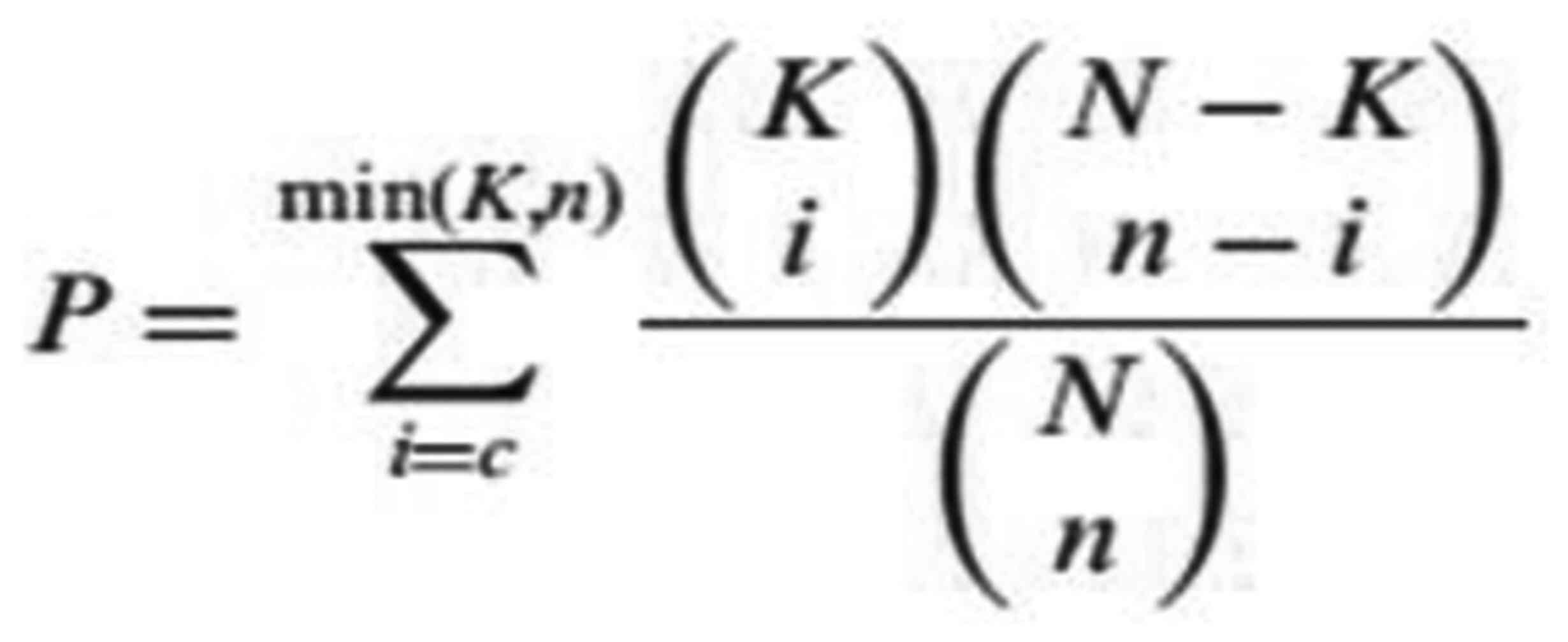Introduction
Thyroid cancer (TC) is the commonest endocrine
malignancy worldwide, accounting for 1–5% of all cancers in females
and <2% of all cancers in males. TC incidence continues to
increase globally and is neither confined to a particular region
nor influenced by underlying rates of TC (1). Papillary thyroid carcinoma (PTC) is
the most frequent type of differentiated thyroid cancer, accounting
for 80% of all pathological types of TC (2,3).
Studies on the incidence trends for TC subtypes and data from
volumes 4–9 of Cancer Incidence Database on five continents (CI5)
show that the incidence of PTC in both sexes has risen steadily in
countries around the world over the past few decades (4–10). As
indicated by a study of PTC in Manitoba, Canada, the
age-standardized rate of PTC increased from 0.93/100,000 to
6.68/100,000 individuals per year over the period 1970–2010,
showing a significant increase (11). At present, the main treatment for
PTC is a combination of surgery, radioactive iodine and
levothyroxine suppression, with a 50% 5-year survival rate
(12–15). Although PTC is less malignant than
other cancer types, the incidence and recurrence rates are still
high (20–40%). The reasons for the apparent increase in PTC
incidence are unclear and overdiagnosis and a small but actual
increase in PTC incidence are indicated as the two main processes
in some studies (16,17). Considering the expanding demands for
targeted therapies, the role of biomarkers in PTC diagnosis and
treatment remains to be elucidated. BRAF mutation is proposed to be
a prognostic biomarker; however, controversy exists regarding the
high positive ratio of BRAF mutation combined with the low ratio of
PTC aggressive biological behaviors (18–20).
Recently, non-coding RNAs and their processing machinery have been
shown to be common hallmarks of cancer regulating tumorigenesis,
progression and metastasis via various mechanisms (21,22).
Micro (mi)RNA dysregulation has been discovered in numerous cancer
types, including miRNA (miR)15a/16-1 cluster dysregulation in
chronic lymphocytic leukemia, miR-127 in primary prostate cancer
and bladder tumors and miR-29 family in lung cancer (23,24).
Significant advances have been made regarding the role of miRNAs in
diagnostics, monitoring and therapy. For example, the expression
levels of miR-145 are inversely correlated with BCR-ABL
rearrangement levels during diagnosis and treatment of chronic
myeloid leukemia, which can be used to modify the clinical
treatment for improved outcomes (25). As potential oncogenes or
oncosuppressor genes, miRNAs can be used as anticancer therapies.
For instance, miR-203 inhibits the spheroid formation and cancer
stem cells marker expression by targeting SOCS3, indicating a novel
miRNA-based clinical treatment for estrogen receptor-positive
breast cancer (26–28). In contrast to miRNAs, circular RNAs
(circRNAs) are a special type of non-coding RNA molecules that are
produced from pre-mRNA by backsplicing mechanisms. Unlike
traditional linear counterparts (containing 5′ and 3′ ends),
circRNAs have a closed circular structure, are not affected by RNA
exonucleases in eukaryotes and exhibit distinct advantages in their
stability over canonical linear RNAs (29,30).
Without free ends, circRNAs are expressed in a cell type-specific
manner, display high stability and function in gene regulation in
various pathologies. Recent studies have indicated that circRNAs
can serve as miRNA sponges to regulate gene expression and
sequester or recruit RNA/proteins; they even encode peptides or
proteins (31,32).
Increasing evidence shows that circRNAs are stably
expressed in exosomes, plasma and saliva, indicating their high
capacity for detection and clinical utility in cancer (33,34).
Evidence suggests that circRNA profiles are cancer specific and
related to tumor occurrence and progression (35–37).
The present study screened circRNA expression in PTC tumors and
verified the dysregulated circRNAs in whole blood from patients
with PTC to evaluate their potential value as PTC biomarkers. The
possible functional associations of these circRNAs with PTC were
also analyzed using bioinformatics.
Materials and methods
Ethics
The present study was approved by the Medical Ethics
Committee of Harbin Medical University (approval no. IRB3011619).
All participants involved in this study were informed of the
research objectives and signed consent forms before the study.
Subjects
Patients who underwent thyroidectomy were recruited
at The First, Second and Fourth Clinical Hospitals Affiliated with
Harbin Medical University between January 2017 and July 2019.
Patients with PTC were diagnosed and evaluated by neck ultrasonic
imaging and pathological diagnosis. Histopathological analysis and
diagnostic immunohistochemistry of all thyroid tissue specimens
were independently performed by two licensed pathologists. The
immunohistochemistry of NF-κB1 was used to confirm the diagnostic
accuracy. Briefly, the morphological and pathological
identification of PTC was performed by H&E staining at room
temperature for 10 min. Paraformaldehyde-fixed (4°C overnight) and
paraffin embedded thyroid tissue slides were dewaxed in xylene and
rehydrated in ethanol. The endogenous peroxidase was blocked by 3%
H2O2 at room temperature for 10 min. Heated
slides in a microwave submersed in 1X citrate unmasking solution
(Cell Signaling Technology, Inc.) until boiling was initiated,
followed by 10 min at sub-boiling temperature (95–98°C). The slides
were cooled on bench top for 30 min. The slides were then incubated
with a 1:100 dilution of NF-κB1 primary antibody (cat. no.
bs-1194R; Beijing Biosynthesis biotechnology Co., Ltd.) at 4°C
overnight. After washing, the slides were incubated with goat
anti-rabbit IgG (cat. no. PV-6001; OriGene Technologies, Inc.) for
20 min, followed by staining with DAB for 5 min at room
temperature. The slides were counterstained with hematoxylin at
room temperature for 20 sec, and then dehydrated with ethanol and
xylene. The morphological changes were observed by a light
microscope (magnification, ×40 or ×100; BM2000; Nanjing Jiangnan
Yongxin Optics Co., Ltd.). Images were captured in three random
fields. The number of cells was analyzed by ImageJ software
(v1.8.0.112, National Institutes of Health). Ultimately, 57 cases
of PTC and age- and sex-matched healthy controls were recruited for
this study (Table I). Among these
subjects, three pairs of PTC tumors and matching paracancerous
tissues were collected for microarray analysis. In total, 57 pairs
of whole-blood samples were obtained from all subjects and
refrigerated at −80°C for further studies.
 | Table I.Correlation between hsa_circ_IPCEF1
expression and clinicopathological characteristics in 57 patients
with papillary thyroid carcinoma. |
Table I.
Correlation between hsa_circ_IPCEF1
expression and clinicopathological characteristics in 57 patients
with papillary thyroid carcinoma.
|
Characteristics | Number of patients
(%) | Relative
expression | P-value |
|---|
| Age (years) |
|
|
|
|
<45 | 26 (45.6) | 0.75±0.16 |
0.1933 |
|
≥45 | 31 (54.4) | 0.52±0.09 |
|
| Sex |
|
|
|
|
Male | 17 (29.8) | 0.76±0.20 |
0.1571 |
|
Female | 40 (70.2) | 0.45±0.11 |
|
| Tumor size
(cm) |
|
|
|
|
<2 | 49 (86.0) | 0.70±0.14 |
0.1246 |
| ≥2 | 8
(14.0) | 0.17±0.08 |
|
| TNM stage |
|
|
|
|
I–II | 42 (73.7) | 0.77±0.20 |
0.1515 |
|
III–IV | 15 (26.3) | 0.28±0.11 |
|
| Multifocality |
|
|
|
|
Yes | 30 (52.6) | 0.61±0.09 |
0.4660 |
| No | 27 (47.4) | 0.48±0.16 |
|
| Extrathyroidal
extension |
|
|
|
|
Yes | 12 (21.1) | 0.69±0.26 |
0.8503 |
| No | 45 (78.9) | 0.63±0.18 |
|
| LNM |
|
|
|
|
Yes | 32 (56.1) | 0.82±0.13 |
<0.001a |
| No | 25 (43.9) | 0.25±0.05 |
|
RNA isolation and quality control
Total RNA was extracted from the tissue samples and
whole blood with TRI Reagent BD, which was supplied by
Sigma-Aldrich (Merck KGaA) in accordance with the manufacturer's
protocol. RNA was precipitated with isopropanol, washed twice with
75% ethanol and resuspended in RNase/DNase-free water. The RNA
concentration and purity were assessed with a Nano-200 system
(Hangzhou Allsheng Instruments Co., Ltd.). RNA samples with
OD260/OD280 ratios ≥1.8 and ≤2.1 and OD260/OD230 ratios >1.8
were used for further studies.
circRNA array
circRNA array analysis was performed by KangChen
Bio-tech. Briefly, three pairs of PTC tumors and matching
paracancerous tissues were used for Arraystar Human circRNA Array
analysis. The linear RNAs were removed by RNase R (Epicenter
Biotechnologies) to enrich circRNAs. The enriched circRNAs were
amplified and transcribed into fluorescent complementary RNA (cRNA)
by the random priming method [Quick Amp Labeling Kit, One-Color
(Agilent p/n 5190-0442)], which was performed by KangChen Bio-tec.
