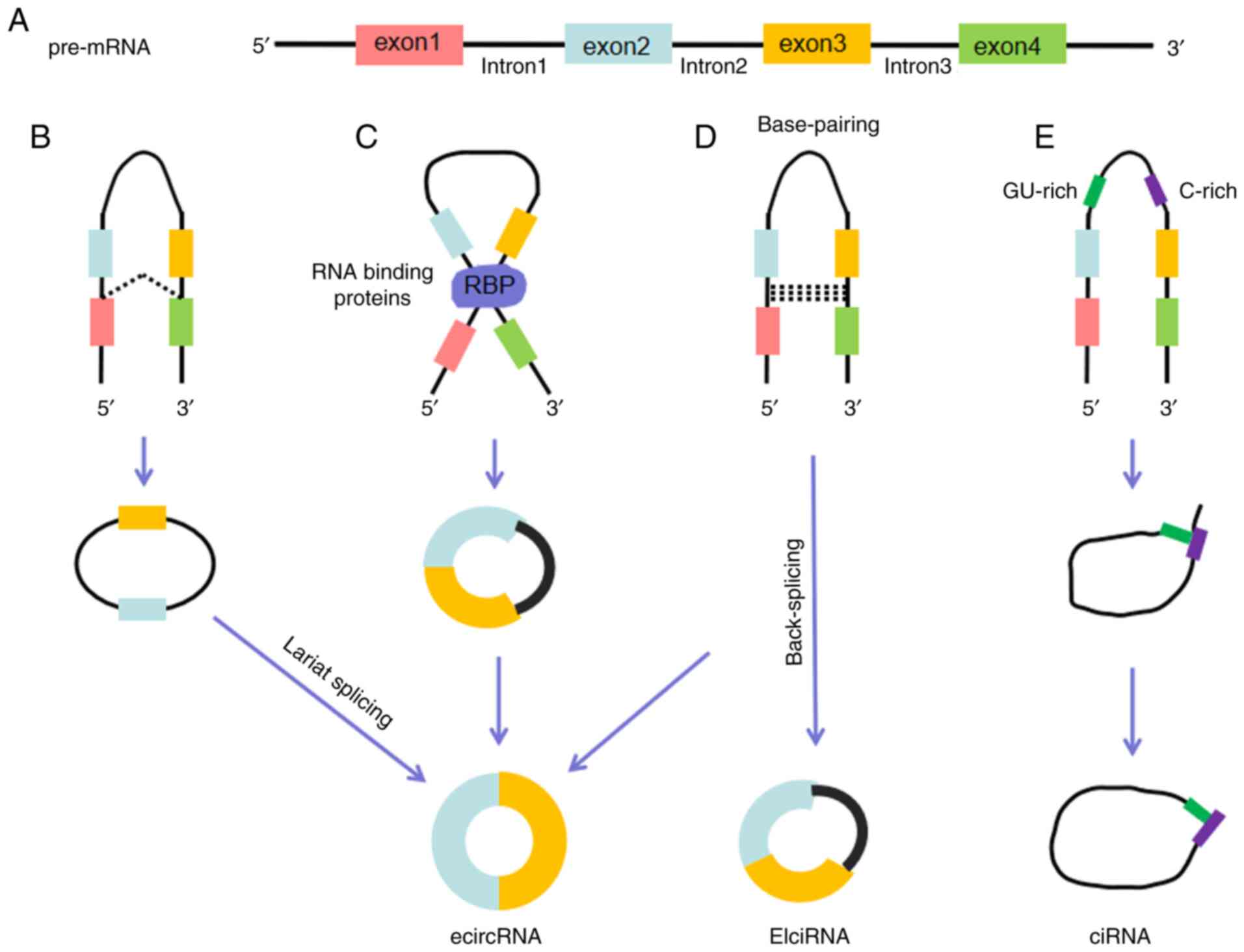
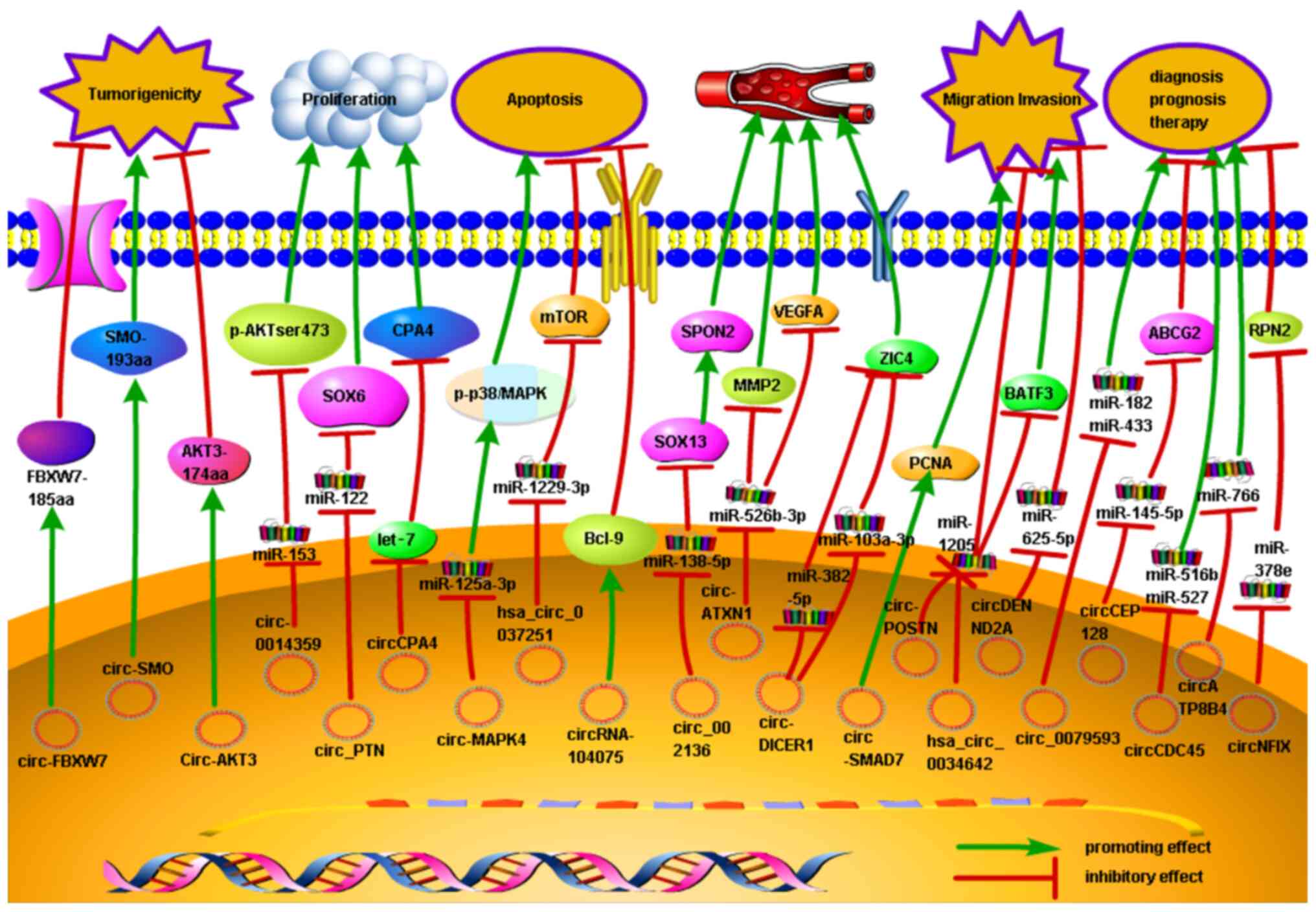
|
1
|
Ng S, Zouaoui S, Bessaoud F, Rigau V, Roux
A, Darlix A, Bauchet F, Mathieu-Daudé H, Trétarre B,
Figarella-Branger D, et al: An epidemiology report for primary
central nervous system tumors in adolescents and young adults: A
nationwide population-based study in France, 2008–2013. Neuro
Oncol. 22:851–863. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sminia P, van den Berg J, van Kootwijk A,
Hageman E, Slotman BJ and Verbakel W: Experimental and clinical
studies on radiation and curcumin in human glioma. J Cancer Res
Clin Oncol. 147:403–409. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maccallini C, Gallorini M, Cataldi A and
Amoroso R: Targeting iNOS As a valuable strategy for the therapy of
glioma. ChemMedChem. 15:339–344. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Daisy Precilla S, Kuduvalli SS and
Thirugnanasambandhar Sivasubramanian A: Disentangling the
therapeutic tactics in GBM: From bench to bedside and beyond. Cell
Biol Int. 45:18–53. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Catalano M, D'Alessandro G, Trettel F and
Limatola C: Role of infiltrating microglia/macrophages in glioma.
