Introduction
Periodontitis, which is a representative
inflammatory disease in the gingival tissues, is generally
triggered by various inflammation-induced factors derived from
bacteria and host cells. If left untreated, the disease eventually
causes tissue breakdown and alveolar bone loss. In addition, it may
increase a risk of developing serious systemic complications such
as diabetes (1–3). Inflammation is a protective immune
response to injury and infection and responsible for maintaining
homeostasis (1). However, excess
and prolonged inflammation can lead to cellular damage and organ
dysfunction (1,3). There are numerous molecules that can
affect the progression of periodontal inflammation. Among them,
TNF-α and interleukins such as IL-1, IL-6 and IL-8 are the central
modulator of immune response and critically involved in
inflammatory events (4,5). These molecules can activate the
transcription factor NF-κB and this activation occurs through
phosphorylation of p65 and translocation of the p65/p50 dimer from
the cytoplasm into the nuclei. Since interleukins and TNF-α are the
targets of NF-κB, this process results in a positive feedback loop
promoting further inflammation (6).
Thus, the precise regulation of this inflammatory signaling pathway
is important to control and treat a number of inflammatory
disorders. At present, however, preventive and therapeutic means
for inflammatory oral diseases still need to be explored.
A microRNA (miRNA) is a small non-cording RNA that
modulates gene expression at the post-transcriptional level via
binding to the complementary sequences in 3′-untranslated region
(3′-UTR) of target mRNAs (7).
Growing evidence has suggested that miRNAs are involved in the
regulation of diverse physiological and pathological events, such
as cell differentiation, development, migration and apoptosis. A
number of studies have demonstrated that miRNAs are often
dysregulated and closely associated with solid and blood cell
cancer and infectious inflammatory diseases (7–13).
Furthermore, circulating extracellular miRNAs are stable and
detectable in body fluid specimens from patients and their contents
in the specimens may reflect or be attributed to a certain disease
status (7–9,13–15).
On the basis of these findings, miRNAs have emerged as an
attractive and powerful tool for diagnostic biomarkers and
therapeutic targets.
Among miRNAs, miR-429, a member of miR-200 family,
is studied as an epithelial mesenchymal transition-associated miRNA
and is often found to be dysregulated in various types of malignant
tumors including oral squamous cell carcinoma (16–19).
Previous studies have reported that miR-429 and its family miRNAs
suppress the expression of multiple genes involved in inflammatory
signaling and production of interleukins, particularly IL-8
(16–18,20–27).
The evidence for the relevance and function of miR-429 in
pathological and physiological events has accumulated; however, the
possible association of miR-429 with oral inflammatory processes
remains to be elucidated.
The present study aimed to investigate the molecular
mechanisms of miR-429 action to suppress inflammation in oral
mucosa using a human gingival squamous cell carcinoma line
(Ca9-22). The results demonstrated that miR-429 inhibited IL-8
production through the NF-κB pathway. In addition, miR-429 directly
interacted with the IKKβ, an essential activator of NF-κB in the
canonical pathway. Collectively, the results suggested that miR-429
has anti-inflammatory function in gingival epithelium, providing
new information concerning with the role of miRNAs as a modulator
of inflammatory cascade in oral diseases.
Materials and methods
Cell culture
Ca9-22 (JCRB0625, Lot. 11182016, Biomedical
Innovation, Health and Nutrition Research Institute, authenticated
by Japanese Collection of Research Bioresources Cell Bank), a human
gingival squamous cell carcinoma line, was cultured in Dulbecco's
modified Eagle's medium (DMEM) (cat. no. D5796; Sigma-Aldrich;
Merck KGaA) supplemented with 10% fetal bovine serum (cat. no.
10270; Gibco; Thermo Fisher Scientific, Inc.), 100,000 U/ml
penicillin and 100 µg/ml streptomycin (cat. no. 164-25251, Wako
Pure Chemical Industries, Ltd.) in a humidified atomosphere of 5%
CO2 at 37°C.
Transfection
The Ca9-22 cells were seeded onto a 24-well plate
(5×104 cells per well) and onto a 12-well plate
(1×105 cells per well), respectively, for the evaluation
of mRNA and protein levels. After the incubation for 24 h at 37°C,
the cells were transfected with miR-429 mimic (sense,
5′-GGUUUUACCAGACAGUAUUATT-3′ and antisense,
5′-UAAUACUGUCUGGUAAAACCGU-3′; miRVana miRNA mimic; assay ID
MC10221; Thermo Fisher Scientific, Inc.), negative control mimic
(NCm; miRVana miRNA mimic negative control #1; cat. no. 4464058;
Thermo Fisher Scientific, Inc.), miR-429 inhibitor
(5′-ACGGUUUUACCAGACAGUAUUA-3′; miRVana miRNA inhibitor; assay ID
MH10221; Thermo Fisher Scientific, Inc.), negative control
inhibitor (NCi; miRVana miRNA Inhibitor Negative Control #1; cat.
no. 4464076; Thermo Fisher Scientific, Inc.), small interfering
(si) RNA targeting IKKβ (siRNA-IKKβ; sense,
5′-GGGCAGUCUUUGCACAUCATT-3′ and antisense,
5′-UGAUGUGCAAAGACUGCCCTG-3′; Silencer select validated siRNA; assay
ID s7263; Thermo Fisher Scientific, Inc.) or negative control siRNA
(NCsi; Silencer select negative control No. #1; cat. no. 4390843;
Thermo Fisher Scientific, Inc.) using Lipofectamine®
RNAiMax (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocols, and incubated for 24 or 48 h at 37°C.
Then, the transfected cells were used for further experiments. The
concentrations of miR-429 mimic, NCm, miR-429 inhibitor and NCi
were 50 nM and those of siRNA-IKKβ and NCsi were 30 nM.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from cells stimulated with
TNF-α (5, 10 or 50 ng/ml; cat. no. 207-15261; Wako Pure Chemical
Industries, Ltd.) or the transfected cells using a Direct-zol
miniprep RNA kit (cat. no. R2063; Zymo Research Corp.) with
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.).
Conversion of total RNA into complementary DNA (cDNA) was performed
by using a TaqMan miRNA reverse transcription kit (Thermo Fisher
Scientific, Inc.) for miR-429 and U6 and by using a PrimeScript RT
reagent kit with genomic DNA Eraser (cat. no. RR047A; Takara Bio,
Inc.) for IL-1β, IL-6, IL-8, IKKβ and GAPDH. Both kits were used
according to the manufacturers' protocols. qPCR was performed using
a CFX96 real-time PCR system (Bio-Rad Laboratories, Inc.) and the
relative expression level was normalized with U6 or GAPDH
expression and calculated based on the Cq (ΔΔCq) method
(28). The expression level of
miR-429 and U6 was examined according to the previous RT-qPCR
condition (12), using TaqMan Fast
universal PCR master mix (Thermo Fisher Scientific, Inc.) with
TaqMan miRNA primers of miR-429 (assay ID 001024; Thermo Fisher
Scientific, Inc.) and U6 (assay ID 001973; Thermo Fisher
Scientific, Inc.). As reported previously (29), the mRNA level of IL-1β, IL-6, IL-8,
IKKβ and GAPDH was examined according to the PCR reaction condition
as follows: 30 sec at 95°C and subsequent 40 repeats of 10 sec at
95°C, 10 sec at 63°C and 15 sec at 72°C. The reaction was conducted
using a SYBR Premix Ex Taq II (cat. no. RR820A; Takara Bio, Inc.)
with specific primers as follows: Human IL-1β (forward,
5′-AAACAGATGAAGTGCTCCTTCC-3′ and reverse,
5′-AAGATGAAGGGAAAGAAGGTGC-3′), human IL-6 (forward,
5′-AATCATCACTGGTCTTTTGGAG-3′ and reverse,
5′-GCATTTGTGGTTGGGTCA-3′), human IL-8 (forward,
5′-GACATACTCCAAACCTTTCCACC-3′ and reverse,
5′-AACTTCTCCACAACCCTCTGC-3′), human IKKβ (forward,
5′-ACTTGGCGCCCAATGACCT-3′ and reverse, 5′-CTCTGTTCTCCTTGCTGCA-3′)
and GAPDH (forward, 5′-CTCATGACCACAGTCCATGC-3′ and reverse,
5′-TTCAGCTCTGGGATGACCTT-3′).
