Introduction
Diabetes is a chronic disease involving metabolic
disorders of sugar, protein and fat that is primarily caused by
insufficient insulin in the body. β cells are endocrine cells that
secrete insulin. Insulin secretion by islet β cells is mainly
influenced by blood glucose levels (1,2). A
sharp decrease in the number of islet β cells results in
insufficient insulin secretion by cells, which in turn induces
metabolic disorders of proteins, glucose and other substances.
Among others, amino acids, hormones and hyperglycemia are all
inducers of the abnormal function of islet β cells, among which
hyperglycemia is the most important one (3,4). It is
of great significance to investigate the underlying mechanism of
damaged islet cells in a high glucose (HG) environment.
MicroRNAs (miRNAs/miRs) are an important regulator
of numerous physiological and pathophysiological processes, and
serve a key role in a number of biological processes, such as cell
proliferation, differentiation, apoptosis and carcinogenesis
(5). A previous study demonstrated
that miRNAs promote insulin secretion and regulate insulin
resistance by acting on multiple pathways, and abnormal miRNA
expression may be the underlying pathogenesis of diabetes (6). It has been reported that miR-532-5p
expression is downregulated in the plasma of patients with type 2
diabetes (7). miR-532-5p expression
is downregulated in H9C2 cells exposed to hypoxia, and in the
myocardium of rats with acute myocardial infarction, reducing the
apoptosis of H9C2 cells induced by hypoxia (8). However, to the best of our knowledge,
the specific role of miR-532-5p in diabetes has not been
reported.
By querying the StarBase website, it was identified
that miR-532-5p could target cyclin D1 (CCND1). CCND1, a member of
the cyclin family, is a regulator of cyclin-dependent kinase. CCND1
expression has been reported to be upregulated in diabetic islets
(9). Shen and Zhu (10) analyzed gene expression in type 2
diabetes and identified 124 upregulated differentially expressed
genes, including CCND1, in the GSE15653 dataset, and these were
associated with fatty acid and glucose metabolic pathways, and
oxidation/reduction reactions, and may be involved in the
development of obesity-related type 2 diabetes. In addition, in
multiple myeloma, CCND1 can control the redox metabolism by
producing reactive oxygen species (ROS) to disrupt redox balance
(11). Furthermore, CCND1 silencing
can damage the repair of DNA double-strand breaks, induce
G0/G1 phase cell cycle arrest and inhibit
ovarian cancer cell proliferation (12).
The apoptotic activation gene p53 induces cell cycle
stagnation at the G0 stage, as well as apoptosis. A
previous study demonstrated that p53 serves an important role in
the initiation of apoptosis under different physiological
conditions (13). Following
treatment with HG, the expression levels of p53 in INS-1 cells
increased in a concentration-dependent manner, and overexpression
of p53 induces apoptosis and reduces insulin secretion (14). Adaptive EGF expression sensitizes
pancreatic cancer cells to ionizing radiation via activation of the
CCND1/p53/poly(ADP-ribose) polymerase (PARP) signaling pathway
(15). Therefore, it was speculated
that miR-532-5p may regulate oxidative stress and insulin secretion
damage of pancreatic β cells induced by HG by regulating
CCND1/p53.
The present study examined the regulatory effects of
miR-532-5p on diabetes and explored the underlying mechanisms by
inducing islet cells with HG. The present study provided a
theoretical basis for the investigation of the underlying
mechanisms of diabetes and potential drug targets.
Materials and methods
Cell lines and culture conditions
The β cell line of the pancreas (MIN6) was obtained
from The Cell Bank of Type Culture Collection of the Chinese
Academy of Sciences and incubated in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C with 5% CO2. In the present
experiment, the normal glucose control (5 mM glucose, NG group) was
set as a control group, the mannitol group (MA) was set up to
exclude the osmotic pressure effects of HG on cells. The MIN6 cells
were incubated in complete medium containing 25 mM glucose (final
concentration in the medium) for 24 h, which was called the HG
group (16). The HG + mimic-NC
group was transfected with a mimic-NC and then induced with 25 mM
glucose. The HG + miR-532-5p mimic group was transfected with the
miR-532-5p mimic and then induced with 25 mM glucose. The HG +
miR-532-5p mimic + pcDNA-NC group were transfected with miR-532-5p
mimic and the pcDNA-NC plasmid, and were then induced with 25 mM
glucose. The HG + miR-532-5p mimic + pcDNA-CCND1 group was
transfected with miR-532-5p mimic and the pcDNA-CCND plasmid, and
were then induced with 25 mM glucose.
Database selection and analysis
StarBase (http://starbase.sysu.edu.cn/) was used to identify the
binding sites of miR-532-5p and CCND1.
MTT assay
Cells were seeded at a density of 1×104
cells/well in 96-well plates. Following treatment of the cells, 20
µl MTT solution (5 mg/ml; Gen-view Scientific, Inc.) was added to
each well and cells were incubated at 37°C with 5% CO2
for 4 h. Subsequently, 100 µl DMSO was added to dissolve the
formazan crystals at 37°C for 10 min. The optical density at 490 nm
was measured the following day to determine the quantities of
formazan formed by cleaving of MTT in living cells.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and then reverse transcribed into cDNA using the First-Strand
cDNA Synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.).
