Introduction
Head and neck squamous cell carcinoma (HNSCC)
accounts for ~6% of systemic malignancies, and there are ~600,000
new cases and 350,000 mortalities worldwide each year (1,2). The
past decade has seen an improved survival of patients with HNSCC
due to the application of new drugs and treatments; however, the
overall survival rate has remained almost unchanged (3). The main reason for treatment failure
is distant metastasis or local recurrence of the tumor (4).
In recent years, previous studies both in China and
abroad have reported that the existence of stem cells is the main
cause of tumor metastasis and recurrence (5). Stem cells are a group of cell
subpopulations that self-renewal, differentiation and passage
abilities (6). These cells have
been discovered and confirmed to be present in numerous blood
tumors and solid tumors, such as leukemia, prostate cancer, breast
cancer and head and neck tumors, amongst others (7–9). In
laboratory and clinical studies, these cells demonstrated tolerance
to traditional therapies, including surgery, radiotherapy and
chemotherapy (10). The
identification of tumor stem cell biomarkers may help to develop
individualized chemotherapy drugs for targeted treatment of tumor
stem cells.
CD44, aldehyde dehydrogenase (ALDH), Oct-4 and Nanog
have all been proved to be useful for the isolation and
identification of tumor stem cells in HNSCC (11). Chen et al (12) revealed that ALDH1+ and
CD44+ cells collected from patients with HNSCC can
resist radiation therapy and maintain tumor stem-like properties in
HNSCC cells, serving a vital role in tumor maintenance and growth.
Moreover, Prince et al (13)
reported that cells exhibiting low levels of ALDH can induce tumors
with the same morphology and heterogeneity as the primary tumor,
and cells with high expression of ALDH can be passaged in animal
models (13,14). These characteristics are consistent
with the nature of tumor stem cells, thus indicating that ALDH is a
highly selective and specific marker for HNSCC stem cells.
According to the literature, microRNA
(miRNA/miR)-26a serves a role in multiple diseases (15,16).
In animal experiments, miR-26a has been shown to inhibit the
release of inflammatory factors and the activation of the NF-κB
pathway by targeting transient receptor potential cation channel
subfamily C member 3, thereby slowing the progression of
atherosclerosis in mice (17). In
bladder cancer, miR-26a inhibits cell invasion by acting on the
high mobility group AT-hook 1 protein (18). miR-26a can also act on its direct
target gene fibroblast growth factor 9 to inhibit the proliferation
and migration of gastric cancer cells (19). Furthermore, studies have reported
that miR-26a has an inhibitory effect on HNSCC (20), while the long non-coding (lnc)RNA
non-coding RNA activated by DNA damage (NORAD) has been found to
interact with miR-26a through bioinformatics, and exert a
cancer-promoting effect in various tumors (21).
Therefore, the aim of the present study was to
investigate whether NORAD affects the proliferation, apoptosis,
migration, invasion and epithelial-mesenchymal transition (EMT) of
HNSCC cells by inhibiting the expression of miR-26a.
Materials and methods
Cells and culture
As recommended by the manufacturer, H357 cell lines
(cat. no. 06092004) purchased from The European Collection of
Authenticated Cell Cultures were cultured in a 37°C, 5%
CO2 environment in DMEM:F12 medium (cat. no. 30-2006)
containing 2 mM glutamine (cat. no. 30-2214), 10% FBS (cat. no.
30-2020; all from the American Type Culture Collection), and 0.5
µg/ml sodium hydrocortisone succinate (cat. no. 125-04-2; Pure
Chemistry Scientific, Inc.). HSC-3 cell lines (cat. no. JCRB0623)
were purchased from the Japanese Collection of Research
Bioresources Cell Bank, and cultured in Eagle's minimum essential
medium (cat. no. 30-2003) with 10% newborn calf serum (cat. no.
16010; both Gibco; Thermo Fisher Scientific, Inc.).
ALDH− cells and ALDH+ cells
from H357 and HSC-3 cell lines were sorted and identified using an
ALDEFLUOR stem cell detection kit (cat. no. 01700; Stemcell
Technologies, Inc.), according to the manufacturer's instructions,
and a flow cytometer (FACScan; BD Biosciences) with BD CellQuest
Pro software V5.1 (BD Biosciences) (22).
Self-renewal ability assay
ALDH− cells and ALDH+ cells
were cultured in common cell culture flasks in DMEM containing 10%
FBS. Cells in the logarithmic growth phase were selected, digested
and then resuspended in serum-free medium containing 10 ng/ml EGF
and 10 ng/ml basic fibroblast growth factor. Subsequently, the
cells were seeded in a low-adhesion 6-well plate at a density of
2,500 cells/ml. After 3 days of culture, 50% of the culture medium
was renewed. The self-renewal capacity of the cells was evaluated
using the method described in the study by Ghods et al
(23). The cell microspheres were
collected using a 40-µm filter, dispersed into a single-cell
suspension and inoculated into a culture plate at a density of
1,000 cells/ml. After 5–7 days of culture, the morphology of the
cell microspheres was imaged using a light microscope
(magnification, ×100). Cells that could form cell microspheres
again were considered to be self-renewing.
Cell transfection
To investigate the function of NORAD in the
development of HNSCC stem cells, NORAD overexpression and small
interfering (si)RNA (siNORAD, 5′-AATAGAATGAAGACCAACCGC-3′) vectors
were transfected into H357 and HSC-3 cells, and empty vector
negative control (NC) and siNC (5′-GCGCGATAGCGCGAATATA-3′) were
introduced as the NC. Subsequently, miR-26a-5p was identified as a
potential downstream target of NORAD using ENCORI software
(http://starbase.sysu.edu.cn/index.php). The effects of
miR-26a-5p expression on the tumor growth were measured via the
transfection of miR-26a-5p inhibitor (I,
5′-UUCUCCGAACGUGUCACGUTT-3′) (2 µg) or inhibitor control (IC,
5′-CAGUACUUUUGUGUAGUACAA-3′) (2 µg) using Lipofectamine™ 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature for 15 min. After 48 h of incubation, the subsequent
experiments were conducted. To verify the effects of the
relationship between NORAD and miR-26a-5p on the development of
HNSCC stem cells, miR-26a-5p I was applied for co-transfection with
siNORAD in H357 and HSC-3 cells. All experiments in vitro
contain six groups, including Blank, siNORAD-siNC, siNORAD, IC, I
and siNORAD+I groups.
Western blotting
Western blot analysis was conducted as previously
described (24). Total protein
extraction from HNSCC stem cells was performed before and after
transfection. Specifically, cells in the logarithmic growth phase
were chosen, and after removal of the culture medium in the cell
culture dish, the cells were washed three times with 4°C
pre-chilled PBS solution for ~5 min each time. After the PBS
solution was exhausted, RIPA lysis buffer (cat. no. C05-01001;
BIOSS) was added and incubated on ice for 30 min to fully lyse the
cells. Then, the cells were scraped off with a cell spatula, and
transferred into a 1.5-ml EP tube together with the lysate,
followed by 10 min of centrifugation at 4°C and 2,000 × g. After
centrifugation, the supernatant was collected into an EP tube and
was directly used in western blot analysis. Protein concentration
was determined using a BCA protein assay kit (Bio-Rad Laboratories,
Inc.). The specific steps of the western blotting, namely gel
preparation, electrophoresis, membrane transfer (PVDF membrane;
cat. no. IPVH00010; EMD Millipore), blocking (buffer; cat. no.
37565, Thermo Fisher Scientific, Inc.), incubation with primary
antibodies for 24 h at 4°C) and with secondary antibodies for 1 h
at room temperature, and luminescence (ECL luminous fluid; cat. no.
WBKlS0100; EMD Millipore; gel imaging system; Tanon 2500; Beijing
Solarbio Science & Technology Co., Ltd.), were performed to
obtain the results.
All antibodies in this study were purchased from
Abcam, and were as follows: CD44 (cat. no. ab157107; 81 kDa;
1:2,000); Oct-4 (cat. no. ab181557; 45 kDa; 1:1,000); Nanog (cat.
no. ab109250; 37 kDa; 1:1,000); MMP-2 (cat. no. ab97779; 74 kDa;
1:1,000); MMP-9 (cat. no. ab38898; 92 kDa; 1:1,000); E-cadherin
(cat. no. ab40772; 97 kDa: 1:10,000); N-cadherin (cat. no. ab18203;
100 kDa; 1:10,000); vimentin (cat. no. ab92547; 54 kDa; 1:1,000);
GAPDH (cat. no. ab181602; 36 kDa; 1:10,000); and Goat Anti-Rabbit
lgG H&L (HRP-conjugated; cat. no. ab205718; 1:2,000).