ELISA
The transfected Ca9-22 cells were incubated for 4 h
at 37°C in the fresh culture medium with or without human
recombinant TNF-α (50 ng/ml). Then, the medium was collected and
stored at −20°C until use. The amount of IL-8 in the medium was
measured with a human IL-8 Uncoated enzyme-linked immunosorbent
assay kit (cat. no. 88-8086-22, Thermo Fisher Scientific,
Inc.).
Western blotting
For evaluation of phosphorylated (p-)p65 and p65
levels, the transfected Ca9-22 cells were cultured in the fresh
medium for 6 h at 37°C and then incubated in the medium with or
without TNF-α (50 ng/ml) for 15 min at 37°C prior to protein
extraction. Total protein of the transfected cells was extracted
using RIPA lysis buffer (cat. no. 20-188; EMD Millipore) containing
Halt protease and phosphatase inhibitor (cat. no. 1861280, Thermo
Fisher Scientific, Inc.). The protein extract (10 µg) was separated
on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel and
blotted onto a PVDF membrane. The membrane was blocked with 5%
non-fat dried milk (cat. no. 999S; Cell Signaling Technology, Inc.)
in Tris-buffered saline with 0.1% Tween-20 (TBS-T) for 1 h at room
temperature. Then, the membrane was incubated with the rabbit
anti-IKKβ (cat. no. 2678P; Cell Signaling Technology, Inc.),
anti-p-NF-κB (cat. no. 3033S; Cell Signaling Technology, Inc.) or
anti-NF-κB (cat. no. sc-109X; Santa Cruz Biotechnology, Inc.) in
Can Get Signal Immunoreaction Enhancer Solution 1 (cat. no.
NKB-201; Toyobo Life Science) at a 1:3,000 dilution overnight at
4°C, followed by the incubation with HRP-conjugated anti-rabbit IgG
antibody (cat. no. 7074S; Cell Signaling Technology, Inc.) diluted
3,000 times in Can Get Signal Immunoreaction Enhancer Solution 2
(cat. no. NKB-301; Toyobo Life Science) for 1 h at room
temperature. For the determination of GAPDH level, the membrane was
incubated with the HRP-conjugated anti-GAPDH antibody (cat. no.
015-25473, Wako Pure Chemical Industries, Ltd.) in TBS-T buffer
containing 5% non-fat dry milk at a 1:10,000 dilution for 1 h at
room temperature. GAPDH was used for normalization of
phosphorylated p65 (p-p65), p65 and IKKβ and the p-p65 level was
normalized relative to the p65 level. The immunoreactive proteins
were detected with Immobilon Forte Western HRP Substrate (cat. no.
WBLUF0100; EMD Millipore) and the bands were visualized and
analyzed on a V3 Western Workflow (Bio-Rad Laboratories, Inc.)
using Image Lab software (version 6.0.1; Bio-Rad Laboratories,
Inc.).
In silico miRNA target gene
prediction
TargetScan (version 7.1; http://www.targetscan.org/) and DIANA-microT-CDS
(version 5.0; http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index)
were used to predict the putative target genes of miR-429.
Dual luciferase assay
For the measurement of NF-κB activity, a plasmid
possessing five tandem repeats of NF-κB transcription responsive
elements (TGGGGACTTTCCGC) inserted into pGL3-promoter vector (cat.
no. E1761; Promega Corporation) was constructed, as previousely
reported (30). The Ca9-22 cells
were seeded onto a 48-well plate at 2.5×104 cells per
well and incubated for 24 h at 37°C. The plasmid (100 ng) was
co-transfected with pRL Renilla luciferase reporter vector
(50 ng; cat. no. E2241; Promega Corporation) and miR-429 mimic,
miR-429 inhibitor or the corresponding NC using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. After
co-transfection for 24 h, the cells were cultured in fresh medium
overnight and then treated with or without TNF-α (50 ng/ml) for 6 h
at 37°C. The firefly and Renilla luciferase activities were
measured using a Dual Luciferase Reporter Assay kit (cat. no.
E1910; Promega Corporation). The firefly luciferase activity was
normalized to Renilla luciferase activity. To determine
whether miR-429 binds to its potential target, a plasmid with IKKβ
3′-UTR wild type (WT; IKKβ-WT) was prepared by cloning the 3′-UTR
of IKKβ into pmirGLO plasmid (cat. no. E1330; Promega Corporation).
A plasmid with IKKβ 3′-UTR mutant type (MUT; IKKβ-MUT), which has
five mutated nucleotides (5′-CAGTATT) in the predicted target site
of miR-429 in 3′-UTR of IKKβ (5′-GACATTA), was prepared by using
inverse PCR with In-Fusion HD Cloning kit (cat. no. 639648; Takara
Bio, Inc.) with IKKβ-WT as a template. The Ca9-22 cells were seeded
onto a 48-well plate at 2.5×104 cells per well and
incubated for 24 h at 37°C. Then, either WT or MUT luciferase
plasmid (200 ng) was co-transfected with miR-429 mimic, miR-429
inhibitor or the corresponding NC using Lipofectamine®
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The luciferase assay was performed as
described above.
Statistical analysis
The data were expressed as the mean ± standard
deviation (SD) of three or four independent experiments.
Statistical differences were determined using Student's t-test,
one-way or two-way ANOVA followed by Bonferroni's multiple
comparison test. Statistical analyses were performed using WinSTAT
(R. Fitch Software). P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-429 downregulates IL-8 mRNA level
and inhibits its secretion in gingival epithelial cells
To evaluate the biological function of miR-429 in
oral inflammation, Ca9-22 cells with over- and under-expressed
miR-429 were prepared through transient transfection with
chemically modified oligonucleotides, miR-429 mimic and miR-429
inhibitor, respectively. As a control, Ca9-22 cells transfected
with modified oligonucleotides having scramble sequences, NCm for
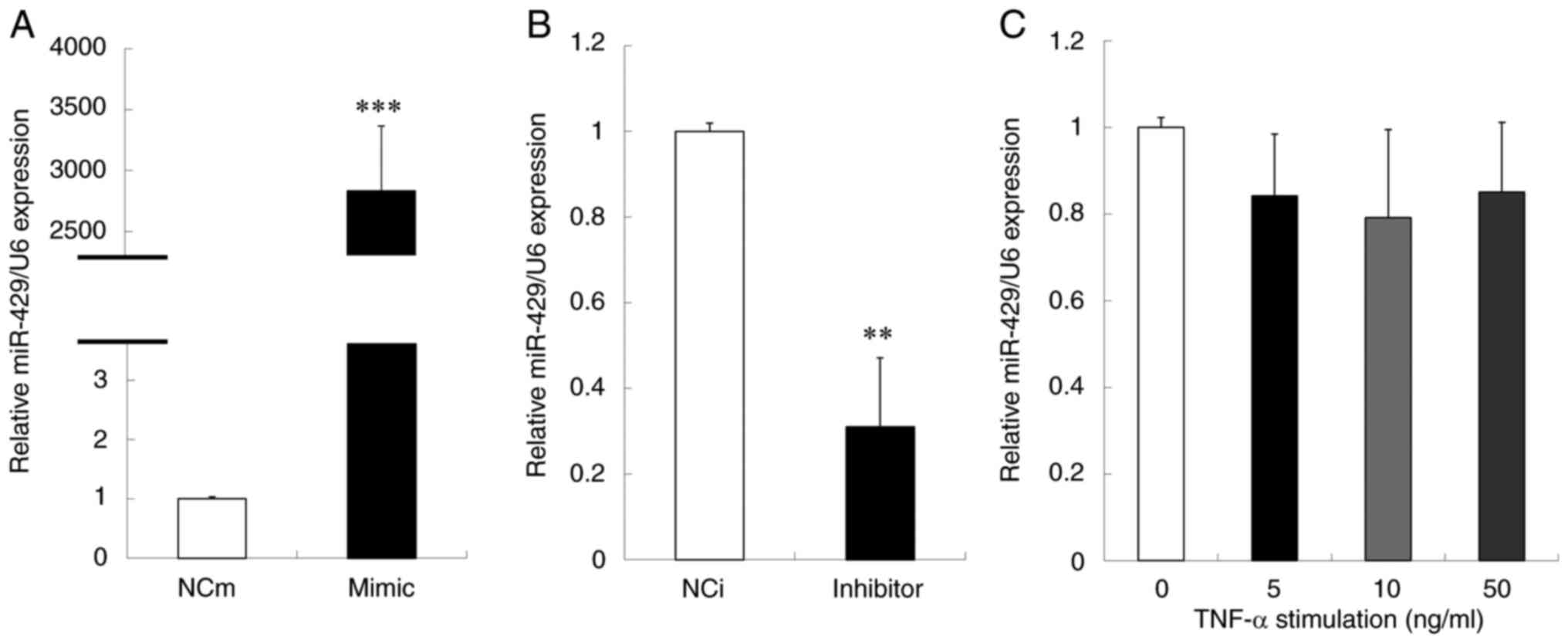
miR-429 mimic and NCi for miR-429 inhibitor, were used. Successful
over- and under-expressions of miR-429 in the cells were confirmed
by the analysis using RT-qPCR (Fig. 1A
and B). In addition, the change in the level of miR-429 in
inflammatory conditions were examined. Under TNF-α stimulation,
there was a tendency of decrease in miR-429 expression in Ca9-22
cells (Fig. 1C). Next, the effect
of miR-429 on the mRNA level of IL-1β, IL-6 and IL-8 was evaluated
using the transfected cells. As shown in Fig. 2A-C, the expression of IL-1β and IL-8
mRNAs was significantly decreased by the over-expression of
miR-429, whereas the IL-6 mRNA level was not changed significantly.