The SuperScript™ III Platinum™ SYBR Green One-Step qRT-PCR kit
(Invitrogen; Thermo Fisher Scientific, Inc.) was used according to
the manufacturer's protocols as follows: 95°C for 10 min, 40 cycles
of 95°C for 10 sec, 55°C for 10 sec and 72°C for 30 sec. Relative
expression levels were calculated according to the
2−ΔΔCq method (17). The
following primers were used: miR-532-5p forward,
5′-GCGCGCATGCCTTGAGTGTAG-3′ and reverse,
5′-ATCCAGTGCAGGGTCCGAGG-3′; CCND1 forward,
5′-ACGAAGGTCTGCGCGTGTT-3′ and reverse, 5′-CCGCTGGCCATGAACTACCT-3′;
p53 forward, 5′-GGCCCACTTCACCGTACTAA-3′ and reverse,
5′-GTGGTTTCAAGGCCAGATGT-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; Insulin1 forward,
5′-TAGTGACCAGCTATAATCAGAG-3′ and reverse,
5′-ACGCCAAGGTCTGAAGGTCC-3′; Insulin2 forward,
5′-CCCTGCTGGCCCTGCTCTT3-3′ and reverse, 5′-AGGTCTGAAGGTCACCTGCT-3′;
and GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse,
5′-TCCACCACCCTGTTGCTGTA-3′. Insulin1 and Insulin2 were detected as
transcripts of the insulin gene, which are markers of pancreatic β
cells (18). GAPDH was used as the
housekeeping gene for CCND1, Insulin1, Insulin2 and p53. U6 was
used as the housekeeping gene for miR-532-5p.
Cell transfection
Cells (1×105 cells/well) were seeded into
6-well plates and cultured for 24 h at 37°C with 5% CO2.
Cell transfection was performed when the cells reached 80%
confluency. The miR-532-5p mimic and mimic-negative control
(mimic-NC; Invitrogen; Thermo Fisher Scientific, Inc.) were
transfected directly into cells. For overexpression of miR-532-5p,
the cells were transfected with mimic at a final concentration of
25 nM for 48 h at 37°C. For pcDNA-NC (empty vector, Invitrogen;
Thermo Fisher Scientific, Inc.) and pcDNA-CCND1, full length
transcript of CCND1 was amplified from cDNA obtained from 293T
cells by PCR using PrimeSTAR® HS DNA polymerase (Takara
Bio, Inc.), and was transfected at a final concentration of 500 ng
for 48 h at 37°C. The PCR amplification product was inserted into
the KpnI and BamHI sites of the pcDNA vector
(Invitrogen; Thermo Fisher Scientific, Inc.), which was termed
pcDNA-CCND1. Transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Transfection efficiency was determined by RT-qPCR 48 h after
transfection.
ELISA
Cytokine concentration normalized to total insulin
was detected using ELISA kits (ELISA MAX™ Deluxe Set Human IGFALS;
cat. no. 445904 BioLegend, Inc.) (16). After the cells were centrifuged (300
× g, 10 min, 37°C), cell-free supernatant was diluted to 100 µl
(1:20) and incubated with the specific capture antibody and
detection antibody. Measurements were made at an optical density of
450 nm.
ROS assay
ROS levels of cells were detected using the
fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate
(Sigma-Aldrich; Merck KGaA), which could be rapidly oxidized into
the fluorescent 2′,7′-dichlorofluorescein (DCF) in the presence of
intracellular ROS. Fluorescence was monitored with a laser scanning
confocal microscope (Leica Microsystems GmbH) at 488 nm
(magnification, ×200). The amount of ROS was quantified as the
relative fluorescence intensity of DCF per cell in the scanned
area.
TUNEL assay
A total of 3×104 cells were seeded in
24-well plates and incubated overnight. The cells were treated
after fusion. Cells were fixed with 4% paraformaldehyde for 30 min
at room temperature and then washed with PBS. Subsequently, 0.3%
Triton X-100 in PBS was added and incubated for 5 min. The cells
were collected and washed with PBS three times, and treated with 50
µl TUNEL assay solution (Roche Diagnostics GmbH) at 37°C in the
dark for 60 min, followed by the addition of stop solution.
Subsequently, cells were incubated with DAB solution and stained
with hematoxylin and eosin for 5 min at room temperature according
to the manufacturer's protocol. Stained apoptotic cells were
visualized at ×20 magnification under an LSM 710 laser scanning
confocal microscope (Carl Zeiss AG).
Western blotting
Cells were collected, lysed with RIPA lysis buffer
(Beyotime Institute of Biotechnology) and incubated for 30 min on
ice. Subsequently, proteins were detected using a BCA protein assay
kit (Bio-Rad Laboratories, Inc.). A total of 40 µg protein was
loaded onto 10% SDS-polyacrylamide gels to separate proteins, which
were subsequently transferred to PVDF membranes. The membranes were
blocked with 10% skimmed milk for 2 h at room temperature, followed
by incubation overnight at 4°C with the following primary
antibodies: Anti-Bax (1:1,000; cat. no. 14796S; Cell Signaling
Technology, Inc.), anti-caspase-3 (1:1,000; cat. no. 700182; Thermo
Fisher Scientific, Inc.), anti-cleaved caspase-3 (1:1,000; cat. no.
PA5-17913; Thermo Fisher Scientific, Inc.), anti-Bcl-2 (1:1,000;
cat. no. 15071S; Cell Signaling Technology, Inc.), anti-CCND1
(1:1,000; cat. no. MA5-14512; Thermo Fisher Scientific, Inc.),
anti-P53 (1:1,000; cat. no. MA5-12557; Thermo Fisher Scientific,
Inc.) and anti-GAPDH (1:1,000; cat. no. 5174S; Cell Signaling
Technology, Inc.). Subsequently, the membranes were incubated with
goat anti-rabbit horseradish peroxidase-conjugated IgG secondary
antibodies (1:5,000; cat. nos. A32731 and A11032; Thermo Fisher
Scientific, Inc.) at room temperature for 1 h. The signals were
detected using enhanced chemiluminescence reagent (GE Healthcare),
and ImageJ software (version 146; National Institutes of Health)
was used to analyze the fold changes of protein levels.