Lentivirus transfection
NORAD was cloned into a pCDNA3 plasmid (Invitrogen;
Thermo Fisher Scientific, Inc.). The construction of NORAD
overexpression and NC lentivirus vectors, and the packaging of
lentiviruses were all conducted by Shanghai GenePharma Co., Ltd.
The sorted ALDH+/− HNSCC cells
(1×103 cell) were seeded on a 96-well plate for 24 h of
regular culture. Then, when reaching 70% confluence,
ALDH− cells were infected with viruses (MOI, 10), and
ALDH+ cells were transfected with 10 µg siNORAD or NORAD
overexpression vector. The transfection of cells was carried out
using Lipofectamine 3000. After 48 h, subsequent experimentations
were carried out.
Cell viability assay
An MTT cell proliferation and cytotoxicity assay kit
(cat. no. C0009; Beyotime Institute of Biotechnology) was used to
determine the effects of siNORAD and miR-26a-5p I on the viability
of HNSCC stem cells. According to the experimental requirements,
HNSCC stem cells (1×104 cells/ml) were cultured at 37°C
for 24, 48 and 72 h. Subsequently, 10 µl MTT solution was added to
each well, and the incubation was continued at 37 °C for 4 h. Next,
100 µl formazan lysis solution was added to each well. After
mixing, the cells were incubated at 37°C for 3–4 h to dissolve
formazan. The absorbance was measured at 570 nm using an iMark
microplate reader (Bio-Rad Laboratories, Inc.).
Apoptosis assay
An Annexin V-FITC Apoptosis Detection kit (cat. no.
C1062M; Beyotime Institute of Biotechnology) was used to analyze
the apoptosis of HNSCC stem cells. Specifically, after 48 h of
transfection, the collected cell microspheres were digested with
trypsin and pipetted into a single cell suspension. Then, the cell
suspension was centrifuged at 1,000 × g for 5 min at room
temperature and the supernatant was discarded. Next, 195 µl Annexin
V-FITC binding solution was added to gently resuspend the cells.
Subsequently, 5 µl Annexin V-FITC and 10 µl PI staining solution
were added in sequence, and the cells were incubated at room
temperature (20–25°C) for 10–20 min in the dark, and then placed in
an ice bath. A flow cytometer (CytoFLEX V2-B4-R2; Beckman Coulter,
Inc.) and Kaluza analysis V2.0 software (Beckman Coulter, Inc.)
were used to detect the amount of apoptosis (percentage of early
apoptotic cells + percentage of late apoptotic cells) in each
group.
Wound healing assay
HNSCC stem cells of different groups were collected
separately. After digestion, the supernatant was discarded and the
cell suspension was subjected to centrifugation at 1,600 × g for 20
min at room temperature. Then, the cells were resuspended in their
respective culture solutions and subsequently pipetted into a
single cell suspension (3×105 cells/ml). Next, the cell
suspension was seeded on a 6-well cell culture plate, so that the
cells could reach the state of monolayer adherent growth and attain
nearly 100% confluence the next day. When the cells adhered to the
well, serum-free cell culture medium was added to culture the cells
overnight at 37°C. Then, the center of the 6-well plate was
scratched with a 200-µl sterile tip, with even force applied. The
floating cells were washed with PBS and cultured in a serum-free
medium. Images were captured under an inverted microscope
(magnification, ×100; CKX53; Olympus Corporation) at 0 and 48 h
after scratching to observe the healing of cell scratches and to
compare the healing rate of each group, which was calculated with
the measurement values of the scratch distances of five randomly
selected points. In total, three parallel holes were made in each
group of cells, and the experiment was repeated three times.
Transwell assay
The cell suspension was resuspended in a prepared
24-well plate invasion chamber (cat. no. 3455-024-01; Cultrex;
R&D Systems, Inc.) at a density of 5×104 cells/ml.
The upper chamber surface was coated with Matrigel (cat. no.
354234; Corning, Inc.). About 10,000 cells were seeded into the
upper chamber of the inserts supplemented with 70 µl serum-free
DMEM. Then, 750 µl DMEM containing 10% FBS was added as a chemical
attractant to the lower chamber of the 24-well plate. The cell
suspension was incubated for 48 h. At the end of the incubation,
the chamber was removed and the non-invasive cells in the upper
chamber were wiped off with a sterile cotton swab. Invasive cells
were stained with Giemsa stain (cat. no. G5637; Sigma-Aldrich;
Merck KGaA) at room temperature for 10 min, and the number of
transmembrane cells, which were randomly selected from each field,
was counted under an inverted light microscope (magnification,
×250) for each membrane.
Target gene prediction and
verification
The binding relationship between NORAD and
miR-26a-5p was predicted using the Encyclopedia of RNA Interactomes
website (http://starbase.sysu.edu.cn/).
The binding relationship between NORAD and
miR-26a-5p was verified using a dual luciferase assay according to
the following experimental steps: Luciferase reporter gene vectors
(pmirGLO; cat. no. E1330; Promega Corporation) containing NORAD
wild-type or mutant sequences were constructed. After screening
positive clones and sequencing, the recombinant luciferase reporter
gene vector was extracted, and then co-transfected with 200 ng
miR-26a-5p I or IC into HNSCC stem cells using Lipofectamine 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions, for 48 h at 37°C. After
transfection for 36 h, the dual-luciferase system (cat. no.
D0010-100T; Beijing Solarbio Science & Technology Co., Ltd.)
and a detector (GloMax 20/20; Promega Corporation) were used to
analyze the fluorescence intensity of the reporter gene. Luciferase
activity of genes was normalized to that of Renilla
luciferase.
Reverse transcription-quantitative
(RT-q)PCR
To detect the expression levels of NORAD, miR-26a-5p
and EMT-related genes, TRIzol® reagent (cat. no.
15596018; Invitrogen; Thermo Fisher Scientific, Inc.) was used to
extract mRNA and miRNA, according to the manufacturer's
instructions, and a NanoDrop system (cat. no. ND-LITE-PR; NanoDrop;
Thermo Fisher Scientific, Inc.) was used to determine RNA
concentration and purity. The RNA was then used to generate cDNA
using a reverse transcription kit (OneStep RT-PCR kit; cat. no.
210210; Qiagen GmbH) at 37°C for 15 min and 85°C for 5 sec. The
RT-qPCR experiment was performed as follows: Initial denaturation
at 95°C for 10 min; denaturation at 95°C for 15 sec and annealing
at 60°C for 1 min for a total of 40 cycles, and a final extension
step at 60°C for 1 min using the SYBR Green mixture (cat. no.
A25742; Applied Biosystems; Thermo Fisher Scientific, Inc.) and a
PCR Bio-Rad Chromo 4 system (Bio-Rad Laboratories, Inc.). The
reference gene was either GAPDH or U6. The reaction was repeated
three times in total, and data were statistically analyzed using a
modified 2−ΔΔCq method (25). The primers used were as follows
(5′-3′): NORAD, [forward (F): TGATAGGATACATCTTGGACAT GGA, reverse
(R): AACCTAATGAACAAGTCCTGACAT ACA]; MMP-2, (F:
TACAGGATCATTGGCTACACACC, R: GGTCACATCGCTCCAGACT); MMP-9, (F:
TCTATGGT CCTCGCCCTGAA, R: CATCGTCCACCGGACTCAAA); E-cadherin, (F:
CGAGAGCTACACGTTCACGG, R: GGGTG TCGAGGGAAAAATAGG); N-cadherin, (F:
TCAGGCGTCT GTAGAGGCTT, R: ATGCACATCCTTCGATAAGACTG); Vimentin, (F:
GACGCCATCAACACCGAGTT, R: CTTTGTC GTTGGTTAGCTGGT); miR-26a-5p, (F:
TTCAAGTAATCC AGGATAGGCT, R: TATCCAGTGCGTGTCGTGGA); GAPDH, (F:
AGCTCCCAAAAATAGACGCAC, R: TTCATA GCAGTAGGCACAAAGG); and U6, (F:
CTCGCTTCGGCA GCACA, R: AACGCTTCACGAATTTGCGT).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 8.0 (GraphPad Software, Inc.) and SPSS 20.0 (IBM Corp.). The
measurement data are presented as the mean ± SD of three or more
experiments, and were analyzed using an unpaired t-test. The
independent-sample t-test was used for comparison between two
groups of data, one-way ANOVA was used for analysis among multiple
groups followed by Tukey's or Dunnett's tests for pairwise
comparison between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Isolation and identification of tumor
stem cells from HNSCC cell lines
In order to study the possible mechanism in HNSCC,
ALDH− and ALDH+ cells were sorted from H357
and HSC-3 cell lines using flow cytometry (Fig. 1A). Then, the self-renewal capacity
of ALDH− and ALDH+ cells was separately
examined. It was found that ALDH+ cells could form cell
microspheres again, suggesting that ALDH+ cells had a
self-renewal capacity (Fig. 1B).