Subsequently, these labeled cRNAs were purified by an RNeasy Mini
kit (Qiagen GmbH) and were hybridized to an Arraystar human
circular RNA array V2 (8×15K, Arraystar, Inc.). The arrays were
then scanned with an Agilent G2505C scanner (Agilent Technologies,
Inc.) followed by washing. Array images and data analysis were
performed using Agilent Feature Extraction software (version
11.0.1.1; Agilent Technologies, Inc.) and the R software package
version 3.1.2 (R Foundation for Statistical Computing; http://www.R-project.org/) Differentially expressed
circRNAs with P-values <0.05 between the two groups were
visualized in a volcano plot and with hierarchical clustering. The
relevant datasets have been submitted to National Center for
Biotechnology Information Gene Expression Omnibus (GEO). The GEO
accession no. is GSE173299 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173299).
Reverse transcription-quantitative
(RT-q) PCR validation
The top 20 most differentially expressed circRNAs
from array analysis were further validated by RT-qPCR on 57 pairs
of whole blood and three pairs of tissue samples from subjects.
Total RNA (1 µg) was reverse-transcribed into cDNA using a First
Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.) in
accordance with the manufacturer's protocol. The candidate circRNAs
were amplified by Power SYBR Green PCR Master Mix (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with various primers.
The primers for each circRNA are presented in Table II. RT-qPCR was performed on an ABI
7300 Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) in a 10 µl PCR mixture. The thermocycling
conditions were as follows: 95°C for 10 min, 40 cycles of 95°C for
15 sec and 60°C for 60 sec. β-actin was used as an internal
control. The relative expression of each group was analyzed using
the 2−ΔΔCq method (38).
 | Table II.Primers fortop 20 dysregulated
circRNAs. |
Table II.
Primers fortop 20 dysregulated
circRNAs.
| circRNA | Regulation | Forward | Reverse |
|---|
|
hsa_circ_0000277 | Up |
5′CAGTCTTCAAGGTGGGATCGTAA3′ |
5′TGGAAGGCTTGGATCAGTCAG3′ |
|
hsa_circ_0044556 | Up |
5′CTGGTCCTGATGGCAAAACTG3′ |
5′GGGGTCCTTGAACACCAACA3′ |
|
hsa_circ_0014234 | Up |
5′CCAGAGCTATGCTTTAGGTCTCA3′ |
5′AGTGGGAAGTGGGAGGTGTC3′ |
|
hsa_circ_0040773 | Up |
5′AAGTATTACCCCGTCTTTAAGCAG3′ |
5′TTCCAGACACGCCCATCAC3′ |
|
hsa_circ_0074530 | Up |
5′ATGAGCAGGCACTCCTTGGA3′ |
5′TCAGTGGCGGGTACACCTTC3′ |
|
hsa_circ_0074595 | Up |
5′GCCTATAAGGAGGACTATCACAAG3′ |
5′CTGCGGTGCGTGATGATA3′ |
|
hsa_circ_0001681 | Up |
5′AGAGGTGGCATCTGTGAACTGTC3′ |
5′GGGAAGGCGTATGTTCAAGGTA3′ |
|
hsa_circ_0049237 | Up |
5′CCATAGGCTCACAACACCACA 3′ |
5′CCCTGCGTGTCCACCTCTA3′ |
|
hsa_circ_0058230 | Up |
5′TGGATGGGGAGCCCTACAAG3′ |
5′CCAGGTGCGGGTGTACAGG3′ |
|
hsa_circ_0057691 | Up |
5′AGCAACCAAGTGCCAGGAGT3′ |
5′CGGGTGCATCTGTCACATAACT3′ |
| hsa_circ_FGFR2 | Down |
5′GAATACGGGTCCATCAATCACA3′ |
5′ATCACGGCGGCATCTTTC3′ |
|
hsa_circ_0095448 | Down |
5′CCCCTGGAATAACATACAAACC3′ |
5′GTAGCTGCTCCCGTAAACTGAT3′ |
|
hsa_circ_0031968 | Down |
5′GTTTGGTGTCTCCCCGCTAT3′ |
5′GCCTTCTGCAACTGGAATCA3′ |
|
hsa_circ_IPCEF1 | Down |
5′AGATAAGCCTGCTGGATCAAAG3′ |
5′ACAGTGAAATCAGGCAGGTTG3′ |
|
hsa_circ_0021549 | Down |
5′TTCGGAGGTAACAGTGAAGGGA3′ |
5′AGGCATCTGGATACCATCTGTTCT3′ |
|
hsa_circ_0070098 | Down |
5′GCTCAGTGTCAGCCTCTATTTTG3′ |
5′GCATTGGTTGGCAGCTATTTG3′ |
|
hsa_circ_0079891 | Down |
5′GAATTGCTTTTGATGCTGAGTCTG3′ |
5′TTTCCATGAGTTTGGGGTAGG3′ |
|
hsa_circ_0001938 | Down |
5′CATCGTTCTTACAGTTCTGCACA3′ |
5′CAGCCACGAAGCCAAAGC3′ |
|
hsa_circ_0020396 | Down |
5′GCTTGATCGAAATCGTCCACA3′ |
5′GAAAGTTCATCCGCTCCTCTG3′ |
|
hsa_circ_0021550 | Down |
5′GATTCGGAGGTAACAGTGAAGG3′ |
5′TACAGAGCAAAAGATGGAAGCA3′ |
| β-actin |
|
5′GTGGCCGAGGACTTTGATTG3′ |
5′CCTGTAACAACGCATCTCATATT3′ |
Circularization of hsa_circ_IPCEF1
validation
To confirm the splicing junction of hsa_circ_IPCEF1,
RT-PCR followed by sequencing was performed in whole blood from PTC
subjects. Briefly, total RNA was extracted from whole blood from
patients with PTC as described above. After reverse transcription,
the cDNA was amplified using PrimeSTAR® Max DNA
Polymerase (Takara Biotechnology Co., Ltd.). The thermocycling
conditions were 35 cycles of 95°C for 10 sec, 55°C for 15 sec and
72°C for 10 sec. Using divergent primers (forward:
5′-GTTTGTCTGCTGCTGAAGATGAG-3′; reverse
5′-CCATCAGCTTTCTCTGCCTTTGTC-3′) to amplify the hsa_circ_IPCEF1
product containing a head-to-tail splicing junction site. The PCR
product was purified by 1.5% agarose gel electrophoresis,
visualized by ethidium bromide and identified by Sanger
sequencing.
Competing endogenous RNA (ceRNA)
network analysis
An interaction analysis of circRNA/miRNA was
performed by KangChen Bio-tech based on TargetScan (39–41).
Through merging the commonly targeted miRNAs, a circRNA-miRNA-mRNA
interaction network of hsa_circ_IPCEF1 was constructed by Cytoscape
3.7.2 (https://cytoscape.org). Briefly, by
merging the commonly targeted miRNAs, a ceRNA network was
constructed via three conditions (42). First, the relative concentration of
the ceRNAs and their microRNAs is clearly important; second, the
effectiveness of a ceRNA depends on the number of microRNAs that it
can ‘sponge’; and third, not all of the MREs on ceRNAs are equal.
Therefore, only these ceRNA-pair relations that passed some
filtering measures were accepted. In addition to a measure with the
number of common microRNAs, a hypergeometric test was executed for
each ceRNA pair separately, which was defined by four parameters:
i) N is the total number of miRNAs used to predict targets; ii) K
is the number of miRNAs that interact with the chosen gene of
interest; iii) n is the number of miRNAs that interact with the
candidate ceRNA of the chosen gene; and iv) is the common miRNA
number between the two genes (43).
The test calculates the P-value by using the following formula:
Functional group analysis
The molecular functional roles of the circRNA-target
gene profiles were analyzed using Gene Ontology (http://www.geneontology.org), including biological
process (BP), cellular component (CC) and molecular function (MF).
The P-value produced by topGO (http://www.bioconductor.org/packages/release/bioc/html/topGO.html)
denoted the significance of GO term enrichment in the circRNA-miRNA
targeted genes. Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analysis was then performed (https://www.genome.jp/kegg/). The P-value (EASE score,
Fisher P-value, or hypergeometric P-value) denoted the significance
of the pathway relevant to PTC. The P-value cutoff was 0.05. The
ceRNAs of each differentially expressed circRNA were further
annotated in terms of the diseases and pathways using the KEGG
orthology-based annotation system (KOBAS), (version 3.0; http://kobas.cbi.pku.edu.cn/kobas3/?t=1). P<0.05
was considered to indicate a statistically significant
difference.
Statistical analysis
All data are expressed as the means ± standard error
of the mean. circRNAs with fold changes ≥2 and P-values ≤0.05 were
selected as the significantly differentially expressed circRNAs.