By contrast, the under-expression of miR-429 significantly
increased the mRNA level of IL-1β and IL-8 but not that of IL-6
(Fig. 2D-F). These results
indicated that miR-429 inhibited the expression of IL-1β and IL-8
at a transcriptional level.
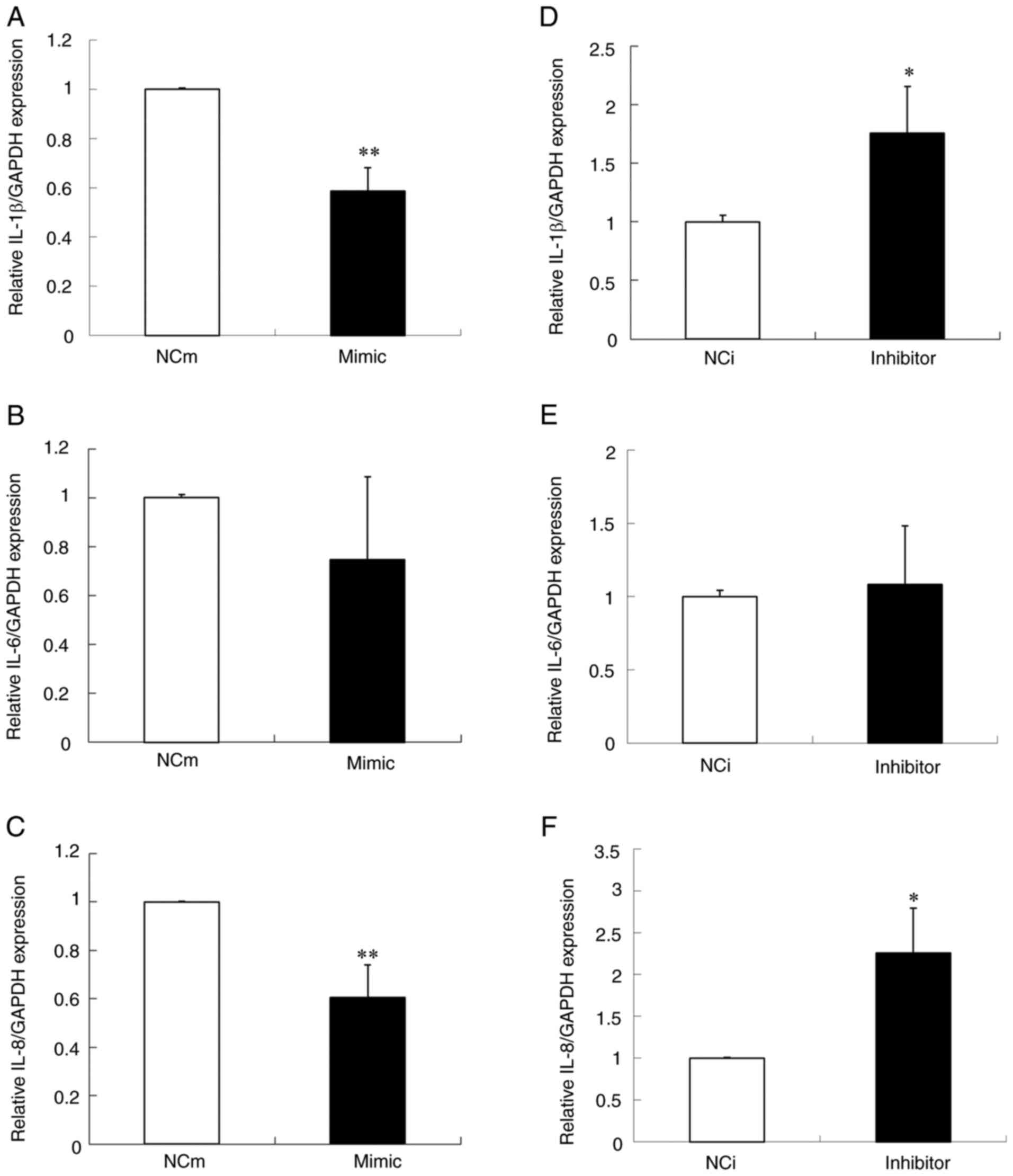 | Figure 2.The effect of over- and
under-expression of miR-429 on the expression of proinflammatory
cytokines in Ca9-22 cells. Expression levels of proinflammatory
cytokines, (A) IL-1β, (B) IL-6 and (C) IL-8, in Ca9-22 cells
transfected with miR-429 mimic or NCm were determined by reverse
transcription-quantitative PCR. Expression levels of
proinflammatory cytokines, (D) IL-1β, (E) IL-6 and (F) IL-8, in
Ca9-22 cells transfected with miR-429 inhibitor or NCi were also
determined. GAPDH was used as a housekeeping gene control. Data are
presented as the means ± standard deviation (n=3). The t-test was
used for the calculation of P-values. *P<0.05, **P<0.01 vs.
the control group. miR, microRNA; mimic, miR-429 mimic; NCm,
negative control mimic; inhibitor, miR-429 inhibitor; NCi, negative
control inhibitor. |
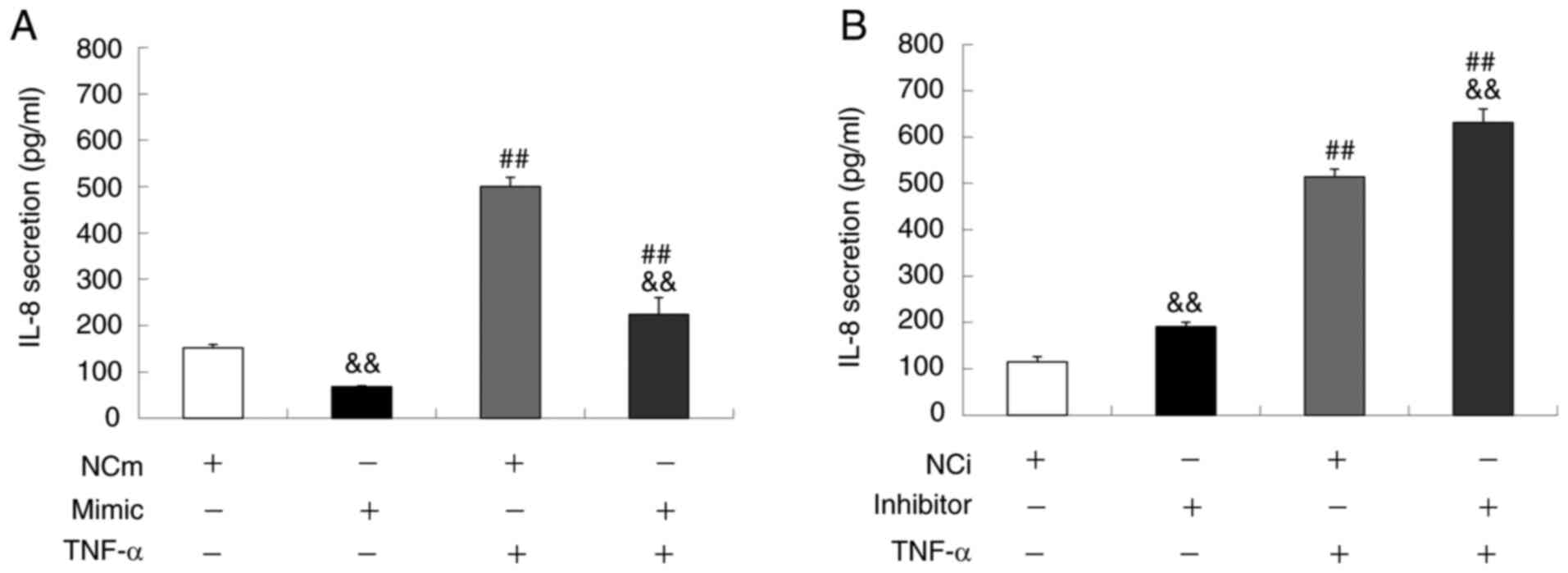
Next, the concentration of IL-8 in the conditional
medium of Ca9-22 cells was measured by ELISA (Fig. 3A and B). Under the basal condition,
the miR-429 mimic significantly reduced IL-8 secretion from the
cells whereas the miR-429 inhibitor significantly increased the
secretion when compared to the corresponding NC group. Similarly,
under TNF-α stimulated condition where the IL-8 secretion was
significantly increased, the miR-429 mimic significantly inhibited
the IL-8 secretion whereas the miR-429 inhibitor further enhanced
the secretion when compared to the cells treated with NCm or NCi.