Moreover, comparisons of ALDH protein expression levels among
control, ALDH− and ALDH+ cells, were
conducted and the results indicated that expression of ALDH in
ALDH− cells was significantly lower compared with that
in control (P<0.01) and ALDH+ groups (P<0.001);
ALDH protein expression levels were highest in the ALDH+
group (Fig. 1C and D).
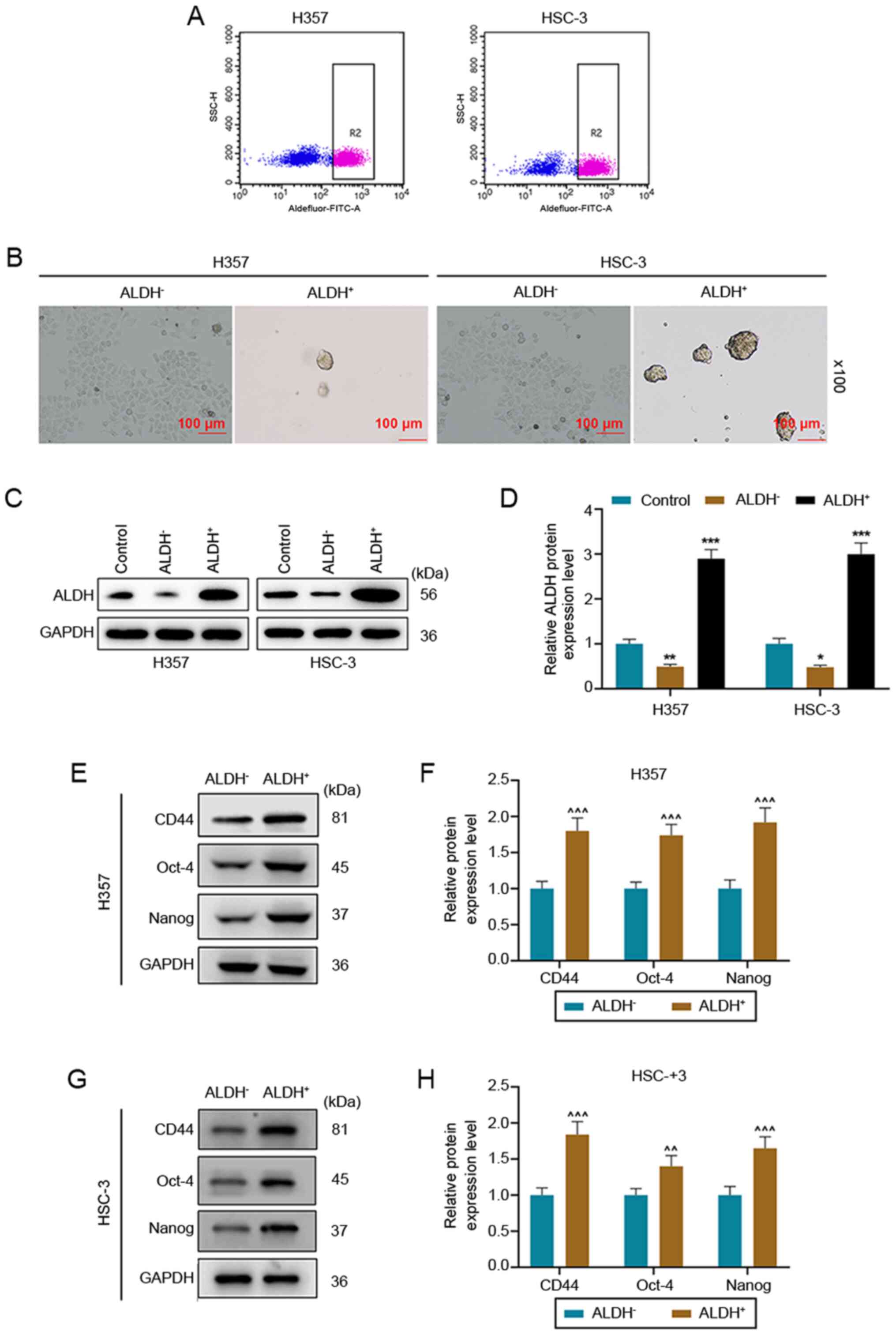 | Figure 1.Isolation and identification of tumor
stem cells from HNSCC cell lines. (A) Flow cytometry was used to
sort ALDH− and ALDH+ cells from HNSCC cell
lines H357 and HSC-3. (B) ALDH+ cells suspended in
serum-free medium formed cell microspheres (magnification, ×100).
(C) Protein expression levels of ALDH in control, ALDH−
and ALDH+ cells were detected using western blotting,
(D) and the results were semiquantified. (E) Western blotting
results of the (F) expression levels of tumor stem cell marker
genes CD44, Oct-4 and Nanog in ALDH− and
ALDH+ H357 cells. (G) Western blotting results of the
(H) expression levels of tumor stem cell marker genes CD44, Oct-4
and Nanog in ALDH− and ALDH+ HSC-3 cells.
*P<0.05, **P<0.01, ***P<0.001 vs. Control;
^^P<0.01, ^^^P<0.001 vs.
ALDH−. ALDH, aldehyde dehydrogenase; HNSCC, head and
neck squamous cell carcinoma. |
The positive expression of CD44, Oct-4 and Nanog is
considered to be a marker for tumor stem cells (11). The results demonstrated that the
expression levels of CD44, Oct-4 and Nanog were significantly
higher in ALDH+ cells compared with those in
ALDH− cells (P<0.01; Fig.
1E-H).
lncRNA NORAD affects the tumor
characteristics of HNSCC tumor stem cells
To investigate the specific role of NORAD in HNSCC
tumor stem cells, the effects of overexpression or knockdown of
NORAD on tumor stem cell marker genes in ALDH− and
ALDH+ cells were examined. The expression of NORAD was
significantly increased in ALDH− (H357 and HSC-3) cells
after transfection with overexpression plasmid (P<0.01; Fig. 2A), while the mRNA expression of
NORAD in ALDH+ cells transfected with siNORAD was
significantly decreased, compared with the siNORAD-siNC and Blank
groups (P<0.01; Fig. 2B). In
ALDH− cells, overexpression of NORAD slightly increased
the expression levels of stem cell markers, but there was no
significant statistical significance (P>0.05; Fig. 2C-F). On the contrary, knockdown of
NORAD significantly downregulated the expression levels of CD44,
Oct-4 and Nanog in ALDH+ cells (P<0.05; Fig. 2G-J). Therefore, ALDH+,
CD44+, Oct-4+ and Nanog+ cells
were selected for subsequent experiments.