Receiver operating characteristic (ROC) curves and the area under
the curve (AUC) value and 95% confidence intervals (CIs) were
established using GraphPad Prism 8.0 (GraphPad Software, Inc.) to
predict their potential association with PTC. Comparisons of data
were acquired by one-way ANOVA followed by Bonferroni post hoc or
Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
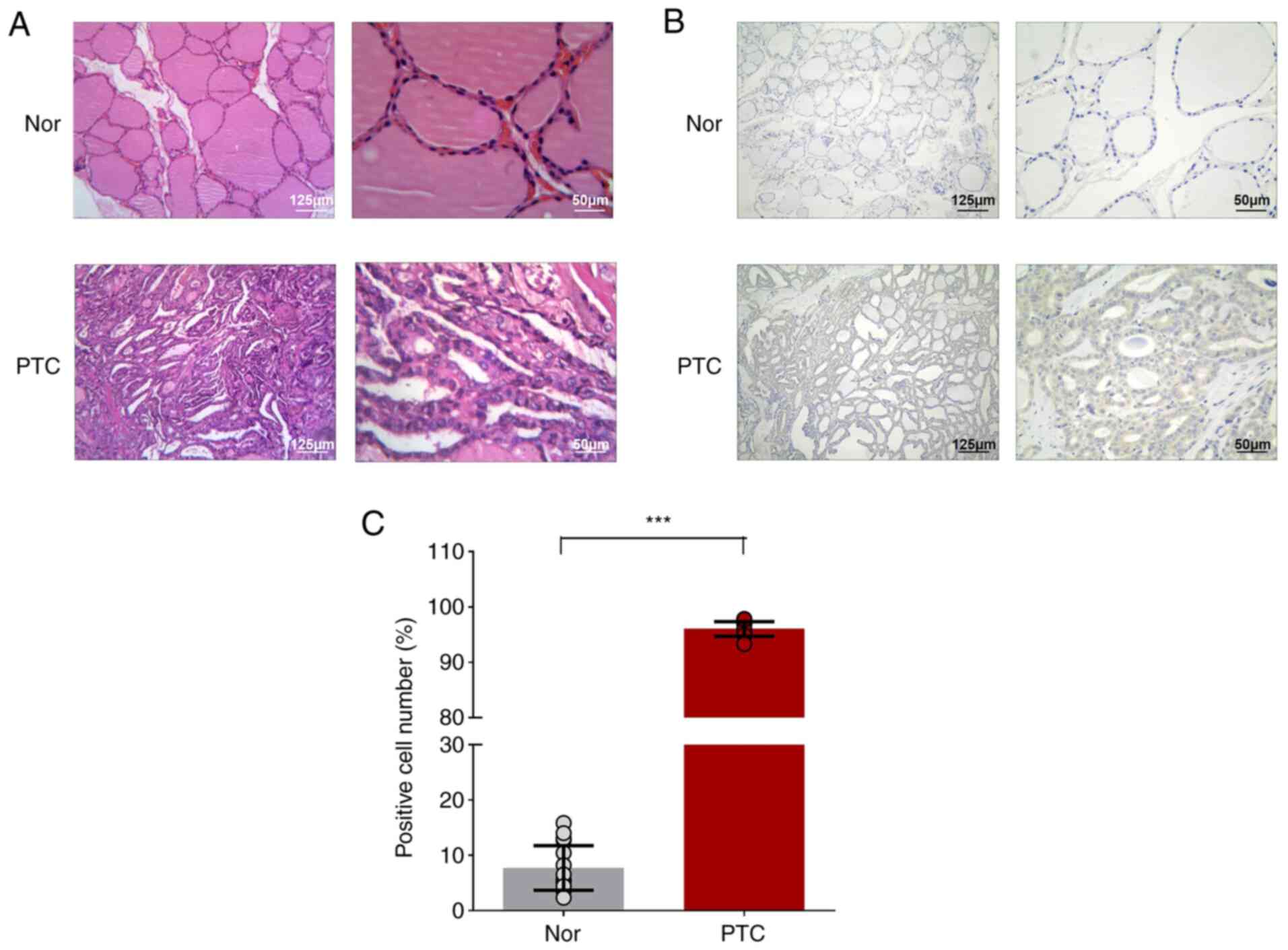
Characterization of human thyroid
samples
A total of three pairs of PTC tumors and matching
paracancerous tissues were collected for microarray. The PTC tumor
showed the typical papillary architecture with branching and the
enlarged irregular nuclei were oval-shaped and overlapping, showing
ground-glass appearance with prominent nuclear grooves and pink
cytoplasmic invaginations and intra-nuclear pseudoinclusion
(Fig. 1A). NF-κB1-related cancer
phenotype (increased expression of NF-κB1 in cell cytoplasm) were
also observed in PTC tumors (Fig. 1B
and C). Through pathological reports, the tumor tissues were
confirmed to be PTC for circRNA chip analysis.
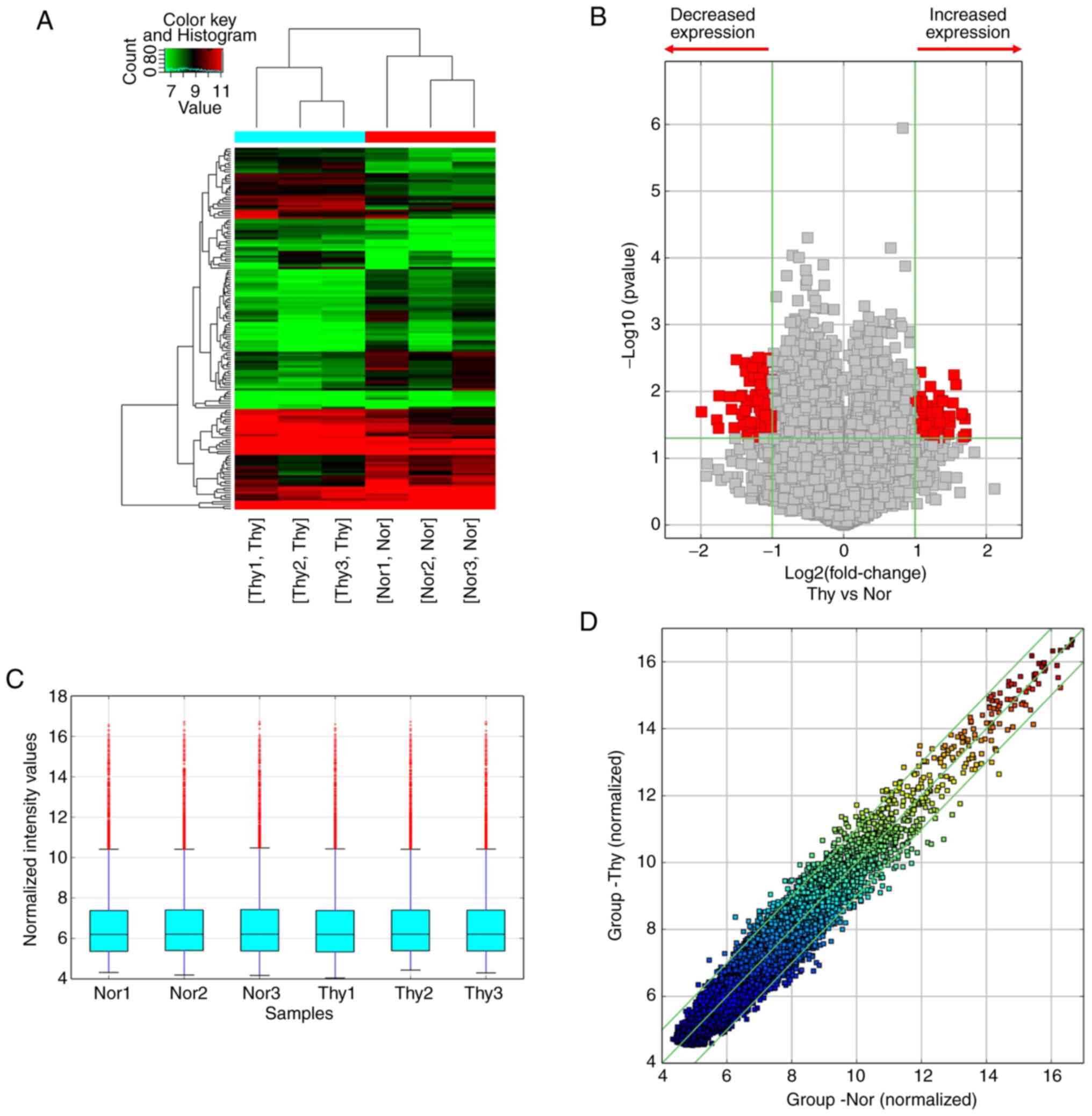
circRNA expression profile in PTC
circRNA profiling was performed with Arraystar Human
circRNA Array in three pairs of PTC tumor tissues and cancer
contralateral normal thyroid tissues. Hierarchical cluster analysis
revealed that the circRNA expression patterns were distinctive
between PTC tumor and tumor-adjacent normal thyroid tissues
(Fig. 2A). A cut-off value for
differential expression fold change was set to 2.0 and 158
significantly dysregulated circRNAs (74 upregulated and 84
downregulated) were found in the PTC tumors (Fig. 2B-D, P<0.05 and false discovery
rate <0.05).
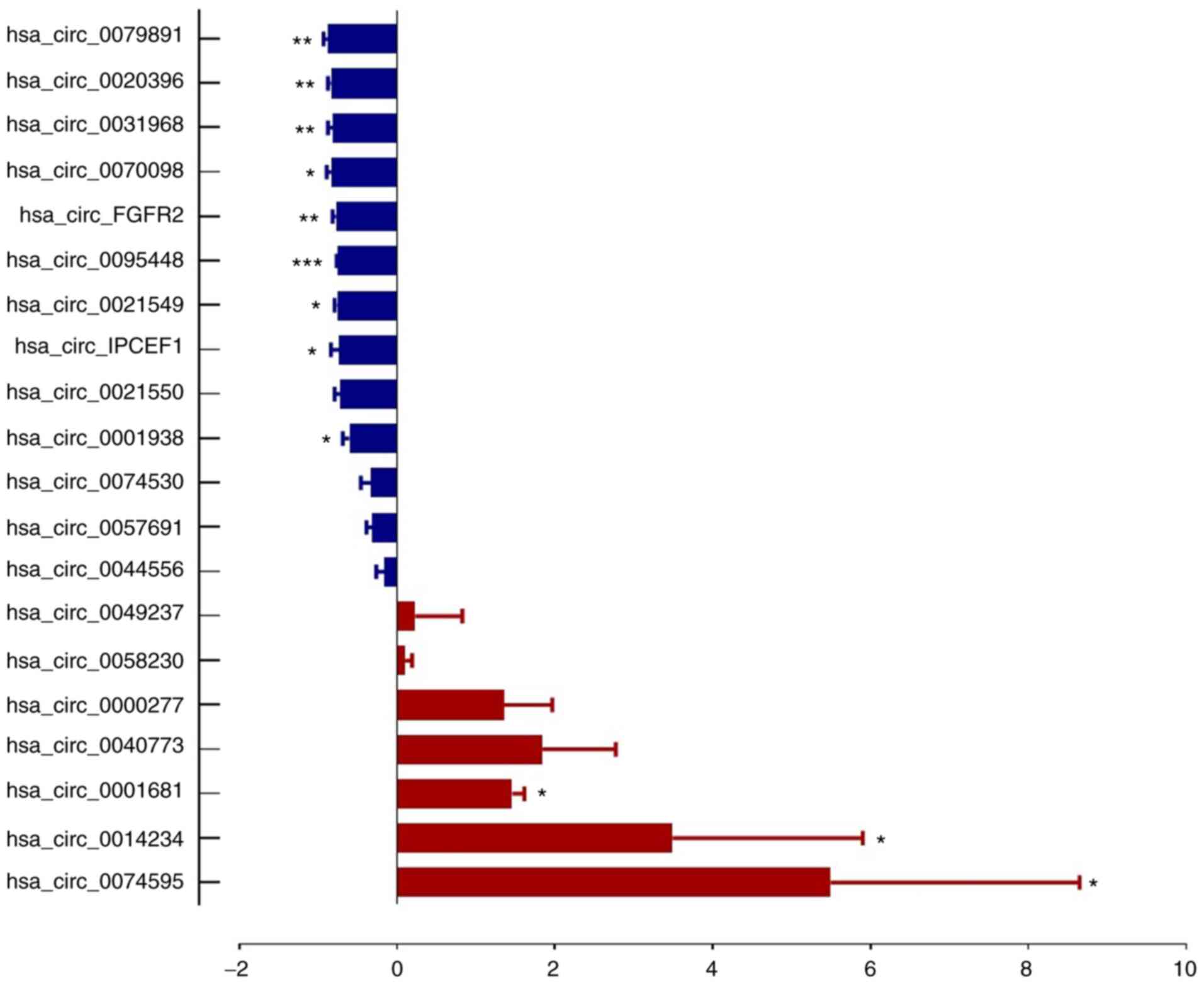
Validation of the differential
expression of circRNAs in PTC
From microarray results, thetop 20 most dysregulated
circRNAs (10 upregulated and 10 downregulated, Table II) were selected for RT-qPCR
validation on three pairs of PTC tumor tissues and cancer
contralateral normal thyroid tissues as well as in whole blood from
57 pair of patients with PTC and healthy controls. The
characteristics of the 57 patients with PTC are presented in
Table I. The RT-qPCR results showed
that 7 circRNA expression trends were consistent with the
microarray assay, which included 3 upregulated circRNAs
(hsa_circ_0000277, hsa_circ_0074530 and hsa_circ_0057691) and 4
downregulated circRNAs (hsa_circ_0020396, hsa_circ_0095448,
hsa_circ_IPCEF1 and hsa_circ_0021549; Figs. 3 and 4).