Adv Exp Med Biol. 1202:281–298. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang L, Liu F, Weygant N, Zhang J, Hu P,
Qin Z, Yang J, Cheng Q, Fan F, Zeng Y, et al: A novel integrated
system using patient-derived glioma cerebral organoids and
xenografts for disease modeling and drug screening. Cancer Lett.
500:87–97. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ballo MT, Urman N, Lavy-Shahaf G, Grewal
J, Bomzon Z and Toms S: Correlation of tumor treating fields
dosimetry to survival outcomes in newly diagnosed glioblastoma: A
large-scale numerical simulation-based analysis of data from the
phase 3 EF-14 randomized trial. Int J Radiat Oncol Biol Phys.
104:1106–1113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cloughesy TF, Mochizuki AY, Orpilla JR,
Hugo W, Lee AH, Davidson TB, Wang AC, Ellingson BM, Rytlewski JA,
Sanders CM, et al: Neoadjuvant anti-PD-1 immunotherapy promotes a
survival benefit with intratumoral and systemic immune responses in
recurrent glioblastoma. Nat Med. 25:477–486. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roura AJ, Gielniewski B, Pilanc P,
Szadkowska P, Maleszewska M, Krol SK, Czepko R, Kaspera W, Wojtas B
and Kaminska B: Identification of the immune gene expression
signature associated with recurrence of high-grade gliomas. J Mol
Med (Berl). 99:241–255. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Eslahi M, Dana PM, Asemi Z, Hallajzadeh J,
Mansournia MA and Yousefi B: The effects of chitosan-based
materials on glioma: Recent advances in its applications for
diagnosis and treatment. Int J Biol Macromol. 168:124–129. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pedretti S, Masini L, Turco E, Triggiani
L, Krengli M, Meduri B, Pirtoli L, Borghetti P, Pegurri L, Riva N,
et al: Hypofractionated radiation therapy versus chemotherapy with
temozolomide in patients affected by RPA class V and VI
glioblastoma: A randomized phase II trial. J Neurooncol.
143:447–455. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saw PE, Xu X, Chen J and Song EW:
Non-coding RNAs: The new central dogma of cancer biology. Sci China
Life Sci. 64:22–50. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ryu J, Ahn Y, Kook H and Kim YK: The roles
of non-coding RNAs in vascular calcification and opportunities as
therapeutic targets. Pharmacol Ther. 218:1076752021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu K, Yu M and Chen Y: Non-coding RNAs
regulating androgen receptor signaling pathways in prostate cancer.
Clin Chim Acta. 513:57–63. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Uddin MN and Wang X: The landscape of long
non-coding RNAs in tumor stroma. Life Sci. 264:1187252021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fort V, Khelifi G and Hussein SMI: Long
non-coding RNAs and transposable elements: A functional
relationship. Biochim Biophys Acta Mol Cell Res. 1868:1188372021.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kalani M, Hodjati H, Ghoddusi Johari H and
Doroudchi M: Memory T cells of patients with abdominal aortic
aneurysm differentially expressed micro RNAs 21, 92a, 146a, 155,
326 and 663 in response to helicobacter pylori and lactobacillus
acidophilus. Mol Immunol. 130:77–84. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bahreini F, Rayzan E and Rezaei N:
microRNA-related single-nucleotide polymorphisms and breast cancer.