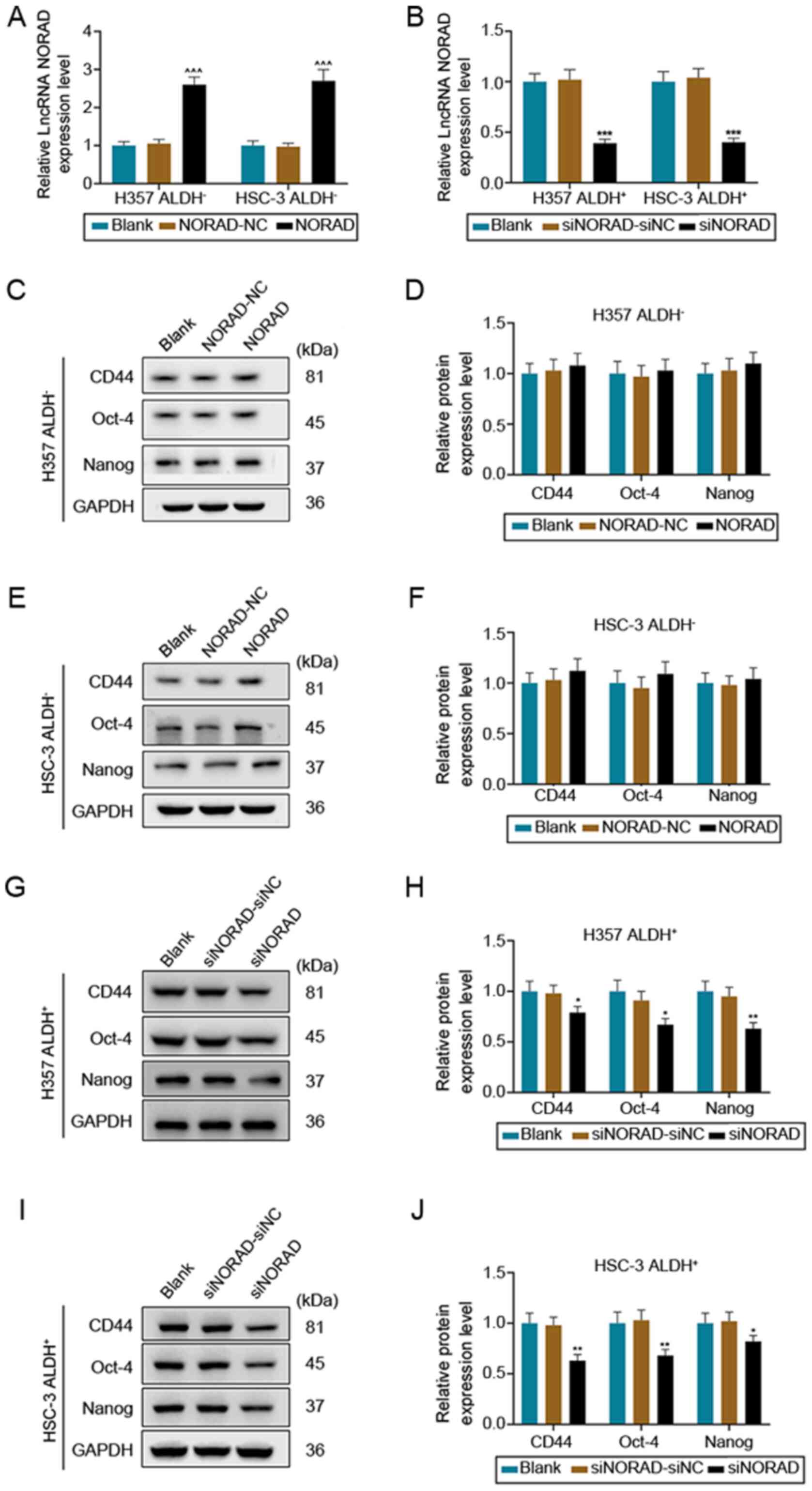 | Figure 2.Overexpression and knockdown of NORAD
affects the expression levels of stem cell markers in H357 and
HSC-3 cell lines. Transfection efficiencies of (A) overexpression
and (B) siRNA vector were measured using reverse
transcription-quantitative PCR. (C) Western blotting was used to
detect the effect of NORAD overexpression on the expression levels
of CD44, Oct-4 and Nanog in from H357 ALDH− cells. (D)
Relative protein expression levels in H357 ALDH− cells.
(E) Western blotting was used to detect the effect of NORAD
overexpression on the expression levels of CD44, Oct-4 and Nanog in
HSC-3 ALDH− cells. (F) Relative protein expression
levels in HSC-3 ALDH− cells. (G) Western blotting was
used to detect the effect of NORAD knockdown on the expression
levels of CD44, Oct-4 and Nanog in H357 ALDH+ cells. (H)
Relative protein expression levels in H357 ALDH+ cells.
(I) Western blotting was used to detect the effect of NORAD
knockdown on the expression levels of CD44, Oct-4 and Nanog in
HSC-3 ALDH+ cells. (J) Relative protein expression
levels in HSC-3 ALDH+ cells. GAPDH was used as a
control. ^^^P<0.001 vs. NORAD-NC; *P<0.05,
**P<0.01, ***P<0.001 vs. siNORAD-siNC. NC, negative control;
siRNA, small interfering RNA; NORAD, non-coding RNA activated by
DNA damage; ALDH, aldehyde dehydrogenase; lncRNA, long non-coding
RNA. |
Using functional experiments, it was identified that
NORAD knockdown significantly decreased cell viability and promoted
apoptosis (P<0.05; Fig. 3A-C).
Additionally, the migratory and invasive rates of the siNORAD group
were lower compared with those of the siNORAD-siNC group
(P<0.05; Fig. 3D-G).
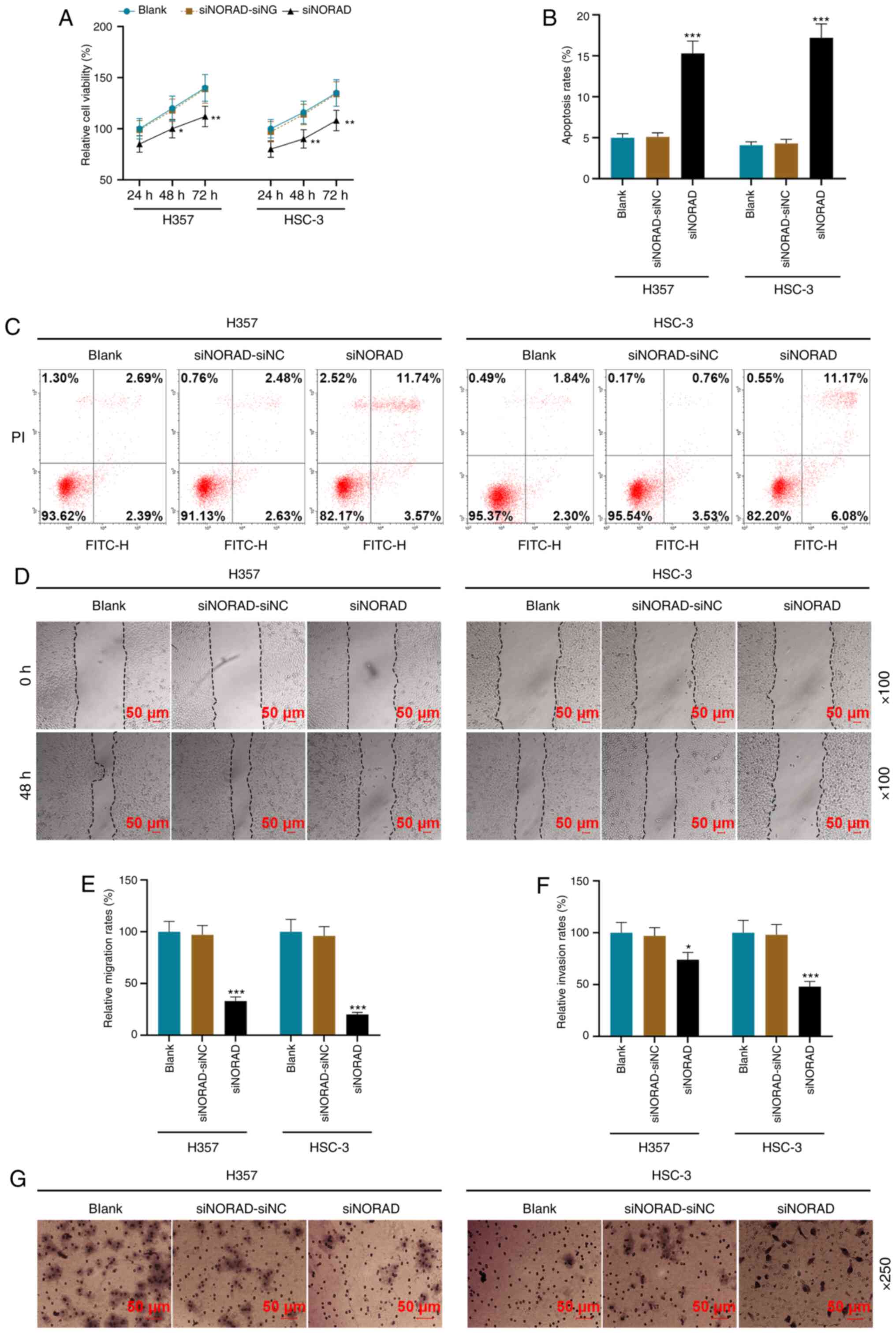 | Figure 3.NORAD knockdown affects the tumor
characteristics of HNSCC tumor stem cells. (A) siNORAD inhibited
the viability of HNSCC tumor stem cells, which was detected using a
MTT assay. (B) Results of the (C) flow cytometry, which was used to
detect the effect of lncRNA NORAD silencing on the apoptosis of
HNSCC tumor stem cells. (D) A wound healing assay was used to
measure the (E) cell migration rate of the Blank, siNORAD-NC and
siNORAD groups; magnification, ×100. (F) Cell invasion rate of each
group was measured using a (G) Transwell assay; magnification,
×250). *P<0.05, **P<0.01, ***P<0.001 vs. siNORAD-siNC.