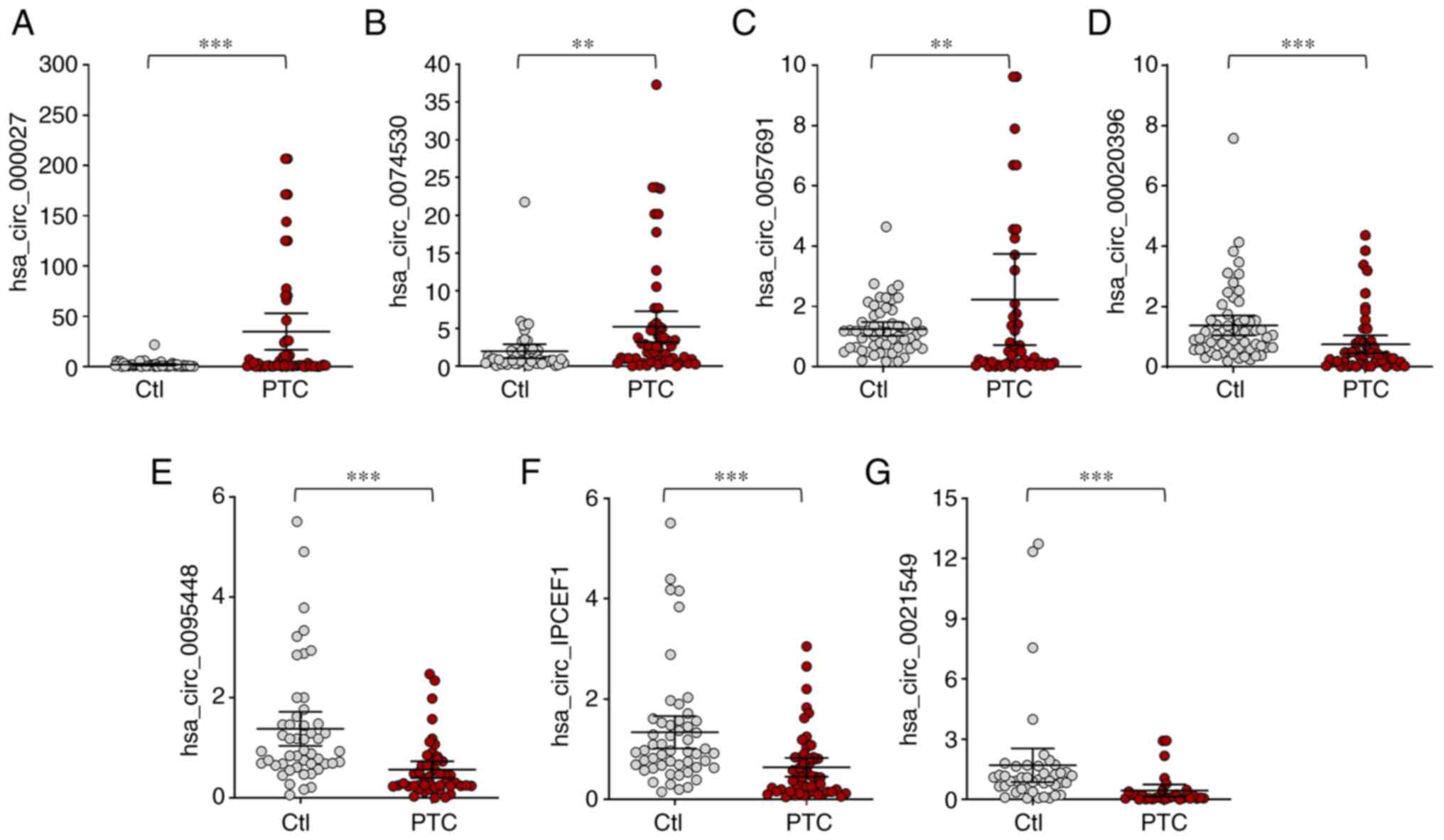 | Figure 4.Confirmation of the differential
expression of circRNAs in whole blood from PTC patients and healthy
controls. (A) hsa_circ_0000277, (B) hsa_circ_0074530, (C)
hsa_circ_0057691 were significantly upregulated, while (D)
hsa_circ_0020396, (E) hsa_circ_0095448, (F) hsa_circ_IPCEF1 and (G)
hsa_circ_0021549 were significantly downregulated in PTC patients.
**P<0.01, ***P<0.001, n=57 pairs of subjects. Comparisons of
data were acquired by one-way ANOVA followed by Bonferroni post
hoc. circRNA/circ, circular RNA; PTC, papillary thyroid cancer;
Ctl, healthy controls. |
Functional annotation of the
dysregulated circRNAs
By targeting miRNA response elements (MREs),
circRNAs can regulate mRNA by acting as miRNA sponges. The
predicted MREs for these seven validated circRNAs are presented in
Table III. Based on TargetScan,
the targets/miRNAs were analyzed to identify the potential targets
of miRNAs (KangChen Bio-Tech). Based on the seven dysregulated
circRNAs and their predicted MREs, the 256 predicted ceRNA genes
are listed in Table SI.
 | Table III.Predicted MREs of seven validated
circRNAs. |
Table III.
Predicted MREs of seven validated
circRNAs.
| circRNA | FC (abs) | chrom | GeneSymbol | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
|
hsa_circ_0057691 | 2.81 | chr2 | SATB2 | hsa-miR-9-5p | hsa-miR-1264 | hsa-miR-628-5p |
hsa-miR-514a-5p |
hsa-miR-30c-1-3p |
|
hsa_circ_0074530 | 2.92 | chr5 | CD74 |
hsa-miR-4677-5p |
hsa-miR-7161-3p | hsa-miR-515-3p |
hsa-miR-1271-3p |
hsa-miR-3663-5p |
|
hsa_circ_0000277 | 2.55 | chr11 | PDE3B |
hsa-miR-138-2-3p | hsa-miR-873-5p | hsa-miR-571 | hsa-miR-205-3p | hsa-miR-421 |
|
hsa_circ_0020396 | 2.68 | chr10 | DOCK1 |
hsa-miR-103a-2-5p | hsa-miR-132-3p |
hsa-miR-196a-5p |
hsa-miR-138-2-3p | hsa-miR-136-5p |
|
hsa_circ_0095448 | 3.02 | chr11 | SOX6 |
hsa-miR-1228-5p | hsa-miR-367-5p |
hsa-miR-6501-5p |
hsa-miR-3692-5p | hsa-miR-22-3p |
|
hsa_circ_IPCEF1 | 2.69 | chr6 | IPCEF1 | hsa-miR-3916 |
hsa-miR-3619-5p | hsa-miR-4314 |
hsa-miR-5589-5p |
hsa-miR-6837-3p |
|
hsa_circ_0021549 | 2.61 | chr11 | MPPED2 | hsa-miR-574-5p | hsa-miR-452-5p | hsa-miR-145-3p |
hsa-miR-29b-1-5p | hsa-miR-553 |
The molecular functional roles of the circRNA-target
gene profiles were analyzed using GO and KEGG analyses. For the
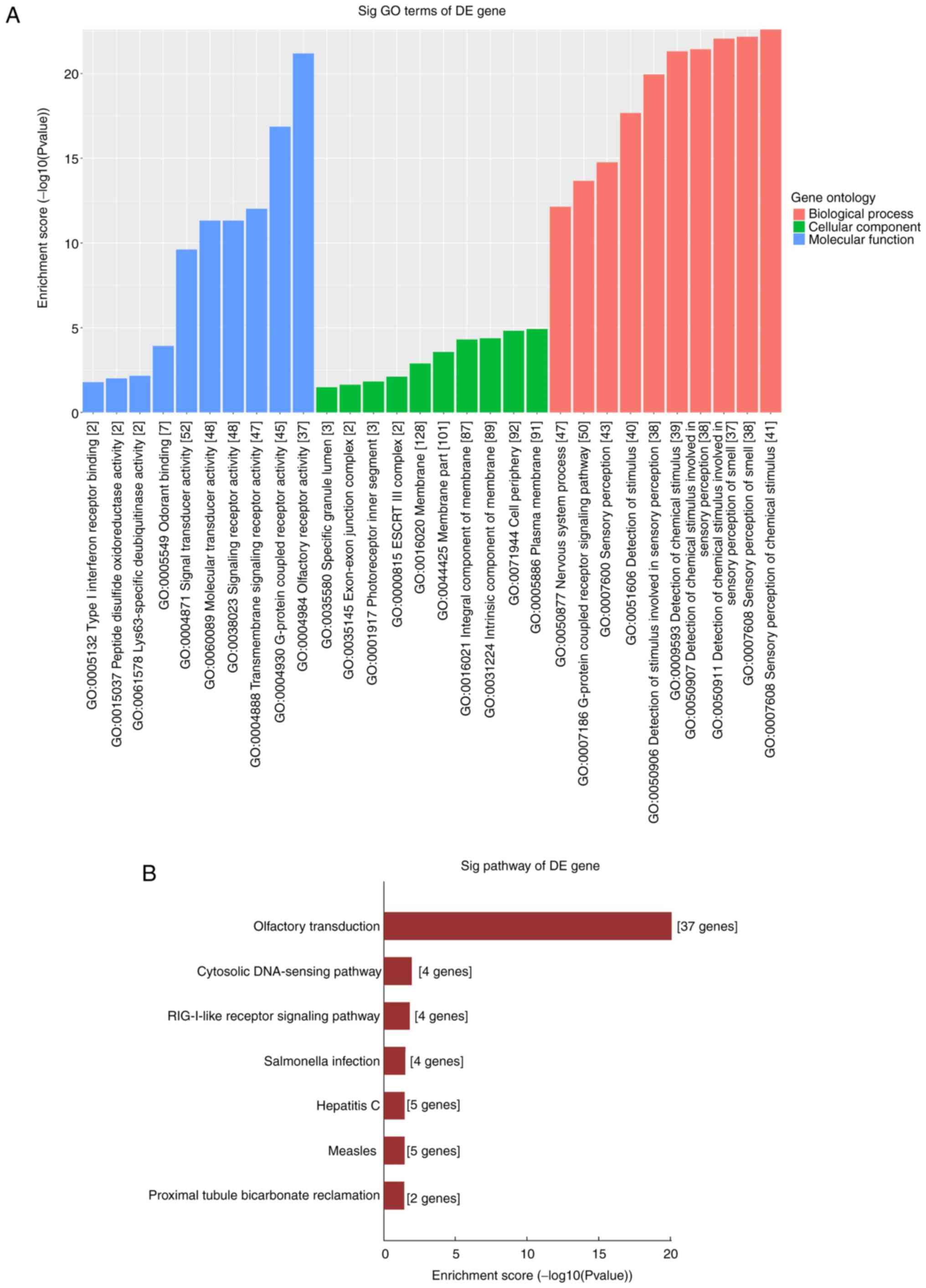
seven dysregulated circRNAs, the top 10 enriched GO terms are shown
and ranked by Enrichment Score [-log10 (P-value)]. Through GO
analysis of the different genes of dysregulated circRNAs, it is
possible to identify genes that may be related to changes in gene
function in PTC pathogenesis. As a result, the enriched MF, BP and
CC terms were determined to be involved in ‘Olfactory receptor
activity’, ‘G-protein coupled receptor signaling pathway’ and
‘Signal receptor activity in the plasma membrane’ (Fig. 5A). In KEGG analysis, the
significantly enriched pathways were involved in the ‘cytosolic
DNA-sensing pathway’, the ‘RIG-I-like receptor signaling pathway’
and the ‘Olfactory transduction’ pathway (Fig. 5B). These results indicate that these
dysregulated circRNAs may cause PTC cells to undergo a series of
molecular and signal changes.