J Cell Physiol. 236:1593–1605. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang S: Perspectives on MicroRNA study in
oncogenesis: Where are we? Neoplasia. 23:99–101. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cao YZ, Sun JY, Chen YX, Wen CC and Wei L:
The roles of circRNAs in cancers: Perspectives from molecular
functions. Gene. 767:1451822021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu Z, Huang Q, Zhang Q, Wu H and Zhong Z:
circRNAs open a new era in the study of cardiovascular disease
(review). Int J Mol Med. 47:49–64. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zaiou M: circRNAs signature as potential
diagnostic and prognostic biomarker for diabetes mellitus and
related cardiovascular complications. Cells. 9:6592020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Sun D, Li X, Yang B and Zhang W:
Identification of key circRNAs in non-small cell lung cancer. Am J
Med Sci. 361:98–105. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y, Nguyen TM, Zhang XO, Wang L, Phan
T, Clohessy JG and Pandolfi PP: Optimized RNA-targeting
CRISPR/Cas13d technology outperforms shRNA in identifying
functional circRNAs. Genome Biol. 22:412021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Koch L: CRISPR-Cas13 targets circRNAs. Nat
Rev Genet. 22:682021. View Article : Google Scholar
|
|
26
|
Li Y, Feng W, Kong M, Liu R, Wu A, Shen L,
Tang Z and Wang F: Exosomal circRNAs: A new star in cancer. Life
Sci. 269:1190392021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luo Q, Li X, Fu B, Zhang L, Fang L, Qing
C, Guo Y, Huang Z and Li J: Expression profile and diagnostic value
of circRNAs in peripheral blood from patients with systemic lupus
erythematosus. Mol Med Rep. 23:12021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang X, Ye T, Liu H, Lv P, Duan C, Wu X,
Jiang K, Lu H, Xia D, Peng E, et al: Expression profiles,
biological functions and clinical significance of circRNAs in
bladder cancer. Mol Cancer. 20:42021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu T, Wang M, Jiang L, Ma L, Wan L, Chen
Q, Wei C and Wang Z: circRNAs in anticancer drug resistance: Recent
advances and future potential. Mol Cancer. 19:1272020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang X, Yang H, Zhao L, Li G and Duan Y:
Circular RNA PRKCI promotes glioma cell progression by inhibiting
microRNA-545. Cell Death Dis. 10:6162019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng H, Qin C, Zhang C, Su J, Xiao Q, Xiao
Y, Xiao K and Liu Q: circCPA4 acts as a prognostic factor and
regulates the proliferation and metastasis of glioma. J Cell Mol
Med. 23:6658–6665. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu J, Hou K, Ji H, Mi S, Yu G, Hu S and
Wang J: Overexpression of circular RNA circ-CDC45 facilitates
glioma cell progression by sponging miR-516b and miR-527 and
predicts an adverse prognosis. J Cell Biochem. 121:690–697. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng Q, Li S, Liu Y, Zhang S, Jin J, Zhang
Y, Guo C, Liu B and Sun Y: Circular RNA circSCAF11 accelerates the
glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol Ther
Nucleic Acids. 17:669–677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Zhang Y, Chen B, Ding L, Mu Z and
Li Y: Elevation of circular RNA circ-POSTN facilitates cell growth
and invasion by sponging miR-1205 in glioma. J Cell Biochem.
120:16567–16574. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng SQ, Qi Y, Wu J, Zhou FL, Yu H, Li L,
Yu B, Chen XF and Zhang W: CircPCMTD1 acts as the sponge of
miR-224-5p to promote glioma progression. Front Oncol. 9:3982019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin S, Tian X, Zhang J, Sun P and Li G:
PCirc: Random forest-based plant circRNA identification software.
BMC Bioinformatics. 22:102021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Goodall GJ and Wickramasinghe VO: RNA in
cancer. Nat Rev Cancer. 21:22–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang D, Ni N, Wang Y, Tang Z, Gao H, Ju
Y, Sun N, He X, Gu P and Fan X: circRNA-vgll3 promotes osteogenic
differentiation of adipose-derived mesenchymal stem cells via
modulating miRNA-dependent integrin α5 expression. Cell Death
Differ. 28:283–302. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang J, Liu Y and Shi G: The
circRNA-miRNA-mRNA regulatory network in systemic lupus
erythematosus. Clin Rheumatol. 40:331–339. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen J, Yang J, Fei X, Wang X and Wang K:
circRNA ciRS-7: A novel oncogene in multiple cancers. Int J Biol
Sci. 17:379–389. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao Z, Zhang Y, Wei S, Zhang X, Guo Y and
Han B: Comprehensive circRNA expression profile and function
network in osteoblast-like cells under simulated microgravity.
Gene. 764:1451062021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu T, Li Y, Liang X, Liu X and Tang M:
Identification of potential circRNA-miRNA-mRNA regulatory networks
in response to graphene quantum dots in microglia by microarray
analysis. Ecotoxicol Environ Saf. 208:1116722021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang Z and Lei X: Identifying the sequence
specificities of circRNA-binding proteins based on a capsule
network architecture. BMC Bioinformatics. 22:192021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou B, Yang H, Yang C, Bao YL, Yang SM,
Liu J and Xiao YF: Translation of noncoding RNAs and cancer. Cancer
Lett. 497:89–99. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fu LY, Wang SW, Hu MY, Jiang ZL, Shen LL,
Zhou YP, Guo JM and Hu YR: Circular RNAs in liver diseases:
Mechanisms and therapeutic targets. Life Sci. 264:1187072021.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wei G, Zhu J, Hu HB and Liu JQ: Circular
RNAs: Promising biomarkers for cancer diagnosis and prognosis.