HNSCC, head and neck squamous cell carcinoma; NC, negative control;
siRNA, small interfering RNA; NORAD, non-coding RNA activated by
DNA damage. |
lncRNA NORAD targeted to miR-26a-5p
regulates the viability and apoptosis of HNSCC tumor stem
cells
Using bioinformatics, it was identified that NORAD
was bound to miR-26a-5p (Fig. 4A).
Furthermore, compared with IC, the miR-26a-5p I increased
luciferase activity, thereby verifying the prediction that NORAD
could bind to miR-26a-5p (P<0.001; Fig. 4B).
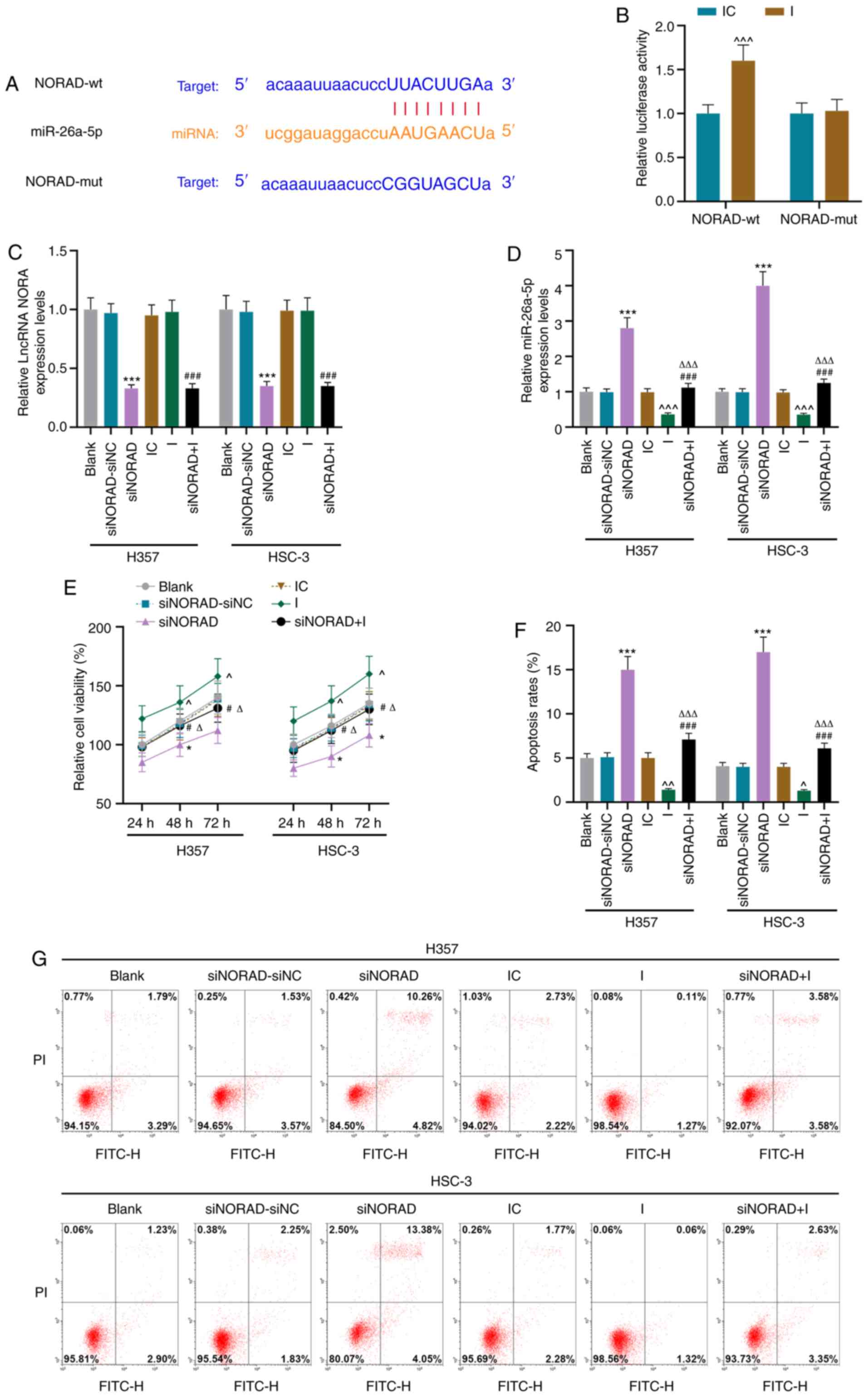 | Figure 4.lncRNA NORAD targeted to miR-26a-5p
regulates the viability and apoptosis of tumor stem cells in HNSCC.
(A) The Encyclopedia of RNA Interactomes website (http://starbase.sysu.edu.cn/) predicted, and (B)
the dual luciferase report assay verified the targeting
relationship between lncRNA NORAD and miR-26a-5p. Reverse
transcription-quantitative PCR was used to determine the expression
levels of (C) lncRNA NORAD and (D) miR-26a-5p in the Blank,
siNORAD-siNC, siNORAD, IC, I and siNORAD+I groups. (E) Viability of
tumor stem cells in each group was detected using the MTT method.
(F) miR-26a-5p partially offset the effect of siNORAD on apoptosis,
as determined via (G) flow cytometry. *P<0.05, ***P<0.001 vs.
siNORAD-siNC; ^P<0.05, ^^P<0.01,
^^^P<0.001 vs. IC; #P<0.05,
###P<0.001 vs. I; ΔP<0.05,
ΔΔΔP<0.001 vs. siNORAD. I, miR-26a-5p inhibitor; IC,
inhibitor control; NC, negative control; siRNA, small interfering
RNA; NORAD, non-coding RNA activated by DNA damage; lncRNA, long
non-coding RNA; wt, wild-type; mut, mutant; miR, microRNA. |
Subsequently, the miR-26a-5p I and siNORAD
lentiviral vectors were separately transfected or co-transfected
into HNSCC tumor stem cells. It was found that the expression of
NORAD was decreased and the expression of miR-26a-5p was increased
in the siNORAD group compared with the siNC group (P<0.001,
Fig. 4C and D). However, compared
with the siNORAD group, the siNORAD+I group displayed a significant
decrease in the expression of miR-26a-5p (P<0.001), while the
expression of NORAD did not markedly change (P>0.05) (Fig. 4C and D). Furthermore, the results of
functional experiments demonstrated that the knockdown of
miR-26a-5p partially counteracted the effect of siNORAD, as it
promoted cell viability and reduced the apoptotic rates (P<0.05;
Fig. 4E-G).
lncRNA NORAD knockdown attenuates the
migration, invasion and expression levels of EMT-related proteins
in HNSCC tumor stem cells via miR-26a-5p
Previous studies have reported that silencing NORAD
inhibits cell migration (26–28),
but whether it serves a role by regulating the downstream gene
miR-26a-5p has not yet been completely determined. Thus, wound
healing assay and Transwell were performed. The results
demonstrated that miR-26a-5p I promoted migration and increased the
number of invading cells. Moreover, the regulation of the migratory
and invasive rates of the siNORAD+I group was reversed compared
with the siNORAD group (P<0.05; Fig.
5A-F). Compared with the siNORAD-siNC group, the siNORAD group
had lower expression levels of MMP-2, MMP-9, N-cadherin and
vimentin, and higher expression of E-cadherin, and the regulation
trend of EMT-related genes in group I was opposite to that in the
siNORAD group (P<0.05; Fig.
6A-F). Furthermore, the miR-26a-5p I partially offset the
regulatory function of siNORAD (P<0.05; Fig. 6A-F).