ceRNA analyses of dysregulated
circRNAs
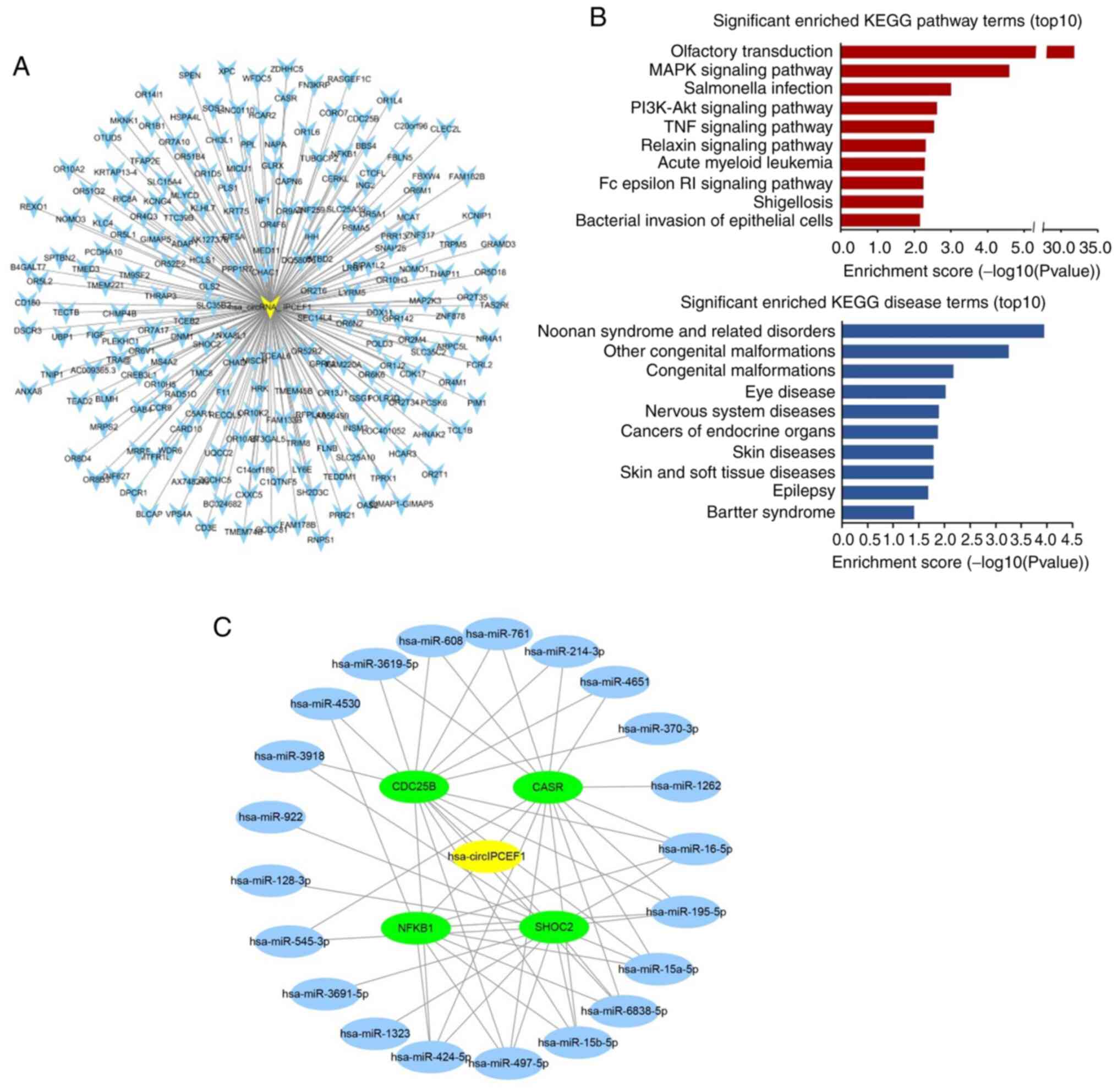
Among these potential circular RNAs, hsa_circ_IPCEF1
was related to 207 ceRNA genes out of the 256 ceRNA genes,
exhibiting the strongest role in gene regulation (Table SII; Fig. 6A). Therefore, 207 ceRNA genes
related to hsa_circ_IPCEF1 were further annotated by KOBAS 3.0,
which were enriched in a variety of pathways and diseases. The top
10 items of KEGG pathway and diseases are presented in Fig. 6B, including ‘MAPK signaling
pathway’, ‘PI3K-AKT signaling pathway’, ‘TNF signaling pathway’,
‘Olfactory transduction’, ‘Salmonella infection’, ‘Relaxin
signaling parhway’, ‘Acute myeloid leukemia’, ‘Fcepsilon RI
signaling pathway’, ‘Shigellosis’, ‘Bacterial invasion of
epithelial cells’ and ‘cancers of endocrine organs’, ‘cancer’,
‘endocrine, metabolic diseases’, respectively, indicating the role
of hsa_circ_IPCEF1 in PTC regulation (Fig. 6B).
Based on a comprehensive search of other studies in
human malignancies, it was found that four ceRNAs (CASR, CDC25B,
NFκB1 and SHOC2) were cancer-related target genes, which were
reported to be regulated by 21 miRNAs (hsa-miR-16-5p,
hsa-miR-195-5p, hsa-miR-15a-5p, hsa-miR-6838-5p, hsa-miR-15b-5p,
hsa-miR-497-5p, hsa-miR-424-5p, hsa-miR-4530, hsa-miR-1323,
hsa-miR-3691-5p, hsa-miR-545-3p, hsa-miR-128-3p, hsa-miR-922,
hsa-miR-3918, hsa-miR-3619-5p, hsa-miR-608, hsa-miR-1262,
hsa-miR-761, hsa-miR-214-3p, hsa-miR-4651 and hsa-miR-370-3p, data
not shown). Therefore, the circRNA-miRNA-mRNA module containing
four mRNAs and 21 miRNAs was constructed in Cytoscape software to
concentrate the ceRNA targets (Fig.
6C).