Luciferase reporter assays
To validate the direct targeting of miR-532-5p, the
3′ untranslated region (3′UTR) of the putative target gene CCND1
was cloned into the psiCHECK2 vector (Promega Corporation),
according to the manufacturer's instructions. The mutated or
wild-type CCND1 cells were divided into mimic-NC + CCND1 group and
miR-532-5p mimic + CCND1 group. Vectors containing the respective
3′UTRs were co-transfected with miRNA mimic
(5′-CAUGCCUUGAGUGUAGGACCGU-3′) into cells (1×106 cells)
at a final concentration of 500 ng for 48 h at 37°C using
Lipofectamine 2000 according to the manufacturer's protocol.
Mutations in each of the predicted target sites in CCND1 3′UTRs
were generated by site-directed mutagenesis using the QuikChange II
Site-Directed Mutagenesis kit (Agilent Technologies, Inc.)
according to the manufacturer's protocols. Subsequently, the cells
were washed with PBS and lysed with cell lysis buffer (Beyotime
Institute of Biotechnology) after transfection. The luciferase
activity was measured using a plate reader (BD Biosciences) and was
normalized to Renilla luciferase activity (pRL-TK) using the
Luc-Screen™ Extended-Glow Luciferase Reporter Gene Assay system
(cat. no. T1033; Thermo Fisher Scientific, Inc.). All procedures
were conducted according to the manufacturers' instructions.
Luciferase gene plasmid (containing WT and Mut UTRs) was
constructed by Thermo Fisher Scientific, Inc.