Gene. 771:1453652021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Varela-Martinez E, Corsi GI, Anthon C,
Gorodkin J and Jugo BM: Novel circRNA discovery in sheep shows
evidence of high backsplice junction conservation. Sci Rep.
11:4272021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li Z, Li X, Xu D, Chen X, Li S, Zhang L,
Chan MTV and Wu WKK: An update on the roles of circular RNAs in
osteosarcoma. Cell Prolif. 54:e129362021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Beilerli A, Gareev I, Beylerli O, Yang G,
Pavlov V, Aliev G and Ahmad A: Circular RNAs as biomarkers and
therapeutic targets in cancer. Semin Cancer Biol. Jan 9–2021.(Epub
ahead of print). doi: 10.1016/j.semcancer.2020.12.026. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang X, Cheng Z, Xu J, Feng M, Zhang H,
Zhang L and Qian L: Circular RNA Arhgap12 modulates
doxorubicin-induced cardiotoxicity by sponging miR-135a-5p. Life
Sci. 265:1187882021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Khanipouyani F, Akrami H and Fattahi MR:
Circular RNAs as important players in human gastric cancer. Clin
Transl Oncol. 23:10–21. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Luo Y and Huang C: circSFMBT2 facilitates
vascular smooth muscle cell proliferation by targeting
miR-331-3p/HDAC5. Life Sci. 264:1186912021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Razavi ZS, Tajiknia V, Majidi S, Ghandali
M, Mirzaei HR, Rahimian N, Hamblin MR and Mirzaei H: Gynecologic
cancers and non-coding RNAs: Epigenetic regulators with emerging
roles. Crit Rev Oncol Hematol. 157:1031922021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li Z, Huang X, Liu A, Xu J, Lai J, Guan H
and Ma J: circ_PSD3 promotes the progression of papillary thyroid
carcinoma via the miR-637/HEMGN axis. Life Sci. 264:1186222021.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang XY and Mao L: Circular RNA
circ_0000442 acts as a sponge of miR-148b-3p to suppress breast
cancer via PTEN/PI3K/Akt signaling pathway. Gene. 766:1451132021.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sun Q, Qi X, Zhang W and Li X: Knockdown
of circRNA_0007534 suppresses the tumorigenesis of cervical cancer
via miR-206/GREM1 axis. Cancer Cell Int. 21:542021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang X, Yang H, Jia Y, Xu Z, Zhang L, Sun
M and Fu J: circRNA_0005529 facilitates growth and metastasis of
gastric cancer via regulating miR-527/Sp1 axis. BMC Mol Cell Biol.
22:62021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li W, Lu H, Wang H, Ning X, Liu Q, Zhang
H, Liu Z, Wang J, Zhao W, Gu Y, et al: Circular RNA TGFBR2 acts as
a ceRNA to suppress nasopharyngeal carcinoma progression by
sponging miR-107. Cancer Lett. 499:301–313. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xing Y, Zha WJ, Li XM, Li H, Gao F, Ye T,
Du WQ and Liu YC: Circular RNA circ-Foxo3 inhibits esophageal
squamous cell cancer progression via the miR-23a/PTEN axis. J Cell
Biochem. 121:2595–2605. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kong Z, Wan X, Lu Y, Zhang Y, Huang Y, Xu
Y, Liu Y, Zhao P, Xiang X, Li L and Li Y: Circular RNA circFOXO3
promotes prostate cancer progression through sponging miR-29a-3p. J
Cell Mol Med. 24:799–813. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zang J, Lu D and Xu A: The interaction of
circRNAs and RNA binding proteins: An important part of circRNA
maintenance and function. J Neurosci Res. 98:87–97. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Okholm TLH, Sathe S, Park SS, Kamstrup AB,
Rasmussen AM, Shankar A, Chua ZM, Fristrup N, Nielsen MM, Vang S,
et al: Transcriptome-wide profiles of circular RNA and RNA-binding
protein interactions reveal effects on circular RNA biogenesis and
cancer pathway expression. Genome Med. 12:1122020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wong CH, Lou UK, Li Y, Chan SL, Tong JH,
To KF and Chen Y: circFOXK2 promotes growth and metastasis of
pancreatic ductal adenocarcinoma by complexing with RNA-binding
proteins and sponging miR-942. Cancer Res. 80:2138–2149. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liang Y, Wang H, Chen B, Mao Q, Xia W,
Zhang T, Song X, Zhang Z, Xu L, Dong G and Jiang F: circDCUN1D4
suppresses tumor metastasis and glycolysis in lung adenocarcinoma
by stabilizing TXNIP expression. Mol Ther Nucleic Acids.