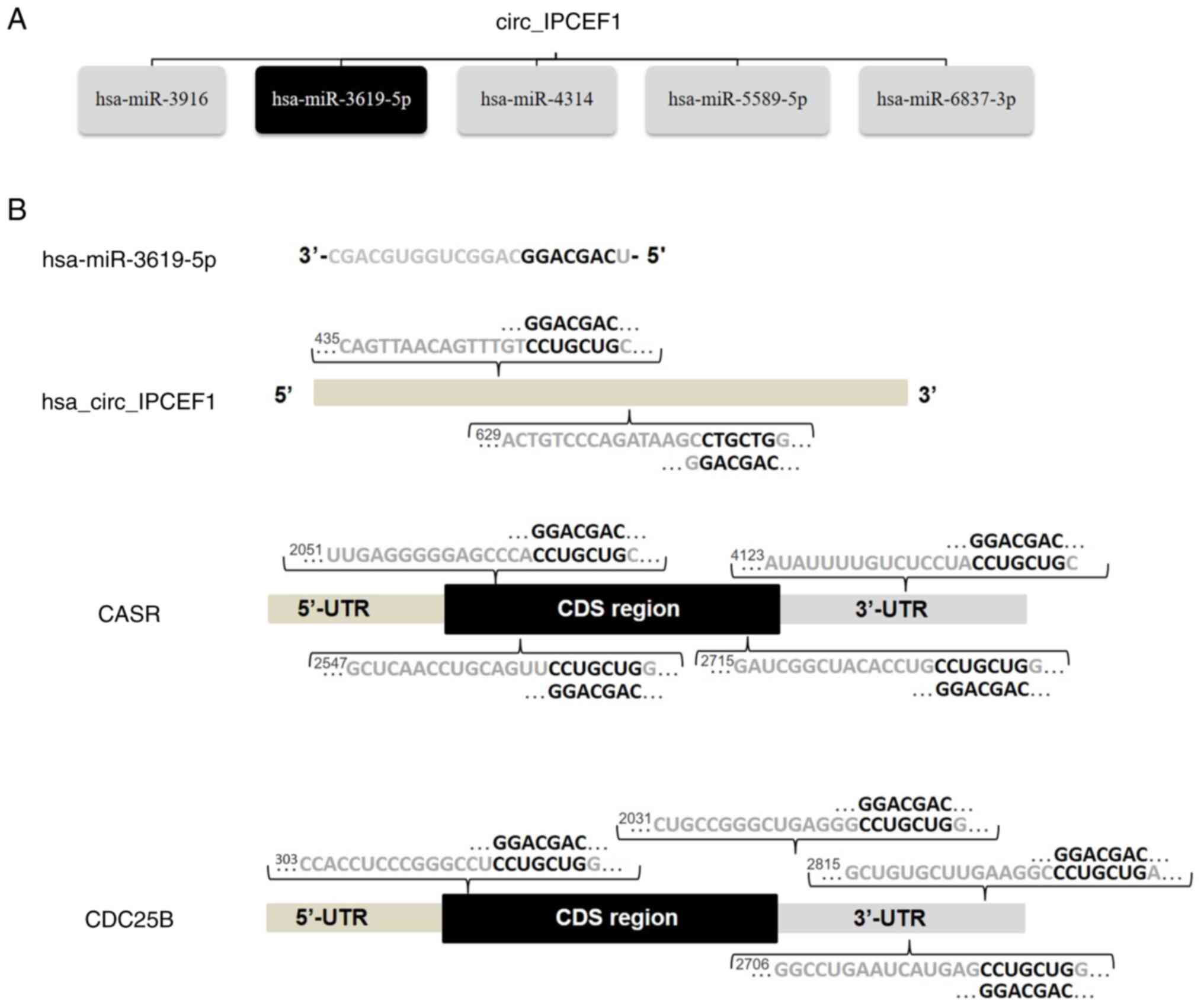
Among these miRNAs, hsa-miR-3619-5p, reported as a
tumor inhibitor, was one of the top MREs of hsa_circ_IPCEF1, with
two seed sequence matching regions (44). Bioinformatics screening suggested
that CDC25B (NM_021873) and CASR (NM_000388) were potential direct
target genes of hsa-miR-3619-5p, with complementary binding sites
located in the 3′-UTRs of CDC25B and CASR mRNAs, respectively
(Fig. 7). These bioinformatics
analyses indicated that hsa_circ_IPCEF1/hsa-miR-3619-5p/target
genes may be involved in PTC pathogenesis.
Prediction value of dysregulated
circRNAs
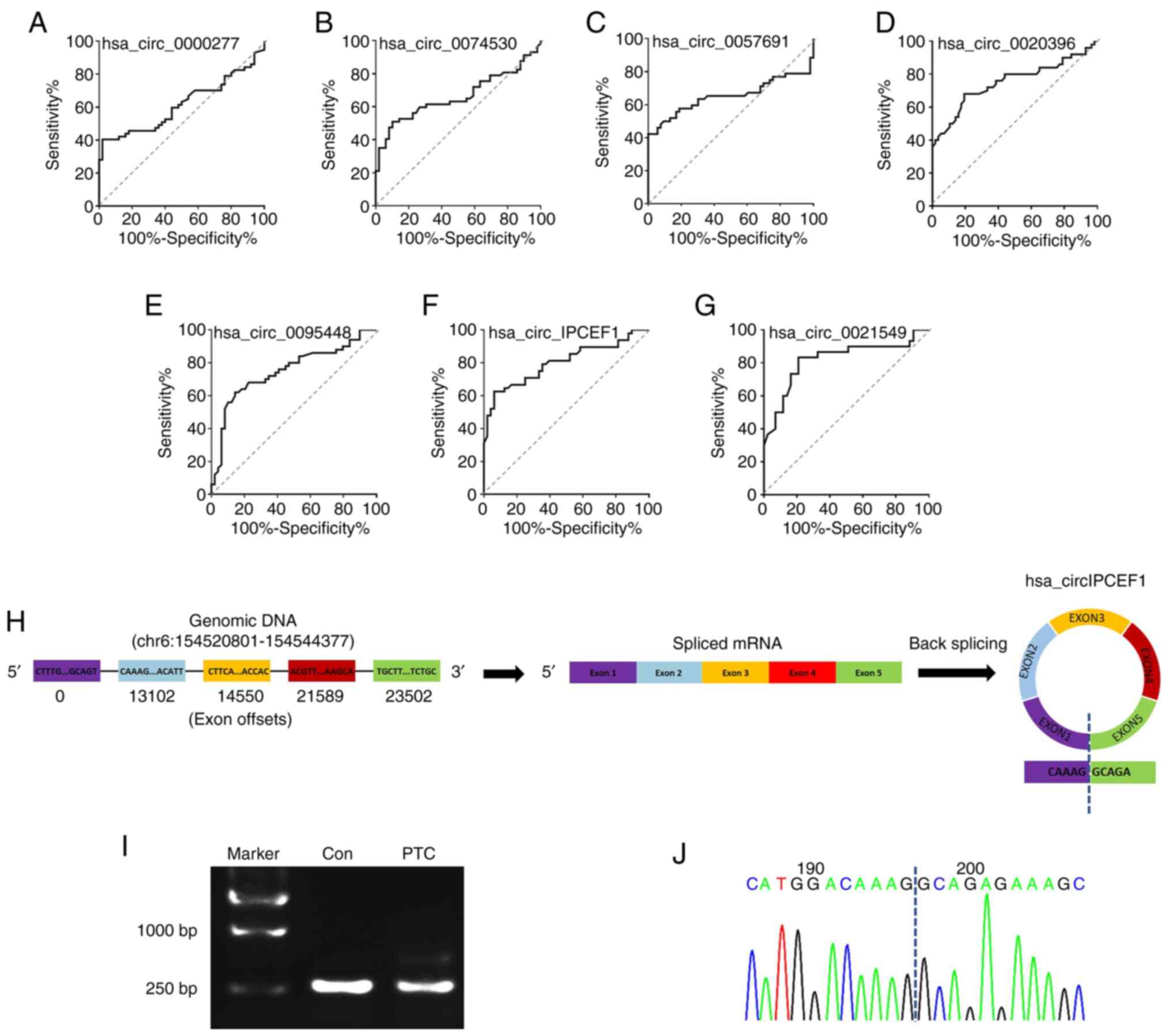
To assess the performance of these seven circRNAs in
predicting their potential association with PTC, their ROC curves
were calculated and the accuracy evaluated with the areas under the
ROC curves (AUCs). The preferable AUC value for circRNAs were
hsa_circ_0021549 (AUC: 0.8194; 95% CI: 0.7117 to 0.9270;
P<0.0001) and hsa_circ_IPCEF1 (AUC: 0.801095% CI: 0.7108 to
0.8912; P<0.0001; Fig. 8A-G).
Instead of other circRNAs, hsa_circ_IPCEF1 exhibited the strongest
functional role in ceRNA analysis, therefore, the correlation
between hsa_circ_IPCEF1 expression and clinicopathological
characteristics was examined in 57 patients with PTC. Statistical
analysis showed that the downregulation of hsa_circ_IPCEF1
expression was associated with tumor lymph node metastasis in
patients with PTC, suggesting its clinical value for PTC diagnosis
and prognosis (Table I). To further
confirm the formation of hsa_circ_IPCEF1 in patients with PTC, the
splicing junction of hsa_circ_IPCEF1 was validated using RT-PCR and
sequencing methods. As shown in Fig.
8H, hsa_circ_IPCEF1 is generated from the IPCEF1 gene
located on human chromosome 6 (154520801–154544377). The 245bp
products of hsa_circ_IPCEF1 in whole blood from subjects were
amplified using divergent primers (Fig.
8I). Sanger sequencing further confirmed that the head-to-tail
junction sites (AG/GC) were consistent with the hsa_circ_IPCEF1
annotation (Fig. 8J).
Discussion
circRNAs, a special novel class of endogenous
non-coding RNAs, have been documented to serve crucial roles in
various diseases by regulating numerous cellular events (45). Although numerous studies have
focused on circRNAs as cancer markers, the diagnostic ability and
understanding of circRNAs in PTC are still unclear. Therefore, the
present study aimed to profile the expression levels of all
circRNAs in patients with PTC, determine their potential roles in
PTC and explore new insights into the pathogenesis of TC.
From microarray analysis, an aberrant expression
pattern of circRNAs in PTC tissues compared to benign thyroid
tissues was found. In liquid biopsy, the usage of whole blood is
the main source of body fluid and circRNAs in blood cells or whole
blood have been proposed as biomarkers for human diseases due to
their high stability, abundance and spatiotemporal specific
expression (46). Therefore, three
upregulated and four downregulated circRNAs were verified in whole
blood from 57 pairs of patients with PTC compared to healthy
subjects and validated by RT-qPCR.
PTC originates from thyroid follicular epithelial
cells, accounting for 89.8% of differentiated thyroid carcinomas,
while only 38% of thyroid cancers show clinical symptoms (47). However, the extensive implementation
of cancer imaging studies in preoperative diagnostics has resulted
in a growing incidence of low-risk PTCs, which has caused a
continuing debate about the adequacy of aggressive surgical
approaches and radioiodine therapy (48). The molecular predictors of
preoperative diagnostics and accurate treatment for PTC are poorly
defined. Although studies have focused on mRNA transcripts and
non-coding RNAs as potential PTC markers, the expression profiles
and roles of circRNAs in PTC remain to be elucidated (49–52).
By contrast with recent studies in circRNA screening in PTC
tissues, the present study validated seven dysregulated circRNAs
(three upregulated and four downregulated) in whole blood from PTC
subjects, which were found to be more suitable for biomarker
detection in the future (53,54).
To explore the mechanisms of these dysregulated circRNAs on the
pathological processes of PTC, GO and KEGG pathway analyses were
performed. KEGG pathway analysis demonstrated that the olfactory
transduction pathway was the most significant pathway for
enrichment, followed by the cytosolic DNA-sensing pathway and the
RIG-I-like receptor signaling pathway. Moreover, according to the
BP, CC and MF terms with substantial enrichment, the genes were
mainly associated with ‘G-protein-coupled receptor activity’,
‘Signal receptor activity in the plasma membrane’ and ‘Olfactory
receptor activity’. These results suggested that the deregulated
circRNAs may participate in the pathological process of PTC.
Recently, circRNAs have been well described as ceRNAs in human
diseases, as has the possibility of using circRNAs as molecular
markers for related diseases (55).