In addition, the IL-1β in the conditional medium was measured, but
it was undetectable under the conditions used in the present study
(data not shown). Taken together, the above results suggested that
miR-429 exerts anti-inflammatory action through the suppression of
IL-8 production in Ca9-22 cells.
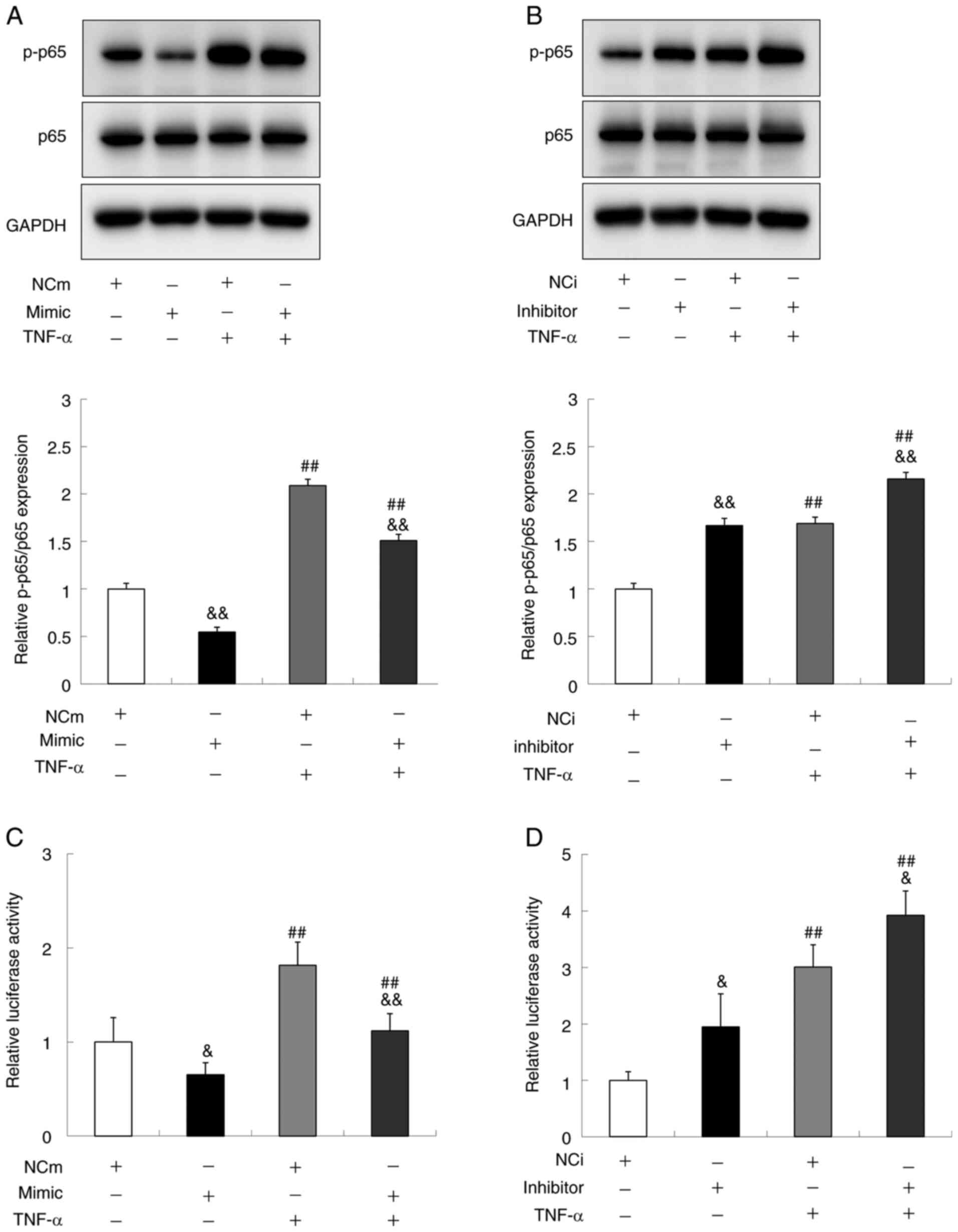
miR-429 suppresses NF-κB activation in
gingival epithelial cells
Since miR-429 inhibited both mRNA expression and
protein secretion of IL-8, the effect of miR-429 on the NF-κB
activation in Ca9-22 cells was next examined. The results
demonstrated that over-expression of miR-429 significantly
decreased the p-p65 level under both the basal and TNF-α treatment
conditions (Fig. 4A). By contrast,
the under-expression of miR-429 resulted in the significant
increase of p-p65 level under those two conditions (Fig. 4B). To further confirm the effect of
miR-429 on NF-κB activity, a luciferase reporter plasmid possessing
NF-κB transcription responsive elements was constructed and the
transcriptional activity of NF-κB was evaluated by performing
luciferase assay. The results demonstrated that the upregulation of
miR-429 decreased the activity whereas its downregulation resulted
in the opposite effect (Fig. 4C and
D), indicating that miR-429 negatively regulates the
transcriptional activity of NF-κB.
miR-429 directly downregulates IKKβ
molecule in gingival epithelial cells
To gain a deeper understanding of miR-429 function
in inflammatory process, experiments were performed to identify
target genes of miR-429 in Ca9-22 cells. In silico
prediction tools (TargetScan and DIANA-microT-CDS) were used to
select the possible target genes of miR-429 and selected IKKβ as a
target candidate as it was predicted by both tools and is indeed
one of the molecules in the NF-κB pathway. As shown in Fig. 5A, the seed sequence of miR-429 has
the complementary site in the 3′-UTR of the IKKβ mRNA, indicating
the direct interaction between miR-429 and IKKβ. Thus, the effect
of miR-429 on the expression of IKKβ in Ca9-22 cells was examined.
As shown in Fig. 5B and D, the
over-expression of miR-429 significantly inhibited the mRNA and
protein expressions of IKKβ. By contrast, the under-expression of
miR-429 demonstrated a tendency to increase the mRNA level of IKKβ,
but did not change its protein level (Fig. 5C and D). The present study further
attempted to optimize the experimental conditions such as the
concentration of miR-429 inhibitor (50 and 100 nM) and the
treatment time and re-examined the effect of miR-429 inhibitor on
the level of IKKβ protein. However, no significant changes in the
protein level were found under all the conditions tested (data not
shown).