Statistical analysis
Data are presented as the mean ± standard deviation.
Each experiment was repeated three times. SPSS version 19.0
software (IBM Corp.) was used to perform statistical analysis.
Comparisons among multiple groups were analyzed using one-way ANOVA
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Overexpression of miR-532-5p improves
the impaired functions of secreted insulin in HG-induced cells
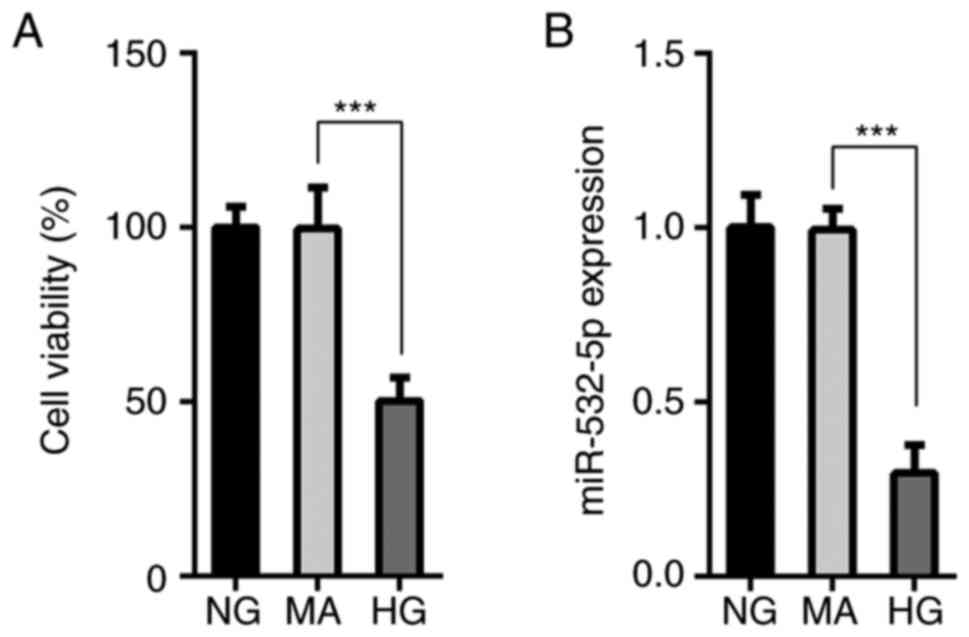
The MTT assay results demonstrated that compared
with that of cells in the NG and MA groups, the survival rate of
cells in the HG group was decreased (Fig. 1A), and this was accompanied by a
decrease in the expression levels of miR-532-5p (Fig. 1B). This suggested that miR-532-5p
served an important role in HG-induced cells. Subsequently,
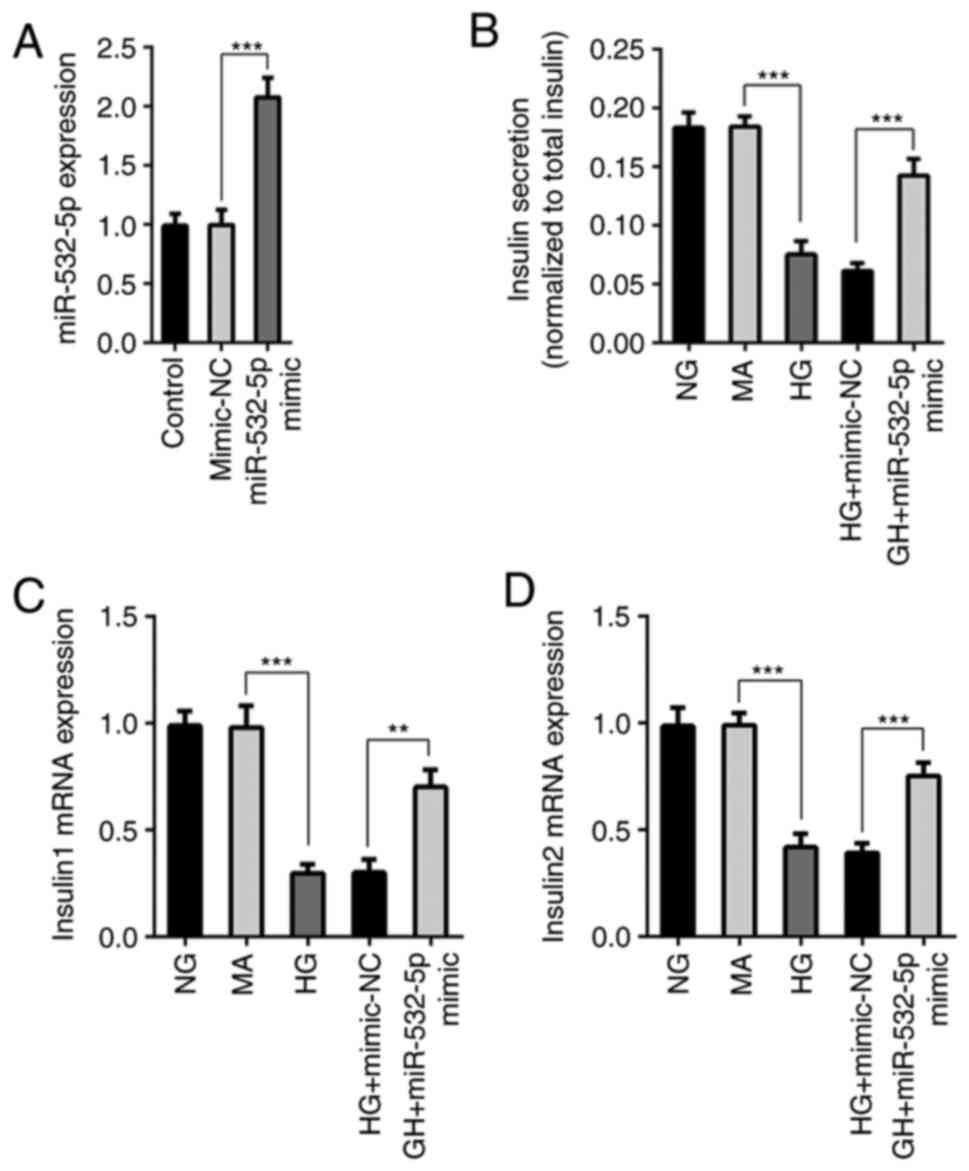
miR-532-5p was overexpressed using the cell transfection technique,
and transfection efficiency was measured by RT-qPCR (Fig. 2A). Furthermore, cells were divided
into NG, MA, HG, HG + mimic-NC and HG + miR-532-5p mimic groups.
ELISA was used to detect the functions of secreted insulin.
Compared with those in the NG and MA groups, the insulin secretion
levels in the HG group were decreased. Compared with those of cells
in the HG + mimic-NC group, the insulin secretion levels of cells
in the HG + miR-532-5p mimic group were increased (Fig. 2B). In addition, RT-qPCR was used to