23:355–368. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Liu Z, Wang Q, Wang X, Xu Z, Wei X and Li
J: Circular RNA cIARS regulates ferroptosis in HCC cells through
interacting with RNA binding protein ALKBH5. Cell Death Discov.
6:722020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu Y, Liu X, Lin C, Jia X, Zhu H, Song J
and Zhang Y: Noncoding RNAs regulate alternative splicing in
cancer. J Exp Clin Cancer Res. 40:112021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Stagsted LVW, O'Leary ET, Ebbesen KK and
Hansen TB: The RNA-binding protein SFPQ preserves long-intron
splicing and regulates circRNA biogenesis in mammals. Elife.
10:e630882021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Conn VM, Hugouvieux V, Nayak A, Conos SA,
Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta
C and Conn SJ: A circRNA from SEPALLATA3 regulates splicing of its
cognate mRNA through R-loop formation. Nat Plants. 3:170532017.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hu Q and Zhou T: EIciRNA-mediated gene
expression: Tunability and bimodality. FEBS Lett. 592:3460–3471.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kong S, Tao M, Shen X and Ju S:
Translatable circRNAs and lncRNAs: Driving mechanisms and functions
of their translation products. Cancer Lett. 483:59–65. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
cancer. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wu X, Xiao S, Zhang M, Yang L, Zhong J, Li
B, Li F, Xia X, Li X, Zhou H, et al: A novel protein encoded by
circular SMO RNA is essential for Hedgehog signaling activation and
glioblastoma tumorigenicity. Genome Biol. 22:332021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z,
Xu B, Wu C, Zhou Q, Hu W, Wu C and Jiang J: A novel protein encoded
by a circular RNA circPPP1R12A promotes tumor pathogenesis and
metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer.
18:472019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao
F, Huang N, Yang X, Zhao K, Zhou H, et al: Novel Role of FBXW7
circular RNA in repressing glioma tumorigenesis. J Natl Cancer
Inst. 110:304–315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H,
Huang N, Yang X, Xiao F, Liu D, et al: A novel tumor suppressor
protein encoded by circular AKT3 RNA inhibits glioblastoma
tumorigenicity by competing with active phosphoinositide-dependent
Kinase-1. Mol Cancer. 18:1312019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
El-Hashash AHK: Histone H3K27M mutation in
brain tumors. Adv Exp Med Biol. 1283:43–52. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
He Z, Yang C, He Y, Gong B, Yin C, Feng J,
Chen L, Tang J and Chen Y: CAMTA1, a novel antitumor gene,
regulates proliferation and the cell cycle in glioma by inhibiting
AKT phosphorylation. Cell Signal. 79:1098822021. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Osama M, Mostafa MN and Alvi MA: Astrocyte
elevated gene-1 as a novel therapeutic target in malignant gliomas
and its interactions with oncogenes and tumor suppressor genes.
Brain Res. 1747:1470342020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang Y, Liu Q and Liao Q: CircHIPK3: A
promising cancer-related circular RNA. Am J Transl Res.
12:6694–6704. 2020.PubMed/NCBI
|
|
83
|
Smyth LCD, Rustenhoven J, Scotter EL,
Schweder P, Faull RLM, Park TIH and Dragunow M: Markers for human
brain pericytes and smooth muscle cells. J Chem Neuroanat.
92:48–60. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jiang Y, Zhou J, Luo P, Gao H, Ma Y, Chen
YS, Li L, Zou D, Zhang Y and Jing Z: Prosaposin promotes the
proliferation and tumorigenesis of glioma through toll-like
receptor 4 (TLR4)-mediated NF-κB signaling pathway. EBioMedicine.
37:78–90. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Shi F, Shi Z, Zhao Y and Tian J: circRNA
hsa-circ-0014359 promotes glioma progression by regulating
miR-153/PI3K signaling. Biochem Biophys Res Commun. 510:614–620.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chen C, Deng L, Nie DK, Jia F, Fu LS, Wan
ZQ and Lan Q: Circular RNA Pleiotrophin promotes carcinogenesis in
glioma via regulation of microRNA-122/SRY-box transcription factor
6 axis. Eur J Cancer Prev. 29:165–173. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ye Z, Zhuo Q, Hu Q, Xu X, Liu M, Zhang Z,
Xu W, Liu W, Fan G, Qin Y, et al: FBW7-NRA41-SCD1 axis
synchronously regulates apoptosis and ferroptosis in pancreatic
cancer cells. Redox Biol. 38:1018072021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Mohamad Anuar NN, Nor Hisam NS, Liew SL
and Ugusman A: Clinical review: Navitoclax as a pro-apoptotic and
anti-fibrotic agent. Front Pharmacol. 11:5641082020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hartman ML and Czyz M: BCL-w: Apoptotic
and non-apoptotic role in health and disease. Cell Death Dis.