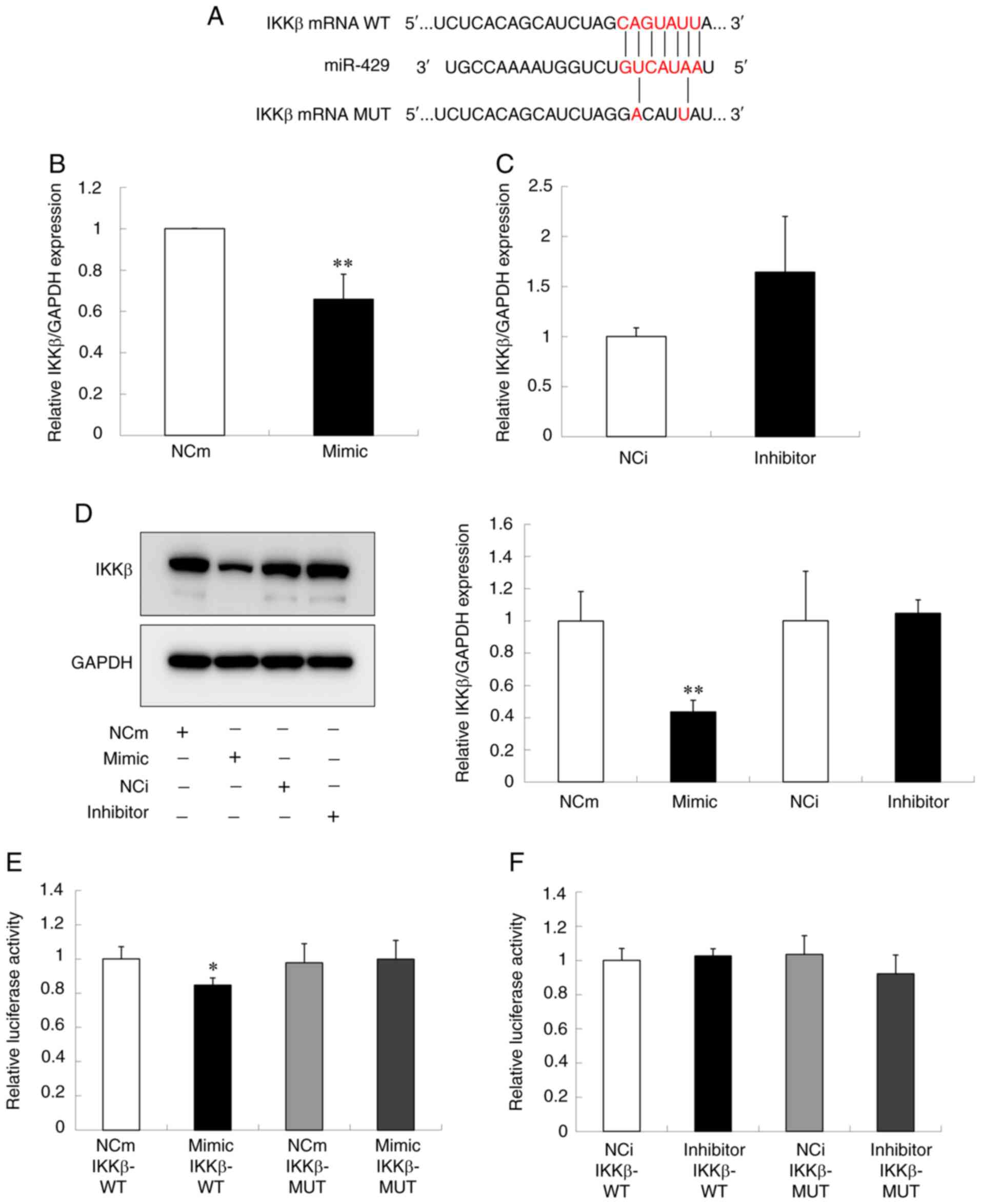 | Figure 5.miR-429 direct binding to IKKβ in
Ca9-22 cells. (A) The putative binding site of miR-429 on IKKβ-WT
and its corresponding site on IKKβ-MUT. The cells were transfected
with miR-429 mimic, miR-429 inhibitor or the corresponding NC. The
levels of IKKβ (B and C) mRNA and (D) protein following the
treatment with miR-429 mimic and with miR-429 inhibitor are shown.
The IKKβ mRNA and protein levels were determined by reverse
transcription-quantitative PCR and western blot analysis,
respectively. GAPDH was used as a housekeeping gene control. (E and
F) The luciferase assay in Ca9-22 cells co-transfected with IKKβ
(IKKβ-WT or IKKβ-MUT) and (E) miR-429 mimic or NCm and (F) miR-429
inhibitor or NCi. The luciferase activity was determined by dual
luciferase reporter assay system. Data are presented as the means ±
standard deviation (n=3). The t-test was used for the calculation
of P-values. *P<0.05, **P<0.01 vs. the control group. miR,
microRNA; WT, wild type; MUT, mutant type; mimic, miR-429 mimic;
NCm, negative control mimic; inhibitor, miR-429 inhibitor; NCi,
negative control inhibitor. |
To further examine whether miR-429 directly bound to
3′-UTR of IKKβ, plasmid vectors that contain either WT 3′-UTR of
IKKβ (IKKβ-WT) or MUT 3′-UTR of IKKβ (IKKβ-MUT) were constructed
(Fig. 5A) and luciferase reporter
assay was conducted. As shown in Fig.
5E and F, the luciferase activity of Ca9-22 cells transfected
with IKKβ-WT was significantly decreased by the co-transfection of
miR-429 mimic, whereas co-transfection of miR-429 inhibitor did not
affect the activity. In addition, either co-transfection of miR-429
mimic or miR-429 inhibitor did not alter the luciferase activity of
the cells transfected with IKKβ-MUT (Fig. 5E and F). Collectively, these results
suggested that miR-429 directly bound to 3′-UTR of IKKβ gene.
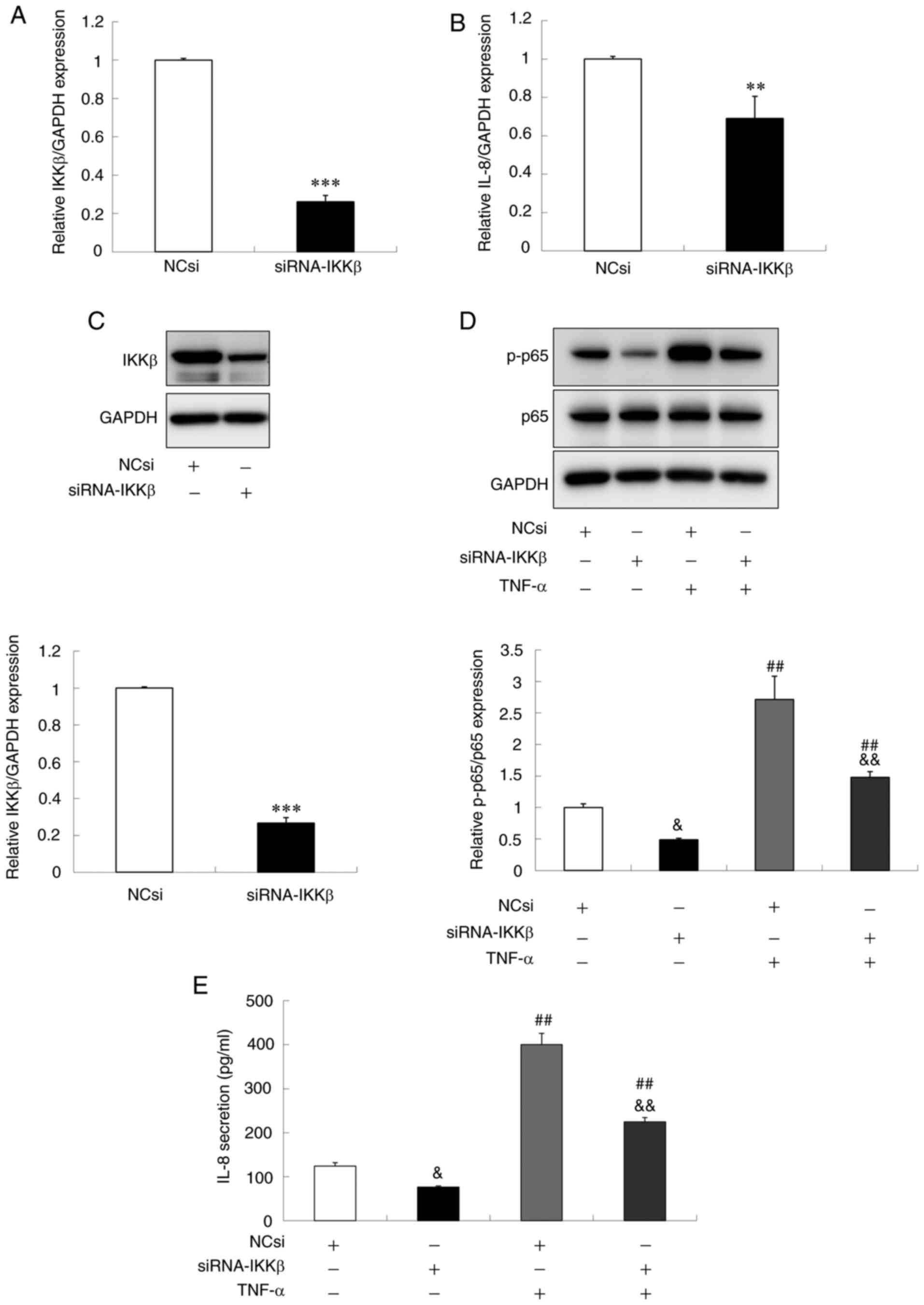
Downregulation of IKKβ leads to the
suppression of IL-8 production in gingival epithelial cells
Previous studies demonstrated that IKKβ knockdown in
some cells causes the attenuation of IL-8 expression and secretion
(31–33). To examine whether the inhibitory
effect of miR-429 on IL-8 secretion and p-p65 level was mediated
via IKKβ downregulation, Ca9-22 cells were transfected with siRNA
targeting IKKβ (siRNA-IKKβ) or NCsi as a control and then the
effect on the level of p-p65 and IL-8 secretion was analyzed. As
shown in Fig. 6A and C, it was
confirmed that the transfection with siRNA-IKKβ inhibited the mRNA
and protein levels of IKKβ expression. In addition, the
downregulation of IKKβ expression was accompanied by the
significant blockade of the phosphorylation of p65 under both the
basal and TNF-α treatment conditions (Fig. 6D). Furthermore, it was found that
the transfection with siRNA-IKKβ significantly decreased the mRNA
expression and protein secretion of IL-8 (Fig. 6B and E). These results indicated
that the downregulation of IKKβ in Ca9-22 cells resulted in the
suppression of IL-8 production via inhibition of NF-κB
signaling.
Discussion
The oral mucosa is a unique tissue, which is
constantly exposed to various types of endogenous and foreign
substances including bacteria and other virulent factors (2,4,5,34).
To protect the host from the pathogens, the immune system in mucosa
initiates a series of inflammatory processes via immune cell
activation, cell-cell communication and the release of
pro-inflammatory mediators such as interleukins. Although
inflammatory responses are essential for host defense against the
pathogens, excess inflammation gives rise to serious cellular
damage, leading to tissue disruption and organ dysfunction. Hence,
the precise control of inflammatory processes is necessary to
maintain oral health; however, the molecular mechanism by which
inflammation is regulated in the gingival mucosa remains to be
fully elucidated.
The miR-200 family, including miR-429, has been
studied for its biological function and aberrant expression in
various diseases, particularly in tumors (16–27,35,36).
Although several studies demonstrate that miR-429 exerted
anti-inflammatory activities in some diseases (35,36),
further investigation is necessary to fully understand its
biological and pathological roles in the inflammatory reaction in
the oral mucosa. Therefore, the present study was performed to
clarify the relationship between action of miR-429 and production
of inflammatory mediator in gingival epithelial cells.
IL-8 is a potent inflammatory mediator and
stimulates neutrophile and lymphocyte recruitment, tissue
remodeling and angiogenesis (2,5). It is
known that NF-κB activation triggers the production of IL-8
(37). A recent meta-analysis