detect the gene transcription levels of Insulin1 and Insulin2, and
the trend was consistent with that of the total insulin level
(Fig. 2C and D). The results
demonstrated that overexpression of miR-532-5p improved the
impaired functions of secreted insulin in HG-induced cells.
Overexpression of miR-532-5p inhibits
oxidative stress levels in HG-induced cells
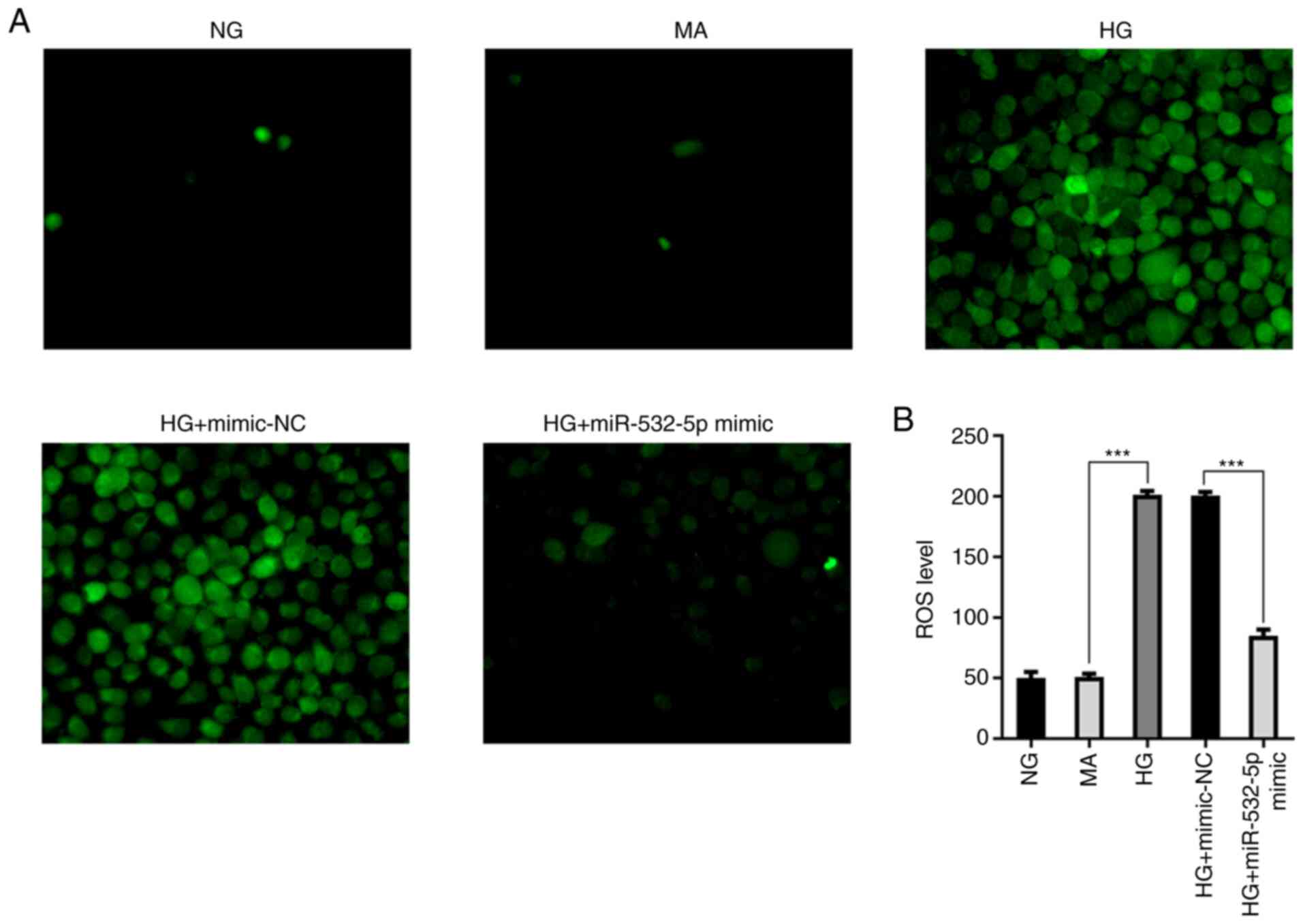
Subsequently, the levels of ROS were detected.
Compared with those in the NG and MA groups, the ROS levels in the
HG group were increased. Additionally, following overexpression of
miR-532-5p, the ROS levels were decreased compared with the HG +
mimic-NC group (Fig. 3A and B).
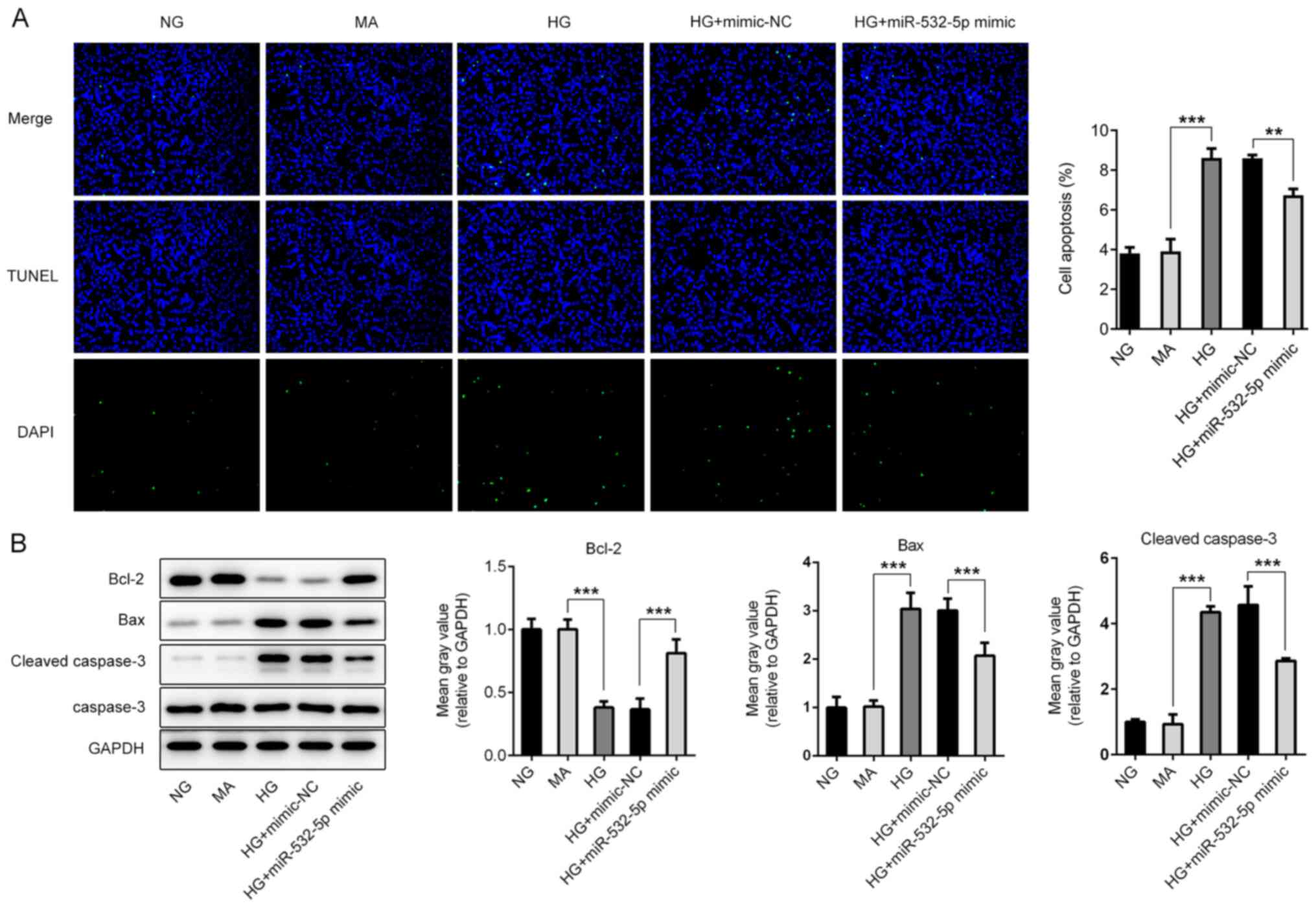
TUNEL staining was subsequently used to detect apoptosis. Compared
with the NG and MA groups, the HG group exhibited an increased
apoptosis rate (Fig. 4A), and this
was accompanied by increased expression levels of Bax and cleaved
caspase-3, and decreased expression levels of Bcl-2 (Fig. 4B). Additionally, compared with that
of cells in the HG + mimic-NC group, the apoptosis rate of cells in
the HG + miR-532-5p mimic group exhibited a significant decline,
and this was accompanied by decreased expression levels of Bax and
cleaved caspase-3, and increased expression levels of Bcl-2. The
results revealed that overexpression of miR-532-5p could inhibit
oxidative stress levels in HG-induced cells.
