators. Adv Hematol. 2013:6957542013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie Y, Li W, Feng J, Wu T and Li J:
MicroRNA-363 and GATA-1 are regulated by HIF-1α in K562 cells under
hypoxia. Mol Med Rep. 14:2503–2510. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Raghuwanshi S, Karnati HK, Sarvothaman S,
Gutti U, Saladi RGV, Tummala PR and Gutti RK: microRNAs: Key
players in hematopoiesis. Adv Exp Med Bio. 887:171–211. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sarakul O, Vattanaviboon P, Tanaka Y,
Fucharoen S, Abe Y, Svasti S and Umemura T: Enhanced erythroid cell
differentiation in hypoxic condition is in part contributed by
miR-210. Blood Cells Mol Dis. 51:98–103. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu Y, Wang D, Wang F, Li T, Dong L, Liu
H, Ma Y, Jiang F, Yin H, Yan W, et al: A comprehensive analysis of
GATA-1-regulated miRNAs reveals miR-23a to be a positive modulator
of erythropoiesis. Nucleic Acids Res. 41:4129–4143. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu B, Sun J, Lei X, Zhu Z, Pei C and Qin
L: MicroRNA-486-5p suppresses TGF-β2-induced proliferation,
invasion and epithelial-mesenchymal transition of lens epithelial
cells by targeting Smad2. J Biosci. 42:575–584. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
AlMegbel AM and Shuler CF: SMAD2
overexpression rescues the TGF-β3 null mutant mice cleft palate by
increased apoptosis. Differentiation. 111:60–69. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheung KS, Sposito N, Stumpf PS, Wilson
DI, Sanchez-Elsner T and Oreffo ROC: MicroRNA-146a regulates human
foetal femur derived skeletal stem cell differentiation by
down-regulating SMAD2 and SMAD3. PLoS One. 9:e980632014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Söderberg SS, Karlsson G and Karlsson S:
Complex and context dependent regulation of hematopoiesis by TGF-β
superfamily signaling. Ann N Y Acad Sci. 1176:55–69. 2009.
View Article : Google Scholar
|
|
40
|
Blank U and Karlsson S: The role of Smad
signaling in hematopoiesis and translational hematology. Leukemia.
25:1379–1388. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shav-Tal Y and Zipori D: The role of
activin A in regulation of hemopoiesis. Stem Cells. 20:493–500.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shiozaki M, Sakai R, Tabuchi M, Nakamura
T, Sugino K, Sugino H and Eto Y: Evidence for the participation of
endogenous activin A/erythroid differentiation factor in the
regulation of erythropoiesis. Proc Natl Acad Sci USA. 89:1553–1556.
1992. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou L, Nguyen AN, Sohal D, Ma JY,
Pahanish P, Gundabolu K, Hayman J, Chubak A, Mo Y, Bhagat TD, et
al: Inhibition of the TGF-β receptor I kinase promotes
hematopoiesis in MDS. Blood. 112:3434–3443. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dussiot M, Maciel TT, Fricot A, Chartier
C, Negre O, Veiga J, Grapton D, Paubelle E, Payen E, Beuzard Y, et
al: An activin receptor IIA ligand trap corrects ineffective
erythropoiesis in β-thalassemia. Nat Med. 20:398–407. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Suragani RN, Cawley SM, Li R, Wallner S,
Alexander MJ, Mulivor AW, Gardenghi S, Rivella S, Grinberg AV,
Pearsall RS and Kumar R: Modified activin receptor IIB ligand trap
mitigates ineffective erythropoiesis and disease complications in
murine β-thalassemia. Blood. 123:3864–3872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Suragani RN, Cadena SM, Cawley SM, Sako D,
Mitchell D, Li R, Davies MV, Alexander MJ, Devine M, Loveday KS, et
al: Transforming growth factor-β superfamily ligand trap ACE-536
corrects anemia by promoting late-stage erythropoiesis. Nat Med.
20:408–414. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Martinez PA, Li R, Ramanathan HN, Bhasin
M, Persall RS, Kumar R and Suragani RNVS: Smad2/3-pathway ligand
trap luspatercept enhances erythroid differentiation in murine
β-thalassaemia by increasing GATA-1 availability. J Cell Mol Med.
24:6162–6177. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wu F, Weigel KJ, Zhou H and Wang XJ:
Paradoxical roles of TGF-β signaling in suppressing and promoting
squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai).
50:730. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tannehill-Gregg SH, Kusewitt DF, Rosol TJ
and Weinstein M: The roles of Smad2 and Smad3 in the development of
chemically induced skin tumors in mice. Vet Pathol. 41:278–282.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Phuah NH, Azmi MN, Awang K and Nagoor NH:
Down-regulation of microRNA-210 confers sensitivity towards
1′s-1′-acetoxychavicol acetate (ACA) in cervical cancer cells by
targeting SMAD4. Mol Cells. 40:291–298. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mizuno Y, Tokuzawa Y, Ninomiya Y, Yagi K,
Yatsuka-Kanesaki Y, Suda T, Fukuda T, Katagiri T, Kondoh Y, Amemiya
T, et al: miR-210 promotes osteoblastic differentiation through
inhibition of AcvR1b. FEBS Lett. 583:2263–2268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Miyazono K, Maeda S and Imamura T: BMP
receptor signaling: Transcriptional targets, regulation of signals,
and signaling cross-talk. Cytokine Growth Factor Rev. 16:251–263.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Maeda S, Hayashi M, Komiya S, Imamura T
and Miyazono K: Endogenous TGF-beta signaling suppresses maturation
of osteoblastic mesenchymal cells. EMBO J. 23:552–563. 2004.
View Article : Google Scholar : PubMed/NCBI
|