revealed that IL-8 level in inflamed gingival tissues is higher
than that in the normal tissues (38), suggesting the involvement of IL-8 in
oral inflammation. Accordingly, the present study examined the
effect of miR-429 on the expression of IL-8 in Ca9-22 cells, a
gingival squamous cell carcinoma line and found that the
over-expression of miR-429 inhibited the IL-8 expression, whereas
its enhanced expression was observed by the under-expression of
miR-429 (Fig. 2C and F). Notably,
several studies by other groups reported that some other members of
miR-200 family suppress the secretion of interleukins, especially
IL-8 (20–24,26,35).
These findings were confirmed by the present study. It was found
that the over-expression of miR-429 attenuated IL-8 secretion in
Ca9-22 cells regardless of TNF-α stimulation (Fig. 3), suggesting that miR-429 negatively
regulated the production of IL-8 in oral mucosa. Next, the present
study investigated the site of action of miR-429 to inhibit IL-8
expression. It has been reported that miR-200c, a member of miR-200
family having the same seed sequence as miR-429, suppresses NF-κB
binding activity in IL-8 promoter in leiomyoma smooth muscle cells,
resulting in downregulation of IL-8 (20). The effect of miR-429 on the
signaling pathway of NF-κB in Ca9-22 cells was examined and it was
found that the over-expression of miR-429 suppressed NF-κB p65
phosphorylation, an indicator of NF-κB activation (Fig. 4).
IL-1β is also known as the inflammatory cyotokine in
the epithelial cells such as Ca9-22. The present study also
demonstrated that miR-429 inhibited the IL-1β expression in Ca9-22
cells (Fig. 2A). However, it was
not possible to examine the effect of miR-429 on the secretion of
IL-1β protein from the cells, because IL-1β was not detectable
under the condition used in this study (data not shown). Nagahama
et al (39) examined the
secretion of IL-8 and IL-1β from Ca9-22 cells and found that the
secreted amount of IL-8 was ~10-fold greater than that of IL-1β.
These data support the importance of IL-8 as a defense molecule in
epithelial cells.
In this study, although the expression of miR-429
was enhanced over 2,000-fold (Fig.
1A), a relatively small effect on the mRNA level of IL-1β
(Fig. 2A) and IL-8 (Fig. 2C) was observed. Grimm et al
(40) demonstrated that the high
abundance of miRNA led to saturated Argonaute (AGO) 2 protein. A
report by Matsui et al (41)
demonstrated that the inhibition of AGO expression by siRNA reduced
the silencing activity of the double-stranded miRNA mimic. These
previous studies indicated the importance of AGO proteins in the
mechanism of miRNA-mediated silencing. Based on these findings, we
speculate that the effect of miR-429 mimic on the level of
interleukins mRNA could be limited by the availability of AGO
proteins.
IL-6 is another cytokine whose expression is
regulated by NF-κB; however, the level of IL-6 mRNA was not changed
significantly in Ca9-22 cells by the treatment of miR-429 mimic and
inhibitor (Fig. 2B and E) while
miR-429 reduced the activity of NF-κB (Fig. 4). It was hypothesized that the
expression of IL-6 in carcinoma cell lines was also regulated by
some other transcriptional factors as previously described
(4,42,43).
It is also possible that IL-6 expression may be regulated at the
post transcriptional level affecting its stability (43).
The inhibitor of NF-κB (IkB) kinase (IKK) complex is
composed of two catalytic subunits (IKKα and IKKβ) and one
regulatory subunit (IKKγ/NEMO) and phosphorylates IkB that blocks
the transcriptional activity of NF-κB. Thus, this phosphorylation
promotes the degradation of IkB and thereby leads to NF-κB
activation (6). It was noteworthy
that, unlike IKKα, IKKβ is essential for inflammation-induced bone
loss (44). In addition, a previous
study demonstrates that oral bacteria and their products caused
IKKβ activation in oral epithelia cells, leading to inflammatory
responses (45). Thus, IKKβ is
known as one of the key mediators of NF-κB activation. The present
study used in silico prediction tools (TargetScan and
DIANA-microT-CDS) to identify a target gene of miR-429 in NF-κB
pathway and selected IKKβ as a candidate. It was demonstrated that
miR-429 suppressed IKKβ at both the transcriptional and
translational levels (Fig. 5B and
D). In addition, miR-429 bound to 3′-UTR of IKKβ in Ca9-22
cells, although the binding between miR-429 and IKKβ 3′-UTR was
weak according to the luciferase assay (Fig. 5E), which is a limitation of the
present study. Some other means such as RNA immunoprecipitation
assay could help strengthen this finding. In addition, the present
study confirmed that the downregulation of IKKβ by siRNA leads to
the decreased production of IL-8 (Fig.