Overexpression of miR-532-5p
downregulates CCND1 expression in HG-induced cells
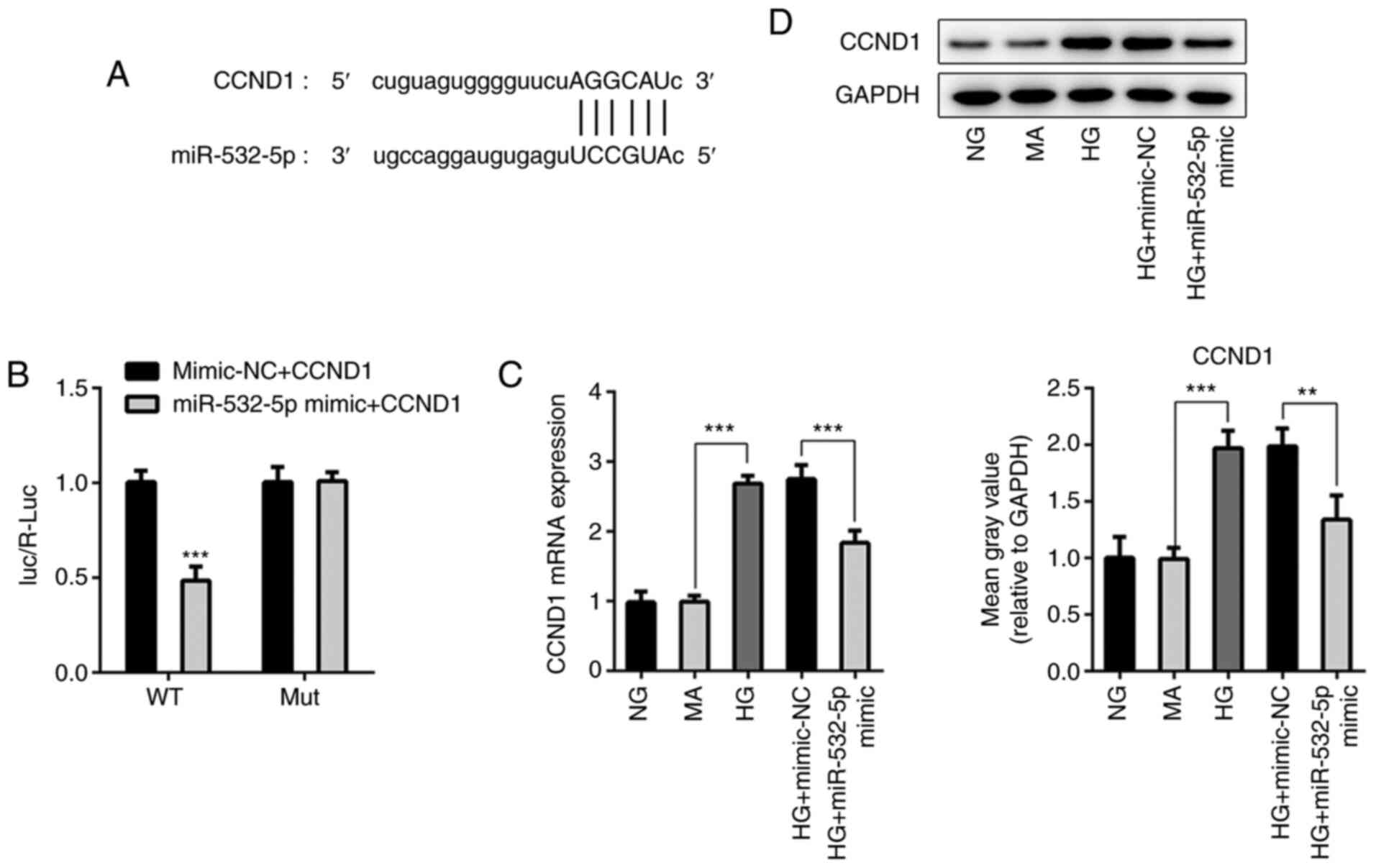
StarBase was used to identify that miR-532-5p could
target the 3′UTR of CCND1 (Fig.