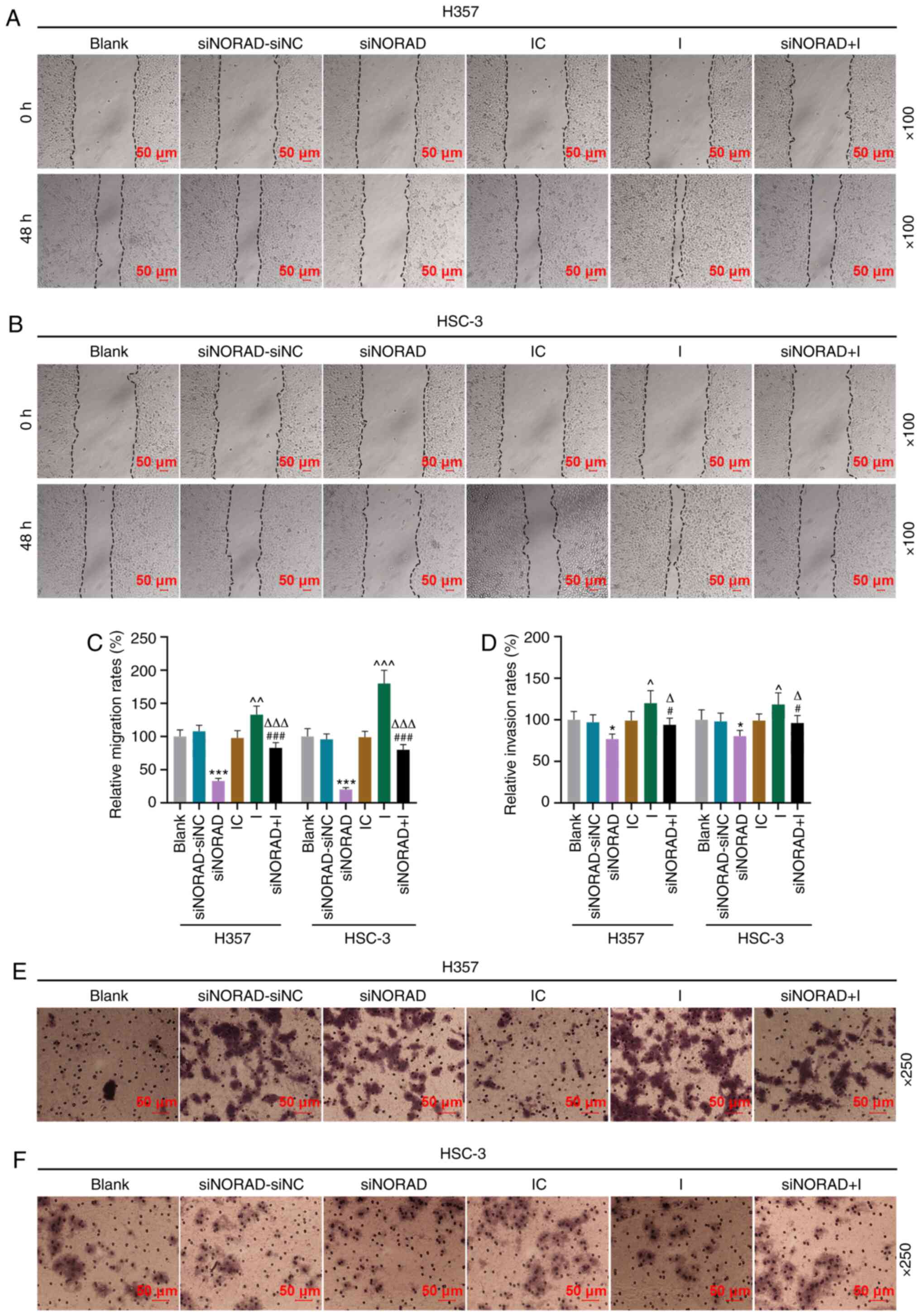 | Figure 5.Long non-coding RNA NORAD knockdown
decreases the migration and invasion of tumor stem cells via
miR-26a-5p in HNSCC. Wound healing assay results from (A) H357 and
(B) HSC-3 cells indicating the migration distances of the Blank,
siNORAD-siNC, siNORAD, IC, I and siNORAD+I groups; magnification,
×100. (C) Relative migration rates are presented as bar diagrams.
(D) Number of invasive tumor stem cells in each group was
determined using Transwell assays in (E) H357 and (F) HSC-3 cells;
magnification, ×250). *P<0.05, ***P<0.001 vs. siNORAD-siNC;
^P<0.05, ^^P<0.01,
^^^P<0.001 vs. IC; #P<0.05,
###P<0.001 vs. I; ΔP<0.05,
ΔΔΔP<0.001 vs. siNORAD. I, microRNA-26a-5p inhibitor;
IC, inhibitor control; NC, negative control; siRNA, small
interfering RNA; NORAD, non-coding RNA activated by DNA damage. |
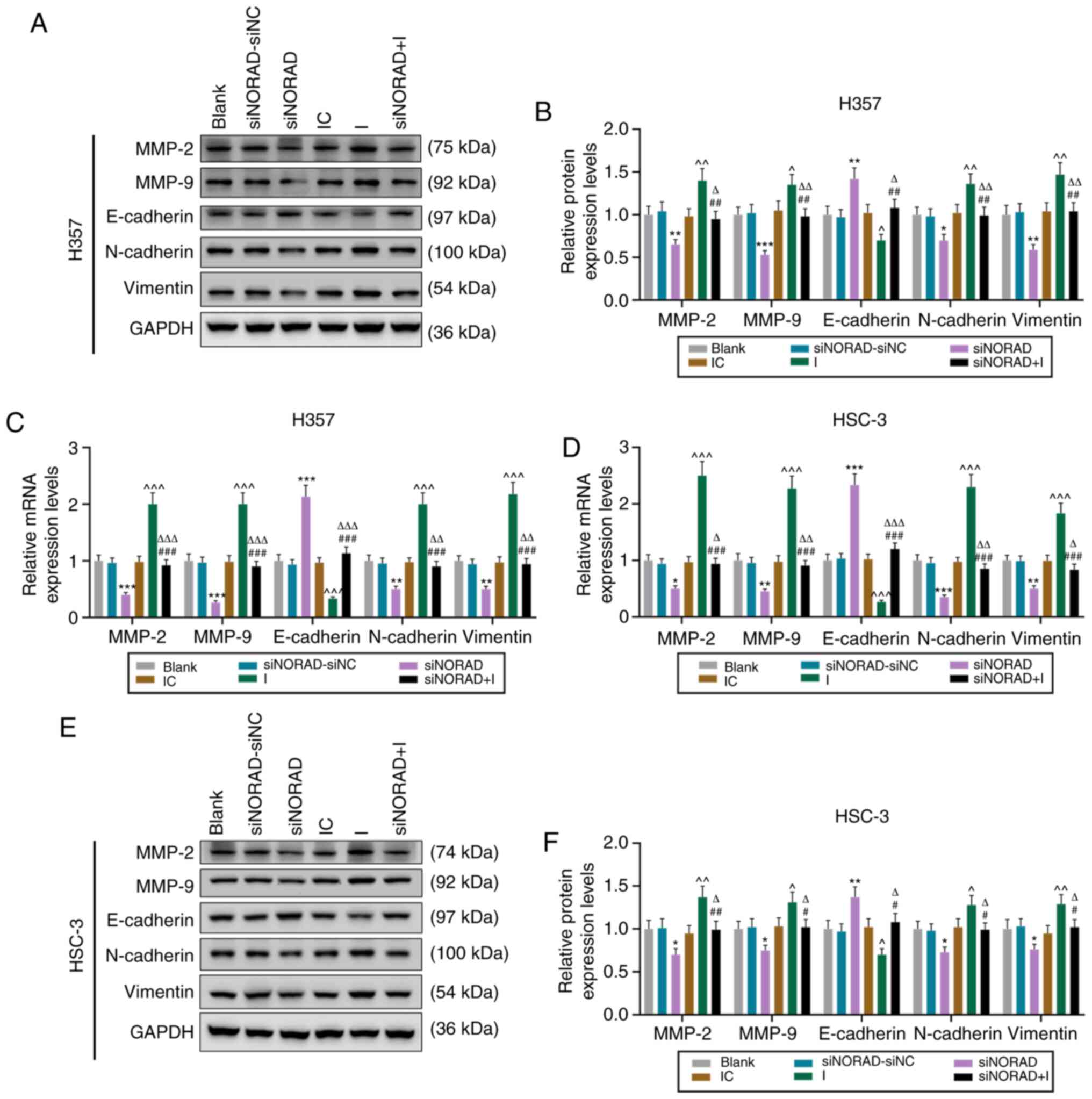 | Figure 6.lncRNA NORAD knockdown attenuates the
expression of EMT-related proteins in HNSCC tumor stem cells via
miR-26a-5p. (A) Western blot analysis was used to detect the
expression of MMP-2, MMP-9, E-cadherin, N-cadherin and Vimentin,
and (B) relative protein expression is presented as bar diagrams in
H357 cells. Reverse transcription-quantitative PCR was used to
determine the mRNA expression of MMP-2, MMP-9, E-cadherin,
N-cadherin and Vimentin in (C) H357 and (D) HSC-3 cells. (E)
Western blot analysis was used to detect the expression of MMP-2,
MMP-9, E-cadherin, N-cadherin and Vimentin, and (F) relative
protein expression is presented as bar diagrams in HSC-3 cells.