6). Taken together, it was demonstrated that miR-429 suppressed
IL-8 production, at least in part, through inhibiting NF-κB
activation via direct binding to IKKβ. It has been reported that
some miRNAs including miR-16, miR-148a, miR-199a, miR-497 and
miR-200 negatively regulate IKKβ in a number of different types of
cells (20,24,26,46–56).
The present study provided the additional information that miR-429
is directly involved in the regulation of IKKβ in gingival
epithelial cells.
The present study was unable to demonstrate that the
loss-of-function of miR-429 caused the significant increase of IKKβ
expression (Fig. 5C and D). This
may be explained by the insufficient inhibition of miR-429 in the
protocol for transient transfection. Another posibility is that the
translation of IKKβ protein may be more complex and thus regulated
by several other miRNAs as suggested in the previous studies
(20,24,26,46–56).
It could be that the inhibition of miR-429 alone may be
insufficient to induce the upregulation of IKKβ protein (57). Other experimental approaches for the
deletion of miR-429 expression such as genome editing may help
clarify this issue (58).
In conclusion, the present study found that miR-429
serves as an anti-inflammatory agent that inhibits the production
of inflammatory cytokines such as IL-8 by inhibiting NF-κB pathway
via binding to IKKβ of the IKK complex. To the best of our
knowledge, this is the first evidence that miR-429 directly
regulates IKKβ in gingival epithelial cells and suggests that
miR-429 could be a useful target for the treatment of inflammatory
disorders in the gingival mucosa.
Acknowledgements
The authors thank Dr Takami Oka of Wakunaga
Pharmaceutical Co. Ltd. (Tokyo, Japan) for his helpful advice,
encouragement and critical reading of the manuscript.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HK and HA designed the experiments, performed the
data analysis and confirm the authencity of all the raw data. HK
performed the experiments and was a major contributor in writing
the manuscript. HA revised the manuscript and gave the final
approval of the version to be submitted. Both authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hasturk H and Kantarci A: Activation and
resolution of periodontal inflammation and its systemic impact.
Periodontol. 69:255–273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Darveau RP: Periodontitis: A polymicrobial
disruption of host homeostasis. Nat Rev Microbiol. 8:481–490. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hajishengallis G: Periodontitis: From
microbial immune subversion to systemic inflammation. Nat Rev
Immunol. 15:30–44. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pan W, Wang Q and Chen Q: The cytokine
network involved in the host immune response to periodontitis. Int
J Oral Sci. 11:302019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fujita T, Yoshimoto T, Kajiya M, Ouhara K,
Matsuda S, Takemura T, Akutagawa K, Takeda K, Mizuno N and Kurihara
H: Regulation of defensive function on gingival epithelial cells
can prevent periodontal disease. Jpn Dent Sci Rev. 54:66–75. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lawrence T: The nuclear factor NF-kappaB
pathway in inflammation. Cold Spring Harb Perspect Biol.
1:a0016512009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Grimaldi A, Zarone MR, Irace C, Zappavigna
S, Lombardi A, Kawasaki H, Caraglia M and Misso G: Non-coding RNAs
as a new dawn in tumor diagnosis. Semin Cell Dev Biol. 78:37–50.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Misso G, Zarone MR, Grimaldi A, Maria TDM,
Lombardi A, Kawasaki H, Paola S, Pierfrancesco T, Pierosandro T and
Caraglia M: Non coding RNAs: A new avenue for the self-tailoring of
blood cancer treatment. Curr Drug Targets. 18:35–55. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takeuchi T, Kawasaki H, Luce A, Cossu AM,
Misso G, Scrima M, Bocchetti M, Ricciardiello F, Caraglia M and
Zappavigna S: Insight toward the MicroRNA profiling of laryngeal
cancers: Biological role and clinical impact. Int J Mol Sci.
21:36932020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Misso G, Zarone MR, Lombardi A, Grimaldi
A, Cossu AM, Ferri C, Russo M, Vuoso DC, Luce A, Kawasaki H, et al:
miR-125b upregulates miR-34a and sequentially activates stress
adaption and cell death mechanisms in multiple myeloma. Mol Ther
Nucleic Acids. 16:391–406. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Maudet C, Mano M and Eulalio A: MicroRNAs
in the interaction between host and bacterial pathogens. FEBS Lett.
588:4140–4147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kawasaki H, Takeuchi T, Ricciardiello F,
Lombardi A, Biganzoli E, Fornili M, De Bortoli D, Mesolella M,
Cossu AM, Scrima M, et al: Definition of miRNA signatures of nodal
metastasis in LCa: miR-449a targets notch genes and suppresses cell
migration and invasion. Mol Ther Nucleic Acids. 20:711–724. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grassia V, Lombardi A, Kawasaki H, Ferri
C, Perillo L, Mosca L, Cave DD, Nucci L, Porcelli M and Caraglia M:
Salivary microRNAs as new molecular markers in cleft lip and
palate: A new frontier in molecular medicine. Oncotarget.
9:18929–18938. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saito A, Horie M, Ejiri K, Akira A,
Katagiri S, Maekawa S, Suzuki S, Kong S, Yamauchi T, Yamaguchi Y,
et al: MicroRNA profiling in gingival crevicular fluid of
periodontitis-a pilot study. FEBS Open Bio. 7:981–994. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gregory PA, Bert AG, Paterson EL, Barry
SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y and Goodall GJ:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors. ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang P, Cao J, Liu S, Pan H, Liu X, Sui A,
Wang L, Yao R, Liu Z and Liang J: Upregulated microRNA-429 inhibits
the migration of HCC cells by targeting TRAF6 through the NF-κB
pathway. Oncol Rep. 37:2883–2890. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arunkumar G, Deva Magendhra Rao AK,
Manikandan M, Prasanna Srinivasa Rao H, Subbiah S, Ilangovan R,
Murugan AK and Munirajan AK: Dysregulation of miR-200 family
microRNAs and epithelial-mesenchymal transition markers in oral
squamous cell carcinoma. Oncol Lett. 15:649–657. 2018.PubMed/NCBI
|
|
20
|
Chuang TD and Khorram O: miR-200c
regulates IL8 expression by targeting IKBKB: A potential mediator
of inflammation in leiomyoma pathogenesis. PLoS One. 9:e953702014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hong L, Sharp T, Khorsand B, Fischer C,
Eliason S, Salem A, Akkouch A, Brogden K and Amendt BA:
MicroRNA-200c represses IL-6, IL-8 and CCL-5 expression and
enhances osteogenic differentiation. PLoS One. 11:e01609152016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pecot CV, Rupaimoole R, Yang D, Akbani R,
Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C, et al:
Tumour angiogenesis regulation by the miR-200 family. Nat Commun.
4:24272013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen Y, Zhou M, Yan J, Gong Z, Xiao Y,
Zhang C, Du P and Chen Y: miR-200b inhibits TNF-α-induced IL-8
secretion and tight junction disruption of intestinal epithelial
cells in vitro. Am J Physiol Liver Physiol. 312:G123–G132.
2017.PubMed/NCBI
|
|
24
|
Matsui S, Zhou L, Nakayama Y, Mezawa M,
Kato A, Suzuki N, Tanabe N, Nakayama T, Suzuki Y, Kamio N, et al:
miR-200b attenuates IL-6 production through IKKβ and ZEB1 in human
gingival fibroblasts. Inflamm Res. 67:965–973. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou X, Lu H, Li F, Hao X, Han L, Dong Q
and Chen X: MicroRNA-429 inhibits neuroblastoma cell proliferation,
migration and invasion via the NF-κB pathway. Cell Mol Biol Lett.
25:52020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu H, Wang G, Wang Z, An S, Ye P and Luo
S: A negative feedback loop between miR-200b and the nuclear
factor-κB pathway via IKBKB/IKK-β in breast cancer cells. FEBS J.