Compared with miRNAs, circRNAs act on miRNA sponges to attenuate
the effect of miRNAs on mRNA expression (42). For example, circCRIM1 promotes the
expression of BTG2 by inhibiting the expression of miR-125b-5p in
LUAC cells, thus affecting the growth of LUAC cells (37). The loss of circ-znf609 inhibits the
proliferation and cell cycle transition and induces the apoptosis
of NPC cells via modulation of the miR-188/elf2 axis (35). In bladder cancer, circRNA-cTFRC is
upregulated and correlated with tumor grade and survival rate,
acting as a miR-107 sponge to regulate TFRC expression, cancer cell
invasion, proliferation, epithelial to mesenchymal transition and
tumor growth (56). These findings
suggest the key roles of abnormal circRNAs and target genes in the
pathogenesis of disease, indicating a new perspective for the early
diagnosis, treatment and prognosis of PTC.
Using a set of bioinformatics tools, the present
study predicted MREs for the seven validated circRNAs and the
potential targets of miRNAs. The present study found one promising
downregulated circRNA (hsa_circ_IPCEF1) in PTC subjects, which was
not indicated in any previously published literature but showed
interactions with four cancer-related ceRNAs (CASR, CDC25B, NF-κB1
and SHOC2). Notably, NF-κB1 is a transcription regulator that
serves important roles in numerous biological processes, such as
survival and proliferation, inflammation and adaptive immune
responses and has been widely implicated in the development and
progression of cancer pathogenesis (57–60).
In cancer cells, chronic activation and nuclear localization result
in NF-κB1 accumulation and NF-κB activation, which stimulate cell
survival by inducing anti-apoptotic gene expression (61). Recently, NF-κB1 has been proved to
be upregulated in a variety of cancers, including subungual
keratoacanthoma, urothelial carcinoma and breast cancer, suggesting
that it is a biomarker for diagnosis and treatment (62–66).
In the results of the present study, NF-κB1 was predicted to be a
ceRNA of hsa_circ_IPCEF1 regulation and the downregulation of
hsa_circ_IPCEF1 in PTC was accompanied by the upregulation of
NF-κB1, suggesting that a combination of hsa_circ_IPCEF1 and NF-κB1
could be used as biomarkers for PTC diagnosis/prognosis. The ROC
analyses of RT-qPCR results, as well as correlation analysis
between hsa_circ_IPCEF1 and clinical parameters, further confirmed
hsa_circ_IPCEF1 as one preferable candidate biomarker for PTC.
Moreover, the circRNA-miRNA-mRNA module suggested that
hsa-miR-3619-5p was a direct target for hsa_circ_IPCEF1 sponging.
As a tumor suppressor, hsa-miR-3619-5p inhibits prostate cancer
cell growth and hampers the proliferation of cutaneous squamous
cell carcinoma and liver cancer cells, exerting tumor inhibitory
effects on human malignancies (67–69).
Bioinformatics analysis further confirmed the interactions between
hsa-miR-3619-5p and CDC25B (an initiator of mitosis) and CASR
(calcium-sensing receptor) mRNAs, which may be direct targets of
hsa_circ_IPCEF1/hsa-miR-3619-5p axis regulation. By controlling the
G2/M cell cycle phase transition and calcium metabolism,
CDC25B and CASR serve important roles in the occurrence and
metastasis of cancer, respectively. These results suggested that
hsa_circ_IPCEF1 may participate in the pathological process of PTC
through sponging hsa-miR-3619-5p. Clearly, the functions and
mechanisms of the hsa_circ_IPCEF1/miR-3619-5p axis need to be
further investigated in PTC cells to conclusively determine which
target genes interact which cellular functions are affected during
tumorigenesis. Taken together, the present study identified
distinctive circRNA expression patterns in PTC. hsa_circ_IPCEF1 may
bind to hsa-miR-3619-5p through sponge action, thus regulating the
target genes of cell proliferation, migration and invasion and
triggering the initiation and progression of PTC. These results
provide insight into the pathogenesis of PTC and suggest the
potential roles of hsa_circ_IPCEF1 in PTC diagnosis and
treatment.
There are still some potential limitations and
shortcomings in the present study. First, in PTC diagnosis,
diagnostic performance was primarily based on neck ultrasonic
diagnosis rather than expensive fine-needle aspiration cytology.
PTC tumors were obtained from aggressive surgical approaches on
patients with high-risk PTC, who were diagnosed and evaluated by
neck ultrasonic imaging and immunohistochemistry. Limited by the
small size of PTC tumors, the present study did not perform
diagnostic assays on these samples to clarify the PTC subtype and
genotype. However, the PTC tumors obtained for microarray were
conventional PTC, exhibiting fibrovascular cores and enlarged
nuclei with nuclear clearing, which were sufficiently accurate for
the present study. Second, the correlation between the expression
level of hsa_circ_IPCEF1 and the clinical parameters in patients
with PTC was analyzed only in 57 pairs of subjects. A new set of
patients with PTC should be recruited to test the predictive value
and the utility of hsa_circ_IPCEF1 as a prognostic biomarker in the
near future. Nevertheless, the present study provided new insight
into the pathogenesis of PTC at the molecular level and proposes
one potential candidate PTC biomarker. Third, the analysis of the
present study was limited to the tissue and whole blood of patients
with PTC and the biological function of the maladjusted circRNAs
was predicted only through bioinformatics. The effects of the
dysregulated circRNAs on the proliferation and migration of PTC
tumor cells still need to be studied in vivo and in
vitro. Also, the construction of the ceRNA network is based on
the sponge function of circRNAs; other regulatory functions still
need to be further studied. The mechanisms and functions of the
hsa_circ_IPCEF1/hsa-miR-3619-5p axis in PTC will be studied in the
near future.
In conclusion, the present study reported seven
significantly differentiated circRNAs in patients with PTC compared
to healthy subjects, suggesting a key role of dysregulated circRNAs
in the pathogenesis of PTC. As a downregulated circRNA,
hsa_circ_IPCEF1 may show promise as a potential biomarker for PTC.
Moreover, the bioinformatics analysis suggested that the
hsa_circ_IPCEF1/hsa-miR-3619-5p axis may be involved in the
pathogenesis of PTC, providing a new theoretical basis for the
future diagnosis and treatment of PTC (Fig. 9).
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (Contract grant no. 81770054); National
Natural Science Foundation of China (Contract grant no.
8197052197); Chinese Natural science foundation of Heilongjiang
province (grant no. LH2019H017); Tianjin Municipal Science and
Technology Project (grant no. 18JCZDJC44900); The Fundamental
Research Funds for the Central Universities-Nankai University
(grant no. 63191150); Spark Research Fund from The Fourth
Affiliated Hospital of Harbin Medical University (grant no.
HYDSYXH201907).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SSL and XJ designed the study and interpreted the
data. MG analyzed and interpreted the patient data regarding
papillary thyroid carcinoma. SSL and YS performed the histological
examination of PTC, molecular studies and were major contributors
in writing the manuscript. JD, YL and SY performed RT-qPCR
experiments. YZ and XJ performed bioinformatics analysis. SSL and
XJ confirm the authenticity of all the raw data. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
The study procedure was approved by Medical Ethics
Committee of Harbin Medical University (approval no.
IRB3011619).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kilfoy BA, Zheng T, Holford TR, Han X,
Ward MH, Sjodin A, Zhang Y, Bai Y, Zhu C, Guo GL, et al:
International patterns and trends in thyroid cancer incidence,
1973–2002. Cancer Causes Control. 20:525–531. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cai X, Zhao Z, Dong J, Lv Q, Yun B, Liu J,
Shen Y, Kang J and Li J: Circular RNA circBACH2 plays a role in
papillary thyroid carcinoma by sponging miR-139-5p and regulating
LMO4 expression. Cell Death Dis. 10:1842019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heidari Z, Mohammadpour-Gharehbagh A,
Eskandari M, Harati-Sadegh M and Salimi S: Genetic polymorphisms of
miRNA let7a-2 and pri-mir-34b/c are associated with an increased
risk of papillary thyroid carcinoma and clinical/pathological
features. J Cell Biochem. Dec 14–2018.(Epub ahead of print). doi:
10.1002/jcb.28152.
|
|
4
|
La Vecchia C, Malvezzi M, Bosetti C,
Garavello W, Bertuccio P, Levi F and Negri E: Thyroid cancer
mortality and incidence: A global overview. Int J Cancer.
136:2187–2195. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Akslen LA, Haldorsen T, Thoresen SO and
Glattre E: Incidence pattern of thyroid cancer in Norway: Influence
of birth cohort and time period. Int J Cancer. 53:183–187. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Colonna M, Grosclaude P, Remontet L,
Schvartz C, Mace-Lesech J, Velten M, Guizard A, Tretarre B, Buemi
AV, Arveux P and Esteve J: Incidence of thyroid cancer in adults
recorded by French cancer registries (1978–1997). Eur J Cancer.
38:1762–1768. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
dos Santos Silva I and Swerdlow AJ:
Thyroid cancer epidemiology in England and Wales: Time trends and
geographical distribution. Br J Cancer. 67:330–340. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Montanaro F, Pury P, Bordoni A and Lutz
JM; Swiss Cancer Registries Network, : Unexpected additional
increase in the incidence of thyroid cancer among a recent birth
cohort in Switzerland. Eur J Cancer Prev. 15:178–186. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pettersson B, Adami HO, Wilander E and
Coleman MP: Trends in thyroid cancer incidence in Sweden,
1958–1981, by histopathologic type. Int J Cancer. 48:28–33. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ferlay J, Colombet M and Bray F: Cancer
incidence in five continents, CI5plus: IARC CancerBase No. 9
[Internet]. Lyon, France: International Agency for Research on
Cancer; 2018
|
|
11
|
Pathak KA, Leslie WD, Klonisch TC and
Nason RW: The changing face of thyroid cancer in a population-based
cohort. Cancer Med. 2:537–544. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jayarangaiah A, Sidhu G, Brown J,
Barrett-Campbell O, Bahtiyar G, Youssef I, Arora S, Skwiersky S and
McFarlane SI: Therapeutic options for advanced thyroid cancer. Int
J Clin Endocrinol Metab. 5:26–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rahmani N, Abbas Hashemi S, Fazli M and
Raisian M: Clinical management and outcomes of papillary,
follicular and medullary thyroid cancer surgery. Med Glas (Zenica).
10:164–167. 2013.PubMed/NCBI
|
|
14
|
Lin JD, Chao TC, Huang MJ, Weng HF and
Tzen KY: Use of radioactive iodine for thyroid remnant ablation in
well-differentiated thyroid carcinoma to replace thyroid
reoperation. Am J Clin Oncol. 21:77–81. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gottlieb JA and Hill CS Jr: Chemotherapy
of thyroid cancer with adriamycin. Experience with 30 patients. N
Engl J Med. 290:193–197. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Long MY, Chen JW, Zhu Y, Luo DY, Lin SJ,
Peng XZ, Tan LP and Li HH: Comprehensive circular RNA profiling
reveals the regulatory role of circRNA_0007694 in papillary thyroid
carcinoma. Am J Transl Res. 12:1362–1378. 2020.PubMed/NCBI
|
|
17
|
Dong S, Xie XJ, Xia Q and Wu YJ:
Indicators of multifocality in papillary thyroid carcinoma
concurrent with Hashimoto's thyroiditis. Am J Cancer Res.