5A). Additionally, a luciferase reporter assay was used to
verify the targeted binding of miR-532-5p and CCND1. The results
demonstrated that before CCND1 mutation, luciferase activity was
significantly decreased in the miR-532-5p mimic + CCND1 group
compared with that in the Vector + CCND1 group. After CCND1
mutation, the luciferase activity was not significantly altered
between the Vector + CCND1 and miR-532-5p mimic + CCND1 groups
(Fig. 5B). Expression levels of
CCND1 in cells were detected following overexpression of
miR-532-5p. As shown in Fig. 5C and
D, compared with those in the NG and MA groups, the expression
levels of CCND1 in the HG group were significantly increased.
Compared with those in the HG + mimic-NC group, the expression
levels of CCND1 in the HG + miR-532-5p mimic group were
decreased.
Overexpression of miR-532-5p improves
the impaired functions of secreted insulin and inhibits oxidative
stress levels in HG-induced cells by downregulating CCND1
expression
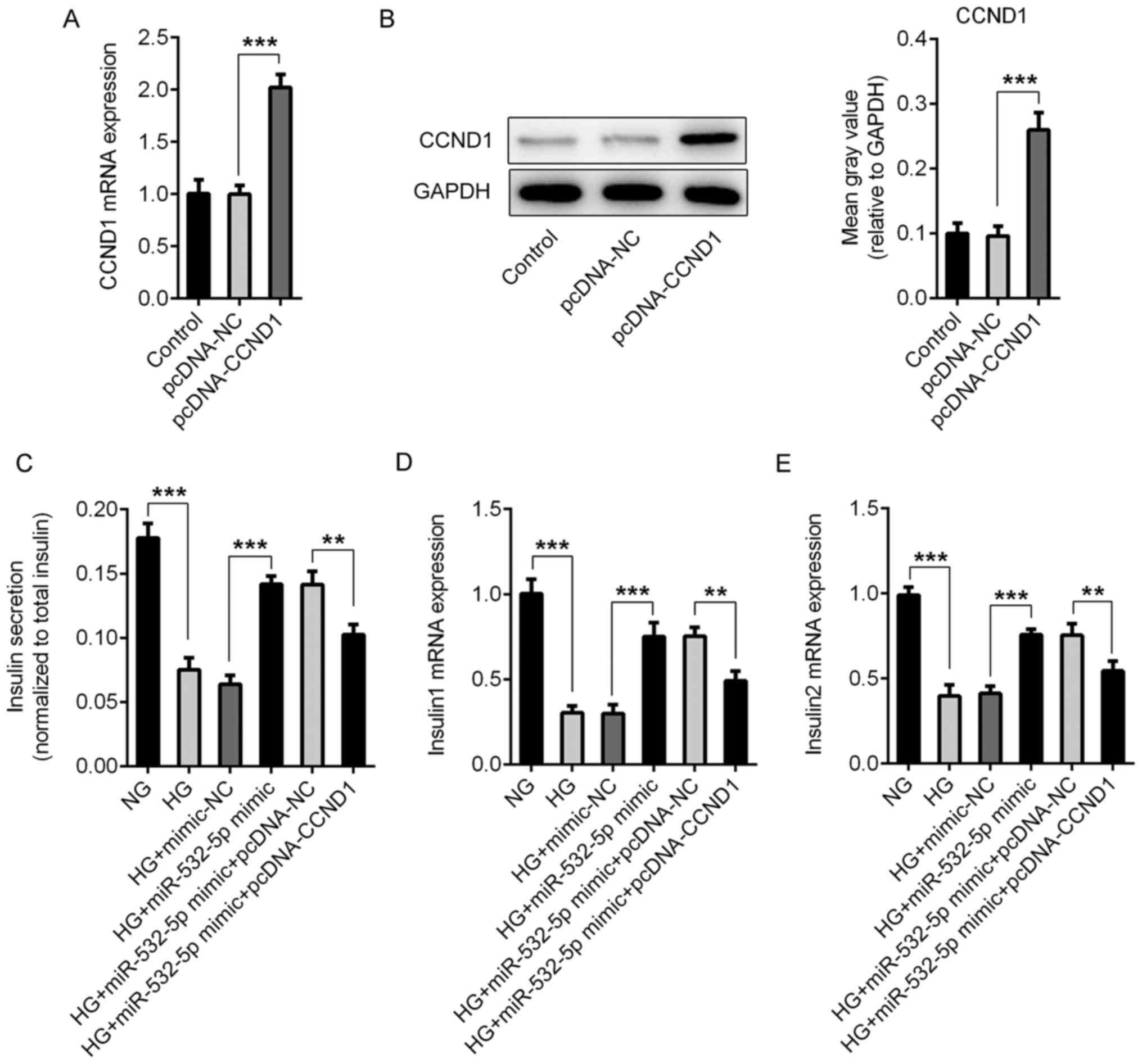
Compared with those in the pcDNA-NC group, the
expression levels of CCND1 in the pcDNA-CCND1 group were
significantly increased, indicating successful overexpression
(Fig. 6A and B). Subsequently, the
cells were divided into NG, HG, HG + mimic-NC, HG + miR-532-5p
mimic, HG + miR-532-5p mimic + pcDNA-NC and HG + miR-532-5p mimic +
PCDNA-CCND1 groups. The secretion of insulin by the cells was
detected using ELISA. Compared with those in the HG + miR-532-5p
mimic + pcDNA-NC group, the total insulin levels in the HG +
miR-532-5p mimic + pcDNA-CCND1 group were decreased (Fig. 6C), as were the levels of Insulin1
(Fig. 6D) and Insulin2 (Fig. 6E). Subsequently, ROS levels were
detected, and the trend was the opposite of that detected for
insulin (Fig. 7A and B). Apoptosis
was detected using a TUNEL assay. Compared with that in the HG +
miR-532-5p mimic + pcDNA-NC group, apoptosis in the HG + miR-532-5p
mimic + pcDNA-CCND1 group increased (Fig. 7C and D), and this was accompanied by
increased expression levels of Bax and cleaved caspase-3, and
decreased expression levels of Bcl-2 (Fig. 7E). These results indicated that
overexpression of miR-532-5p improved the impaired functions of
secreted insulin and inhibited oxidative stress levels in
HG-induced cells by downregulating CCND1 expression.