*P<0.05, **P<0.01, ***P<0.001 vs. siNORAD-siNC;
^P<0.05, ^^P<0.01,
^^^P<0.001 vs. IC; #P<0.05,
##P<0.01, ###P<0.001 vs. I;
ΔP<0.05, ΔΔP<0.01,
ΔΔΔP<0.001 vs. siNORAD. I, microRNA-26a-5p inhibitor;
IC, inhibitor control; NC, negative control; siRNA, small
interfering RNA; NORAD, non-coding RNA activated by DNA damage. |
Discussion
Cancer stem cells have been reported to be closely
associated with tumor metastasis and poor prognosis (5,10). The
existence of tumor stem cells in HNSCC has also been confirmed in
the present research. The present results suggested that the HNSCC
cell line can be sorted into ALDH+ cells, and that
ALDH+ cells can form cell microspheres again, suggesting
that there are tumor stem cells with self-renewal and pluripotent
differentiation in HNSCC cells. Although ALDH+ is
considered to be a highly specific HNSCC stem cell marker (29), whether ALDH alone is sufficient for
the recognition of HNSCC tumor stem cells remains to be further
studied. Therefore, the present study used a variety of stem cell
molecular surface markers, such as CD44, Oct-4 and Nanog, to make
an identification, which is also conducive to obtaining HNSCC tumor
stem cells of higher purity (24).
With the development and application of microarray
chips and other technologies, the number of expression profiles of
HNSCC-related non-coding RNAs is increasing (30). Abnormally expressed miRNA can not
only be used for early diagnosis of HNSCC, but also have certain
guiding significance for tumor metastasis and prognostic
monitoring, which may provide novel ideas for targeted therapy
(31). For example, overexpression
of miR-96-5p leads to increased cell migration and radiation
resistance and chemotherapy resistance in HNSCC cells by targeting
PTEN to activate the PI3K/Akt/mTOR signaling pathway (32). Furthermore, Liu et al
(33) identified 128 abnormally
expressed miRNAs in HNSCC tissues, and suggested that hsa-miR-383,
hsa-miR-615 and hsa-miR-877 may be used as diagnostic markers.
As for miR-26a, which was the primary focus of the
present study, research on this miRNA has achieved certain results;
for example, it has been suggested that miR-26a may have a role as
a biomarker for small extracellular vesicles and as a modulator of
excitatory neurotransmission (34,35).
However, with regards to HNSCC, especially on the regulation of
stem cell stemness and epithelial-mesenchymal function, there is
still a lack of relevant research. The latest report revealed that
miR-26a overexpression inhibited cell migration via PAK1 and
suppressed tumor formation in tongue squamous cell carcinoma
(36). The present study
demonstrated that miR-26a-5p silencing promoted cell viability,
invasion and migration, which is in line with previous results
(37). Unlike the previous study,
however, the present study found that the upstream target gene of
miR-26a-5p may be lncRNA NORAD, according to the prediction of
bioinformatics, suggesting that NORAD mediates the stemness of
HNSCC stem cells via miR-26a-5p.
A previous study revealed that NORAD was abnormally
expressed in gastric cancer samples, and miR-214 interfered with
the proliferation of gastric cancer cells (38). In the current study, as expected,
knockdown of NORAD significantly downregulated the expression
levels of stem cell markers, and siRNA-targeted NORAD had a
significant regulatory effect on the biological characteristics of
HNSCC tumor stem cells. Furthermore, the miR-26a-5p I counteracted
the effect of siNORAD on HNSCC tumor stem cells.
EMT is an important cause of tumor metastasis, and
increasing evidence has shown that EMT and stem cells are also
closely related (39). May et
al (40) reported that when EMT
occurs in human breast epithelial cells, there will be the
subsequent high expression of stem cell-related factors. Moreover,
a study of patients with pancreatic cancer found that the
occurrence of EMT is often accompanied by the activation of stem
cell-related channels (41). A
previous study has also reported that, compared with primary tumor
cells, the expression levels of EMT-related markers in stem cells
are significantly increased. Furthermore, these markers are
downregulated in epithelial cells but have increased expression in
mesenchymal cells (42). EMT not
only promotes the metastasis and invasion of tumor cells, but also
affects the drug resistance and enrichment of stem cells (43). The present results also suggested
that NORAD regulated the expression levels of EMT-related proteins
in stem cells via miR-26a-5p.
However, there is a limitation to the present study.
Although it was demonstrated that the miR-26a-5p I could notably
promote the migration and invasion of HNSCC stem cells, the effects
of miR-26a-5p overexpression on the EMT progression of cancer cells
require further investigation.
In conclusion, the present study demonstrated that
NORAD interfered with the malignant phenotype and EMT of HNSCC stem
cells by inhibiting miR-26a-5p. The present research provides a
novel insight into the molecular regulatory network of HNSCC stem
cells, but these findings should be further clarified in in
vivo experiments and in additional downstream pathway
studies.
Acknowledgements
Not applicable.
Funding
This work was supported by the Zhejiang Provincial
Department of Medicine and Health Project (grant no. 2018247374)
and the Zhejiang Traditional Chinese Medicine Science and
Technology Plan Project (grant no. 201823105013221).
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
Substantial contributions to conception and design:
WH and YZ. Data acquisition, data analysis and interpretation: LS,
ZW, WJ, XJ and ML. Drafting the article or critically revising it
for important intellectual content: WH and YZ. All authors agreed
to be accountable for all aspects of the work in ensuring that
questions related to the accuracy or integrity of the work are
appropriately investigated and resolved. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Svider PF, Blasco MA, Raza SN, Shkoukani
M, Sukari A, Yoo GH, Folbe AJ, Lin HS and Fribley AM: Head and Neck
Cancer. Otolaryngol Head Neck Surg. 156:10–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mendez LC, Moraes FY, Poon I and Marta GN:
The management of head and neck tumors with high technology
radiation therapy. Expert Rev Anticancer Ther. 16:99–110. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Solomon B, Young RJ and Rischin D: Head
and neck squamous cell carcinoma: Genomics and emerging biomarkers
for immunomodulatory cancer treatments. Semin Cancer Biol.
52:228–240. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mermod M, Tolstonog G, Simon C and Monnier
Y: Extracapsular spread in head and neck squamous cell carcinoma: A
systematic review and meta-analysis. Oral Oncol. 62:60–71. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang JC: Cancer stem cells: Role in tumor
growth, recurrence, metastasis, and treatment resistance. Medicine
(Baltimore). 95 (Suppl 1):S20–S25. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nandy SB and Lakshmanaswamy R: Cancer Stem
Cells and Metastasis. Prog Mol Biol Transl Sci. 151:137–176. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou H and Xu R: Leukemia stem cells: The
root of chronic myeloid leukemia. Protein Cell. 6:403–412. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Leão R, Domingos C, Figueiredo A, Hamilton
R, Tabori U and Castelo-Branco P: Cancer Stem Cells in Prostate
Cancer: Implications for Targeted Therapy. Urol Int. 99:125–136.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Curtarelli RB, Gonçalves JM, Dos Santos
LGP, Savi MG, Nör JE, Mezzomo LAM and Rodríguez Cordeiro MM:
Expression of Cancer Stem Cell Biomarkers in Human Head and Neck
Carcinomas: A Systematic Review. Stem Cell Rev Rep. 14:769–784.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dawood S, Austin L and Cristofanilli M:
Cancer stem cells: Implications for cancer therapy. Oncology
(Williston Park). 28:1101–1107, 1110. 2014.PubMed/NCBI
|
|
11
|
Peitzsch C, Nathansen J, Schniewind SI,
Schwarz F and Dubrovska A: Cancer Stem Cells in Head and Neck
Squamous Cell Carcinoma: Identification, Characterization and
Clinical Implications. Cancers (Basel). 11:E6162019. View Article : Google Scholar
|
|
12
|
Chen YC, Chen YW, Hsu HS, Tseng LM, Huang
PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al: Aldehyde
dehydrogenase 1 is a putative marker for cancer stem cells in head
and neck squamous cancer. Biochem Biophys Res Commun. 385:307–313.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prince MEP, Zhou L, Moyer JS, Tao H, Lu L,
Owen J, Etigen M, Zheng F, Chang AE, Xia J, et al: Evaluation of
the immunogenicity of ALDH(high) human head and neck squamous cell
carcinoma cancer stem cells in vitro. Oral Oncol. 59:30–42. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ailles L and Prince M: Cancer stem cells
in head and neck squamous cell carcinoma. Methods Mol Biol.