9:1786–1795. 2019.PubMed/NCBI
|
|
18
|
Xing M, Westra WH, Tufano RP, Cohen Y,
Rosenbaum E, Rhoden KJ, Carson KA, Vasko V, Larin A, Tallini G, et
al: BRAF mutation predicts a poorer clinical prognosis for
papillary thyroid cancer. J Clin Endocrinol Metab. 90:6373–6379.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Basolo F, Torregrossa L, Giannini R,
Miccoli M, Lupi C, Sensi E, Berti P, Elisei R, Vitti P, Baggiani A
and Miccoli P: Correlation between the BRAF V600E mutation and
tumor invasiveness in papillary thyroid carcinomas smaller than 20
millimeters: Analysis of 1060 cases. J Clin Endocrinol Metab.
95:4197–4205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Elisei R, Viola D, Torregrossa L, Giannini
R, Romei C, Ugolini C, Molinaro E, Agate L, Biagini A, Lupi C, et
al: The BRAF(V600E) mutation is an independent, poor prognostic
factor for the outcome of patients with low-risk intrathyroid
papillary thyroid carcinoma: Single-institution results from a
large cohort study. J Clin Endocrinol Metab. 97:4390–4398. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Markopoulos GS, Roupakia E, Tokamani M,
Chavdoula E, Hatziapostolou M, Polytarchou C, Marcu KB,
Papavassiliou AG, Sandaltzopoulos R and Kolettas E: A step-by-step
microRNA guide to cancer development and metastasis. Cell Oncol
(Dordr). 40:303–339. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shukla GC and Gupta S: Hallmarks of
cancer-focus on RNA metabolism and regulatory noncoding RNAs.
Cancer Lett. 420:208–209. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro-RNA genes miR15 and
miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci
USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ferreira AF, Moura LG, Tojal I, Ambrósio
L, Pinto-Simões B, Hamerschlak N, Calin GA, Ivan C, Covas DT,
Kashima S and Castro FA: ApoptomiRs expression modulated by BCR-ABL
is linked to CML progression and imatinib resistance. Blood Cells
Mol Dis. 53:47–55. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 4:143–159. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scholl V, Hassan R and Zalcberg IR:
miRNA-451: A putative predictor marker of Imatinib therapy response
in chronic myeloid leukemia. Leuk Res. 36:119–121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Muhammad N, Bhattacharya S, Steele R and
Ray RB: Anti-miR-203 suppresses ER-positive breast cancer growth
and stemness by targeting SOCS3. Oncotarget. 7:58595–58605. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cardona-Monzonís A, García-Giménez JL,
Mena-Mollá S, Pareja-Galeano H, de la Guía-Galipienso F, Lippi G,
Pallardó FV and Sanchis-Gomar F: Non-coding RNAs and coronary
artery disease. Adv Exp Med Biol. 1229:273–285. 2020. View Article : Google Scholar
|
|
30
|
Zhang Y, Xue W, Li X, Zhang J, Chen S,
Zhang JL, Yang L and Chen LL: The biogenesis of nascent circular
RNAs. Cell Rep. 15:611–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shan C, Zhang Y, Hao X, Gao J, Chen X and
Wang K: Biogenesis, functions and clinical significance of circRNAs
in gastric cancer. Mol Cancer. 18:1362019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mingyan L, Yujie L and Min Y: CircRNA
ZNF609 knockdown suppresses cell growth via modulating miR-188/ELF2
axis in nasopharyngeal carcinoma. Onco Targets Ther. 13:2399–2409.
2020. View Article : Google Scholar
|
|
36
|
Liang HF, Zhang XZ, Liu BG, Jia GT and Li
WL: Circular RNA circ-ABCB10 promotes breast cancer proliferation
and progression through sponging miR-1271. Am J Cancer Res.
7:1566–1576. 2017.PubMed/NCBI
|
|
37
|
Zhang SJ, Ma J, Wu JC, Hao ZZ, Zhang YA
and Zhang YJ: Circular RNA circCRIM1 suppresses lung adenocarcinoma
cell migration, invasion, EMT, and glycolysis through regulating
miR-125b-5p/BTG2 axis. Eur Rev Med Pharmacol Sci. 24:3761–3774.
2020.PubMed/NCBI
|
|
38
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in Drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42((Database Issue)): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Q, Miao S, Han X, Li C, Zhang M, Cui
K, Xiong T, Chen Z, Wang C and Xu H: MicroRNA-3619-5p suppresses
bladder carcinoma progression by directly targeting β-catenin and
CDK2 and activating p21. Cell Death Dis. 9:9602018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Fan H, Sun J, Ni M, Zhang L, Chen C,
Hong X, Fang F, Zhang W and Ma P: Circular RNA expression profile
of Alzheimer's disease and its clinical significance as biomarkers
for the disease risk and progression. Int J Biochem Cell Biol.
123:1057472020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wen G, Zhou T and Gu W: The potential of
using blood circular RNA as liquid biopsy biomarker for human
diseases. Protein Cell. Nov 1–2020.(Epub ahead of print). doi:
10.1007/s13238-020-00799-3. View Article : Google Scholar
|
|
47
|
Rahman ST, McLeod DSA, Pandeya N, Neale
RE, Bain CJ, Baade P, Youl PH and Jordan SJ: Understanding pathways
to the diagnosis of thyroid cancer: Are there ways we can reduce
over-diagnosis? Thyroid. 29:341–348. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Krajewska J, Kukulska A,
Oczko-Wojciechowska M, Kotecka-Blicharz A, Drosik-Rutowicz K,
Haras-Gil M, Jarzab B and Handkiewicz-Junak D: Early diagnosis of
low-risk papillary thyroid cancer results rather in overtreatment
than a better survival. Front Endocrinol (Lausanne). 11:5714212020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bruland O, Fluge Ø, Akslen LA, Eiken HG,
Lillehaug JR, Varhaug JE and Knappskog PM: Inverse correlation
between PDGFC expression and lymphocyte infiltration in human
papillary thyroid carcinomas. BMC Cancer. 9:4252009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Stuchi LP, Castanhole-Nunes MMU,
Maniezzo-Stuchi N, Biselli-Chicote PM, Henrique T, Padovani Neto
JA, de-Santi Neto D, Girol AP, Pavarino EC and Goloni-Bertollo EM:
VEGFA and NFE2L2 gene expression and regulation by MicroRNAs in
thyroid papillary cancer and colloid goiter. Genes (Basel).
11:9542020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lin X, Wu JF, Wang DM, Zhang J, Zhang WJ
and Xue G: The correlation and role analysis of KCNK2/4/5/15 in
human papillary thyroid carcinoma microenvironment. J Cancer.
11:5162–5176. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gugnoni M, Manicardi V, Torricelli F,
Sauta E, Bellazzi R, Manzotti G, Vitale E, de Biase D, Piana S and
Ciarrocchi A: Linc00941 is a novel transforming growth factor β
target that primes papillary thyroid cancer metastatic behavior by
regulating the expression of cadherin 6. Thyroid. 31:247–263. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Peng N, Shi L, Zhang Q, Hu Y, Wang N and
Ye H: Microarray profiling of circular RNAs in human papillary
thyroid carcinoma. PLoS One. 12:e01702872017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ren H, Liu Z, Liu S, Zhou X, Wang H, Xu J,
Wang D and Yuan G: Profile and clinical implication of circular
RNAs in human papillary thyroid carcinoma. PeerJ. 6:e53632018.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Su H, Tao T, Yang Z, Kang X, Zhang X, Kang
D, Wu S and Li C: Circular RNA cTFRC acts as the sponge of
MicroRNA-107 to promote bladder carcinoma progression. Mol Cancer.
18:272019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tang YQ, Jaganath IB, Manikam R and
Sekaran SD: Inhibition of MAPKs, Myc/Max, NFκB, and hypoxia
pathways by Phyllanthus prevents proliferation, metastasis
and angiogenesis in human melanoma (MeWo) cancer cell line. Int J
Med Sci. 11:564–577. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Karin M: Nuclear factor-kappaB in cancer
development and progression. Nature. 441:431–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Pacifico F and Leonardi A: Role of
NF-kappaB in thyroid cancer. Mol Cell Endocrinol. 321:29–35. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Beinke S and Ley SC: Functions of
NF-kappaB1 and NF-kappaB2 in immune cell biology. Biochem J.
382:393–409. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kucharczak J, Simmons MJ, Fan Y and
Gélinas C: To be, or not to be: NF-kappaB is the answer--role of
Rel/NF-kappaB in the regulation of apoptosis. Oncogene.
22:8961–8982. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Fu X, Fang J, Lian M, Zhong Q, Ma H, Feng
L, Wang R and Wang H: Identification of microRNAs associated with
medullary thyroid carcinoma by bioinformatics analyses. Mol Med
Rep. 15:4266–4272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen Z, Yan B and Van Waes C: The role of
the NF-kappaB transcriptome and proteome as biomarkers in human
head and neck squamous cell carcinomas. Biomark Med. 2:409–426.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Durairajan S, Jebaraj Walter CE, Samuel
MD, Palani D, G DJD, C GPD, Pasupati S and Johnson T: Differential
expression of NF-κB heterodimer RelA/p50 in human urothelial
carcinoma. PeerJ. 6:e55632018. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Qadir J, Riaz SK, Sahar NE, Aman D, Khan
MJ and Malik MFA: Transcriptional elucidation of tumor necrosis
factor-α-mediated nuclear factor-κB1 activation in breast cancer
cohort of Pakistan. J Cancer Res Ther. 16:1443–1448.
2020.PubMed/NCBI
|
|
66
|
Honma M, Kato N, Hashimoto M, Takahashi H,
Ishida-Yamamoto A and Iizuka H: Subungual keratoacanthoma: Analysis
of cell proliferation and copy number variation of oncogenes
compared with periungual squamous cell carcinoma. Clin Exp
Dermatol. 36:57–62. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li S, Wang C, Yu X, Wu H, Hu J, Wang S and
Ye Z: miR-3619-5p inhibits prostate cancer cell growth by
activating CDKN1A expression. Oncol Rep. 37:241–248. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang M, Luo H and Hui L: miR-3619-5p
hampers proliferation and cisplatin resistance in cutaneous
squamous-cell carcinoma via KPNA4. Biochem Biophys Res Commun.
513:419–425. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hao P, Yue F, Xian X, Ren Q, Cui H and
Wang Y: Inhibiting effect of MicroRNA-3619-5p/PSMD10 axis on liver
cancer cell growth in vivo and in vitro. Life Sci. 254:1176322020.
View Article : Google Scholar : PubMed/NCBI
|