11:2602020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Yan F, Fan B, Wang J, Wei W, Tang Q, Lu L,
Luo Z, Pu J and Yang SS: circ_0008305-mediated miR-660/BAG5 axis
contributes to hepatocellular carcinoma tumorigenesis. Cancer Med.
10:833–842. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lou J, Hao Y, Lin K, Lyu Y, Chen M, Wang
H, Zou D, Jiang X, Wang R, Jin D, et al: Circular RNA CDR1as
disrupts the p53/MDM2 complex to inhibit gliomagenesis. Mol Cancer.
19:1382020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
He J, Huang Z, He M, Liao J, Zhang Q, Wang
S, Xie L, Ouyang L, Koeffler HP, Yin D and Liu A: Circular RNA
MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling
pathway by sponging miR-125a-3p in gliomas. Mol Cancer. 19:172020.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Cao Q, Shi Y, Wang X, Yang J, Mi Y, Zhai G
and Zhang M: Circular METRN RNA hsa_circ_0037251 promotes glioma
progression by sponging miR-1229-3p and regulating mTOR expression.
Sci Rep. 9:197912019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chi G, Xu D, Zhang B and Yang F: Matrine
induces apoptosis and autophagy of glioma cell line U251 by
regulation of circRNA-104075/BCL-9. Chem Biol Interact.
308:198–205. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zheng X, Liu J, Li X, Tian R, Shang K,
Dong X and Cao B: Angiogenesis is promoted by exosomal DPP4 derived
from 5-fluorouracil-resistant colon cancer cells. Cancer Lett.
497:190–201. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yetkin-Arik B, Kastelein AW, Klaassen I,
Jansen CHJR, Latul YP, Vittori M, Biri A, Kahraman K, Griffioen AW,
Amant F, et al: Angiogenesis in gynecological cancers and the
options for anti-angiogenesis therapy. Biochim Biophys Acta Rev
Cancer. 1875:1884462021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Ahir BK, Engelhard HH and Lakka SS: Tumor
development and angiogenesis in adult brain tumor: Glioblastoma.
Mol Neurobiol. 57:2461–2478. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Carlson JC, Cantu-Gutierrez M, Lozzi B,
Huang-Hobbs E, Turner WD, Tepe B, Zhang Y, Herman AM, Rao G,
Creighton CJ, et al: Identification of diverse tumor endothelial
cell populations in malignant glioma. Neuro Oncol. 23:932–944.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu
L, Ma J, Shao L, Wang D, Shen S, et al:
FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates
angiogenesis in glioma. J Exp Clin Cancer Res. 38:652019.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
He Q, Zhao L, Liu X, Zheng J, Liu Y, Liu
L, Ma J, Cai H, Li Z and Xue Y: MOV10 binding circ-DICER1 regulates
the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4
expression change. J Exp Clin Cancer Res. 38:92019. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Liu X, Shen S, Zhu L, Su R, Zheng J, Ruan
X, Shao L, Wang D, Yang C and Liu Y: SRSF10 inhibits biogenesis of
circ-ATXN1 to regulate glioma angiogenesis via miR-526b-3p/MMP2
pathway. J Exp Clin Cancer Res. 39:1212020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Novikov NM, Zolotaryova SY, Gautreau AM
and Denisov EV: Mutational drivers of cancer cell migration and
invasion. Br J Cancer. 124:102–114. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Yu-Ju Wu C, Chen CH, Lin CY, Feng LY, Lin
YC, Wei KC, Huang CY, Fang JY and Chen PY: CCL5 of
glioma-associated microglia/macrophages regulates glioma migration
and invasion via calcium-dependent matrix metalloproteinase 2.
Neuro Oncol. 22:253–266. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Li X and Meng Y: Analyses of
metastasis-associated genes in IDH wild-type glioma. BMC Cancer.
20:11142020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zuo CY, Qian W, Huang CJ and Lu J:
Circular RNA circ-SMAD7 promoted glioma cell proliferation and
metastasis by upregulating PCNA. Eur Rev Med Pharmacol Sci.