568:175–193. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, Liang Y, Lv H, Meng H, Xiong G, Guan
X, Chen X, Bai Y and Wang K: miR-26a and miR-26b inhibit esophageal
squamous cancer cell proliferation through suppression of c-MYC
pathway. Gene. 625:1–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cabello P, Pineda B, Tormo E, Lluch A and
Eroles P: The Antitumor Effect of Metformin Is Mediated by miR-26a
in Breast Cancer. Int J Mol Sci. 17:E12982016. View Article : Google Scholar
|
|
17
|
Feng M, Xu D and Wang L: miR-26a inhibits
atherosclerosis progression by targeting TRPC3. Cell Biosci.
8:42018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y,
Xu X, Wu J, Li S, Mao Q, et al: miR-26a inhibits proliferation and
motility in bladder cancer by targeting HMGA1. FEBS Lett.
587:2467–2473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deng M, Tang HL, Lu XH, Liu MY, Lu XM, Gu
YX, Liu JF and He ZM: miR-26a suppresses tumor growth and
metastasis by targeting FGF9 in gastric cancer. PLoS One.
8:e726622013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fukumoto I, Kikkawa N, Matsushita R, Kato
M, Kurozumi A, Nishikawa R, Goto Y, Koshizuka K, Hanazawa T,
Enokida H, et al: Tumor-suppressive microRNAs (miR-26a/b,
miR-29a/b/c and miR-218) concertedly suppressed
metastasis-promoting LOXL2 in head and neck squamous cell
carcinoma. J Hum Genet. 61:109–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XM, Wang J, Liu ZL, Liu H, Cheng YF
and Wang T: LINC00657/miR-26a-5p/CKS2 ceRNA network promotes the
growth of esophageal cancer cells via the MDM2/p53/Bcl2/Bax
pathway. Biosci Rep. 40:BSR202005252020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng Y, Wen G, Sun Y, Shen Y, Zeng Y, Du
M, Zhu G, Wang G and Meng X: Osteopontin Promotes Colorectal Cancer
Cell Invasion and the Stem Cell-Like Properties through the
PI3K-AKT-GSK/3β-β/Catenin Pathway. Med Sci Monit. 25:3014–3025.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ghods AJ, Irvin D, Liu G, Yuan X,
Abdulkadir IR, Tunici P, Konda B, Wachsmann-Hogiu S, Black KL and
Yu JS: Spheres isolated from 9L gliosarcoma rat cell line possess
chemoresistant and aggressive cancer stem-like cells. Stem Cells.
25:1645–1653. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kuo SZ, Honda CO, Li WT, Honda TK, Kim E,
Altuna X, Abhold E, Wang-Rodriguez J and Ongkeko WM: Metformin
Results in Diametrically Opposed Effects by Targeting Non-Stem
Cancer Cells but Protecting Cancer Stem Cells in Head and Neck
Squamous Cell Carcinoma. Int J Mol Sci. 20:E1932019. View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang H and Guo H: Long non-coding RNA
NORAD induces cell proliferation and migration in prostate cancer.
J Int Med Res. 47:3898–3904. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu SY, Peng H, Zhu Q, Wu YX, Wu F, Han CR,
Yan B, Li Q and Xiang HG: Silencing the long noncoding RNA NORAD
inhibits gastric cancer cell proliferation and invasion by the
RhoA/ROCK1 pathway. Eur Rev Med Pharmacol Sci. 23:3760–3770.
2019.PubMed/NCBI
|
|
28
|
Huo H, Tian J, Wang R, Li Y, Qu C and Wang
N: Long non-coding RNA NORAD upregulate SIP1 expression to promote
cell proliferation and invasion in cervical cancer. Biomed
Pharmacother. 106:1454–1460. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Toledo-Guzmán ME, Hernández MI,
Gómez-Gallegos ÁA and Ortiz-Sánchez E: ALDH as a Stem Cell Marker
in Solid Tumors. Curr Stem Cell Res Ther. 14:375–388. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sannigrahi MK, Sharma R, Panda NK and
Khullar M: Role of non-coding RNAs in head and neck squamous cell
carcinoma: A narrative review. Oral Dis. 24:1417–1427. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zou AE, Zheng H, Saad MA, Rahimy M, Ku J,
Kuo SZ, Honda TK, Wang-Rodriguez J, Xuan Y, Korrapati A, et al: The
non-coding landscape of head and neck squamous cell carcinoma.
Oncotarget. 7:51211–51222. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vahabi M, Pulito C, Sacconi A, Donzelli S,
D'Andrea M, Manciocco V, Pellini R, Paci P, Sanguineti G, Strigari
L, et al: miR-96-5p targets PTEN expression affecting
radio-chemosensitivity of HNSCC cells. J Exp Clin Cancer Res.
38:1412019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu C, Yu Z, Huang S, Zhao Q, Sun Z,
Fletcher C, Jiang Y and Zhang D: Combined identification of three
miRNAs in serum as effective diagnostic biomarkers for HNSCC.
EBioMedicine. 50:135–143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tormo E, Adam-Artigues A, Ballester S,
Pineda B, Zazo S, González-Alonso P, Albanell J, Rovira A, Rojo F,
Lluch A, et al: The role of miR-26a and miR-30b in HER2+
breast cancer trastuzumab resistance and regulation of the CCNE2
gene. Sci Rep. 7:413092017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lafourcade CA, Fernández A, Ramírez JP,
Corvalán K, Carrasco MÁ, Iturriaga A, Bátiz LF, Luarte A and
Wyneken U: A Role for mir-26a in Stress: A Potential sEV Biomarker
and Modulator of Excitatory Neurotransmission. Cells. 9:E13642020.
View Article : Google Scholar
|
|
36
|
Wei Z, Chang K, Fan C and Zhang Y:
miR-26a/miR-26b represses tongue squamous cell carcinoma
progression by targeting PAK1. Cancer Cell Int. 20:822020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Wang P, Wu LL, Yan J, Pang XY and
Liu SJ: miR-26a-5p Inhibit Gastric Cancer Cell Proliferation and
Invasion Through Mediated Wnt5a. OncoTargets Ther. 13:2537–2550.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tao W, Li Y, Zhu M, Li C and Li P: lncRNA
NORAD Promotes Proliferation And Inhibits Apoptosis Of Gastric
Cancer By Regulating miR-214/Akt/mTOR Axis. OncoTargets Ther.
12:8841–8851. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y and Weinberg RA:
Epithelial-to-mesenchymal transition in cancer: Complexity and
opportunities. Front Med. 12:361–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
May CD, Sphyris N, Evans KW, Werden SJ,
Guo W and Mani SA: Epithelial-mesenchymal transition and cancer
stem cells: A dangerously dynamic duo in breast cancer progression.
Breast Cancer Res. 13:2022011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhou P, Li B, Liu F, Zhang M, Wang Q, Liu
Y, Yao Y and Li D: The epithelial to mesenchymal transition (EMT)
and cancer stem cells: Implication for treatment resistance in
pancreatic cancer. Mol Cancer. 16:522017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Khan MI, Czarnecka AM, Lewicki S,
Helbrecht I, Brodaczewska K, Koch I, Zdanowski R, Król M and
Szczylik C: Comparative Gene Expression Profiling of Primary and
Metastatic Renal Cell Carcinoma Stem Cell-Like Cancer Cells. PLoS
One. 11:e01657182016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|




