283:2259–2271. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song B, Zheng K, Ma H, Liu A, Jing W, Shao
C, Li G and Jin G: miR-429 determines poor outcome and inhibits
pancreatic ductal adenocarcinoma growth by targeting TBK1. Cell
Physiol Biochem. 35:1846–1856. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ohtani M and Nishimura T: Sulfur
containing amino acids in aged garlic extract inhibit inflammation
in human gingival epithelial cells by suppressing intercellular
adhesion molecule-1 expression and IL-6 secretion. Biomed Rep.
12:99–108. 2020.PubMed/NCBI
|
|
30
|
Badr CE, Niers JM, Tjon-Kon-Fat LA, Noske
DP, Wurdinger T and Tannous BA: Real-time monitoring of nuclear
Factor kappaB activity in cultured cells and in animal models. Mol
Imaging. 8:278–290. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Catley MC, Chivers JE, Holden NS, Barnes
PJ and Newton R: Validation of IKK beta as therapeutic target in
airway inflammatory disease by adenoviral-mediated delivery of
dominant-negative IKK beta to pulmonary epithelial cells. Br J
Pharmacol. 145:114–122. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirata Y, Maeda S, Ohmae T, Shibata W,
Yanai A, Ogura K, Yoshida H, Kawabe T and Omata M: Helicobacter
pylori induces IkappaB kinase alpha nuclear translocation and
chemokine production in gastric epithelial cells. Infect Immun.
74:1452–1461. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hernandez L, Hsu SC, Davidson B, Birrer
MJ, Kohn EC and Annunziata CM: Activation of NF-kappaB signaling by
inhibitor of NF-kappaB kinase β increases aggressiveness of ovarian
cancer. Cancer Res. 70:4005–4014. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Moutsopoulos NM and Konkel JE:
Tissue-specific immunity at the oral mucosal barrier. Trends
Immunol. 39:276–287. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo Y and Liao Y: miR-200bc/429 cluster
alleviates inflammation in IgA nephropathy by targeting TWEAK/Fn14.
Int Immunopharmacol. 52:150–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Leng J, Liu W, Li L, Wei FY, Tian M, Liu
HM and Guo W: MicroRNA-429/Cxcl1 axis protective against oxygen
glucose deprivation/reoxygenation-induced injury in brain
microvascular endothelial cells. Dose-Response.
18:15593258209137852020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bezzerri V, Borgatti M, Finotti A,
Tamanini A, Gambari R and Cabrini G: Mapping the transcriptional
machinery of the IL-8 gene in human bronchial epithelial cells. J
Immunol. 187:6069–6081. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Finoti LS, Nepomuceno R, Pigossi SC, Corbi
SC, Secolin R and Scarel-Caminaga RM: Association between
interleukin-8 levels and chronic periodontal disease. Medicine
(Baltimore). 96:e69322017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nagahama Y, Obama T, Usui M, Kanazawa Y,
Iwamoto S, Suzuki K, Miyazaki A, Yamaguchi T, Yamamoto M and Itabe
H: Oxidized low-density lipoprotein-induced periodontal
inflammation is associated with the upregulation of
cyclooxygenase-2 and microsomal prostaglandin synthase 1 in human
gingival epithelial cells. Biochem Biophys Res Commun. 413:566–571.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grimm D, Wang L, Lee JS, Schürmann N, Gu
S, Börner K, Storm TA and Kay MA: Argonaute proteins are key
determinants of RNAi efficacy, toxicity and persistence in the
adult mouse liver. J Clin Invest. 120:3106–3119. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Matsui M, Prakash TP and Corey DR:
Argonaute 2-dependent regulation of gene expression by
single-stranded miRNA mimics. Mol Ther. 24:946–955. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Poplutz MK, Wessels I, Rink L and
Uciechowski P: Regulation of the Interleukin-6 gene expression
during monocytic differentiation of HL-60 cells by chromatin
remodeling and methylation. Immunobiology. 219:619–626. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tanaka T, Narazaki M and Kishimoto T: IL-6
in inflammation, immunity and disease. Cold Spring Harb Perspect
Biol. 6:a0162952014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ruocco MG, Maeda S, Park JM, Lawrence T,
Hsu LC, Cao Y, Schett G, Wagner EF and Karin M: IκB kinase (IKK)β,
but not IKKα, is a critical mediator of osteoclast survival and is
required for inflammation-induced bone loss. J Exp Med.
201:1677–1687. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Groeger S, Jarzina F, Domann E and Meyle
J: Porphyromonas gingivalis activates NFκB and MAPK pathways in
human oral epithelial cells. BMC Immunol. 18:12017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Gao Y and Yu Z: MicroRNA-16 inhibits
interleukin-13-induced inflammatory cytokine secretion and mucus
production in nasal epithelial cells by suppressing the IκB kinase
β/nuclear factor-κB pathway. Mol Med Rep. 18:4042–4050.
2018.PubMed/NCBI
|
|
47
|
Khalife J, Ghose J, Martella M, Viola D,
Rocci A, Troadec E, Terrazas C, Satoskar AR, Gunes EG, Dona A, et
al: miR-16 regulates crosstalk in NF-κB tolerogenic inflammatory
signaling between myeloma cells and bone marrow macrophages. JCI
Insight. 4:e1293482019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Patel V, Carrion K, Hollands A, Hinton A,
Gallegos T, Dyo J, Sasik R, Leire E, Hardiman G, Mohamed SA, et al:
The stretch responsive microRNA miR-148a-3p is a novel repressor of
IKBKB, NF-κB signaling and inflammatory gene expression in human
aortic valve cells. FASEB J. 29:1859–1868. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen R, Alvero AB, Silasi DA, Kelly MG,
Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T and Mor G:
Regulation of IKKβ by miR-199a affects NF-κB activity in ovarian
cancer cells. Oncogene. 27:4712–4723. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wei D, Shen B, Wang W, Zhou Y, Yang X, Lu
G, Yang J and Shao Y: MicroRNA-199a-5p functions as a tumor
suppressor in oral squamous cell carcinoma via targeting the
IKKβ/NF-κB signaling pathway. Int J Mol Med. 43:1585–1596.
2019.PubMed/NCBI
|
|
51
|
Dai L, Gu L and Di W: miR-199a attenuates
endometrial stromal cell invasiveness through suppression of the
IKKβ/NF-κB pathway and reduced interleukin-8 expression. Mol Hum
Reprod. 18:136–145. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yamazaki-Takai M, Takai H, Iwai Y, Noda K,
Mezawa M, Tsuruya Y, Yamaguchi A, Nakayama Y and Ogata Y: miR-200b
suppresses TNF-α-induced AMTN production in human gingival
epithelial cells. Odontology. 109:403–410. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cao Y, Tang S, Nie X, Zhou Z, Ruan G, Han
W, Zhu Z and Ding C: Decreased miR-214-3p activates NF-κB pathway
and aggravates osteoarthritis progression. EBioMedicine.
65:1032832021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Niu D, Gong Z, Sun X, Yuan J, Zheng T,
Wang X, Fan X, Mao Y, Liu X, Tang B and Fu Y: miR-338-3p regulates
osteoclastogenesis via targeting IKKβ gene. In Vitro Cell Dev Biol
Anim. 55:243–251. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kong XJ, Duan LJ, Qian XQ, Xu D, Liu HL,
Zhu YJ and Qi J: Tumor-suppressive microRNA-497 targets IKKβ to
regulate NF-κB signaling pathway in human prostate cancer cells. Am
J Cancer Res. 5:1795–1804. 2015.PubMed/NCBI
|
|
56
|
Mechtler P, Singhal R, Kichina JV, Bard
JE, Buck MJ and Kandel ES: MicroRNA analysis suggests an additional
level of feedback regulation in the NF-κB signaling cascade.
Oncotarget. 6:17097–17106. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen W, He LN, Liang Y, Zeng X, Wu CP, Su
MQ, Cheng Y and Liu JH: TERF1 downregulation promotes the migration
and invasion of the PC3 prostate cancer cell line as a target of
miR-155. Mol Med Rep. 22:5209–5218. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang J, Meng X, Pan J, Jiang N, Zhou C, Wu
Z and Gong Z: CRISPR/Cas9-mediated noncoding RNA editing in human
cancers. RNA Biol. 15:35–43. 2018. View Article : Google Scholar : PubMed/NCBI
|