23:10035–10041. 2019.PubMed/NCBI
|
|
106
|
Yi C, Li H, Li D, Qin X, Wang J, Liu Y,
Liu Z and Zhang J: Upregulation of circular RNA circ_0034642
indicates unfavorable prognosis in glioma and facilitates cell
proliferation and invasion via the miR-1205/BATF3 axis. J Cell
Biochem. 120:13737–13744. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Su H, Zou D, Sun Y and Dai Y:
Hypoxia-associated circDENND2A promotes glioma aggressiveness by
sponging miR-625-5p. Cell Mol Biol Lett. 24:242019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Patil V and Mahalingam K: A four-protein
expression prognostic signature predicts clinical outcome of
lower-grade glioma. Gene. 679:57–64. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Choi J, Kim SH, Ahn SS, Choi HJ, Yoon HI,
Cho JH, Roh TH, Kang SG, Chang JH and Suh CO: Extent of resection
and molecular pathologic subtype are potent prognostic factors of
adult WHO grade II glioma. Sci Rep. 10:20862020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zheng K, Xie H, Wu W, Wen X, Zeng Z and
Shi Y: circRNA PIP5K1A promotes the progression of glioma through
upregulation of the TCF12/PI3K/AKT pathway by sponging miR-515-5p.
Cancer Cell Int. 21:272021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Qu Y, Zhu J, Liu J and Qi L: Circular RNA
circ_0079593 indicates a poor prognosis and facilitates cell growth
and invasion by sponging miR-182 and miR-433 in glioma. J Cell
Biochem. 120:18005–18013. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ghiaseddin AP, Shin D, Melnick K and Tran
DD: Tumor treating fields in the management of patients with
malignant gliomas. Curr Treat Options Oncol. 21:762020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Oldrini B, Vaquero-Siguero N, Mu Q, Kroon
P, Zhang Y, Galán-Ganga M, Bao Z, Wang Z, Liu H, Sa JK, et al: MGMT
genomic rearrangements contribute to chemotherapy resistance in
gliomas. Nat Commun. 11:38832020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Oprita A, Baloi SC, Staicu GA, Alexandru
O, Tache DE, Danoiu S, Micu ES and Sevastre AS: Updated insights on
EGFR signaling pathways in glioma. Int J Mol Sci. 22:5872021.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Hua L, Huang L, Zhang X, Feng H and Shen
B: Knockdown of circular RNA CEP128 suppresses proliferation and
improves cytotoxic efficacy of temozolomide in glioma cells by
regulating miR-145-5p. Neuroreport. 30:1231–1238. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhao M, Xu J, Zhong S, Liu Y, Xiao H, Geng
L and Liu H: Expression profiles and potential functions of
circular RNAs in extracellular vesicles isolated from
radioresistant glioma cells. Oncol Rep. 41:1893–1900.
2019.PubMed/NCBI
|
|
117
|
Ding C, Wu Z, You H, Ge H, Zheng S, Lin Y,
Wu X, Lin Z and Kang D: CircNFIX promotes progression of glioma
through regulating miR-378e/RPN2 axis. J Exp Clin Cancer Res.
38:5062019. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Glazar P, Papavasileiou P and Rajewsky N:
circBase: A database for circular RNAs. RNA. 20:1666–1670. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Panda AC, Dudekula DB, Abdelmohsen K and
Gorospe M: Analysis of circular RNAs using the web tool
circinteractome. Methods Mol Biol. 1724:43–56. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang
J, Cheng H, Yan J, Zhang S, Yang P and Zhao F: Expanded expression
landscape and prioritization of circular RNAs in mammals. Cell Rep.
26:3444–3460.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881.e13. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Fan C, Lei X, Fang Z, Jiang Q and Wu FX:
CircR2Disease: A manually curated database for experimentally
supported circular RNAs associated with various diseases. Database
(Oxford). 2018:bay0442018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Xia S, Feng J, Lei L, Hu J, Xia L, Wang J,
Xiang Y, Liu L, Zhong S, Han L and He C: Comprehensive
characterization of tissue-specific circular RNAs in the human and
mouse genomes. Brief Bioinform. 18:984–992. 2017.PubMed/NCBI
|
|
124
|
Xia S, Feng J, Chen K, Ma Y, Gong J, Cai
F, Jin Y, Gao Y, Xia L, Chang H, et al: CSCD: A database for
cancer-specific circular RNAs. Nucleic Acids Res. 46(D1):
D925–D929. 2018. View Article : Google Scholar : PubMed/NCBI
|