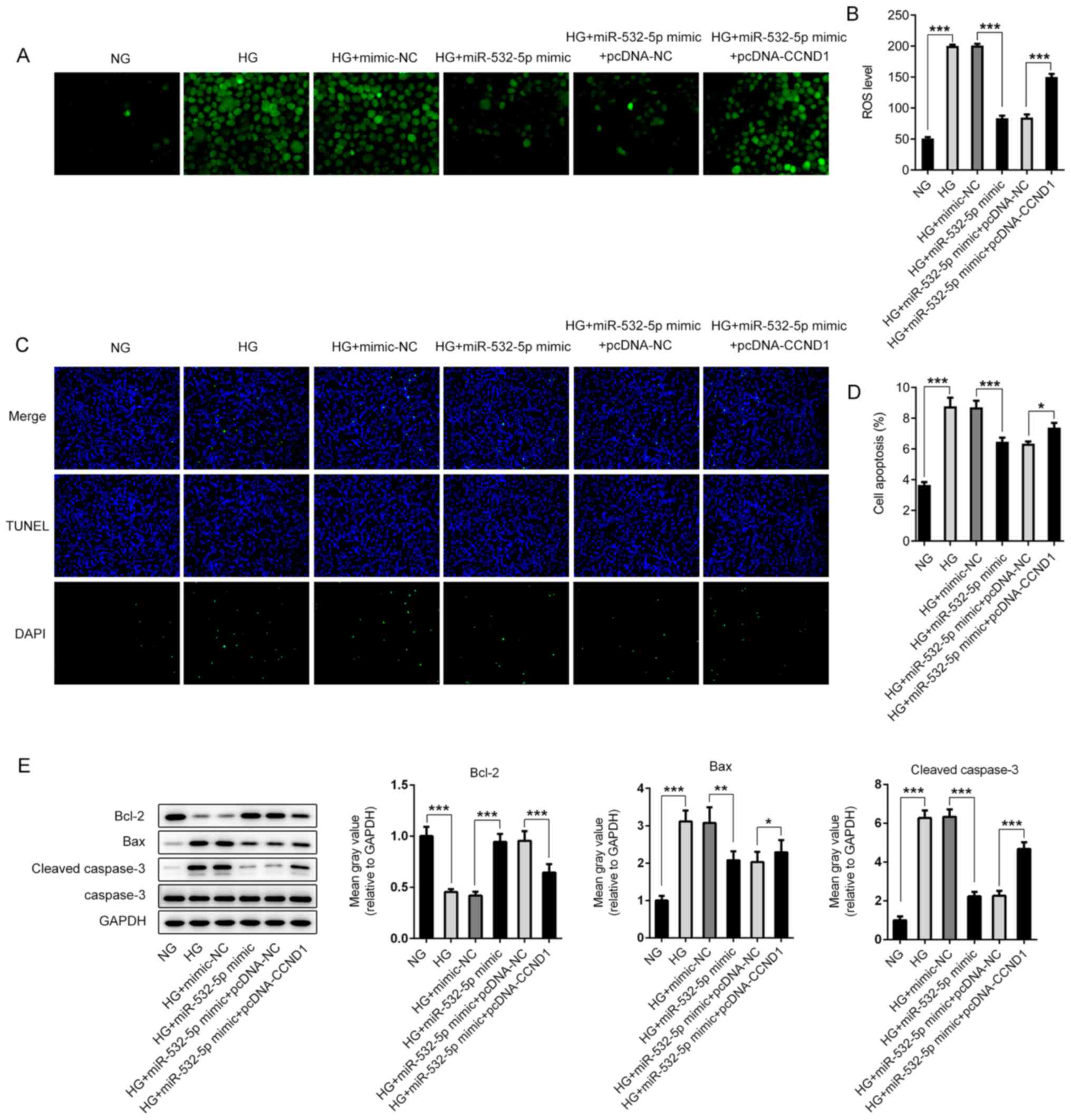 | Figure 7.Overexpression of miR-532-5p inhibits
oxidative stress levels in HG-induced cells by downregulating
CCND1. (A) 2′,7′-dichlorodihydrofluorescein diacetate fluorescence
probe was used to detect ROS expression. (B) Quantification of ROS
expression. (C) TUNEL assay was performed to detect the apoptosis
rate of cells. (D) Quantification of apoptosis rates. (E) The
expression levels of apoptosis-related proteins were determined
using western blotting. *P<0.05, **P<0.01, ***P<0.001. HG,
high glucose; miR, microRNA; MA, mannitol group; NG, normal
glucose; NC, negative control; CCND1, cyclin D1; ROS, reactive
oxygen species. |
Overexpression of miR-532-5p regulates
the expression levels of p53 in HG-induced cells by downregulating
CCND1 expression
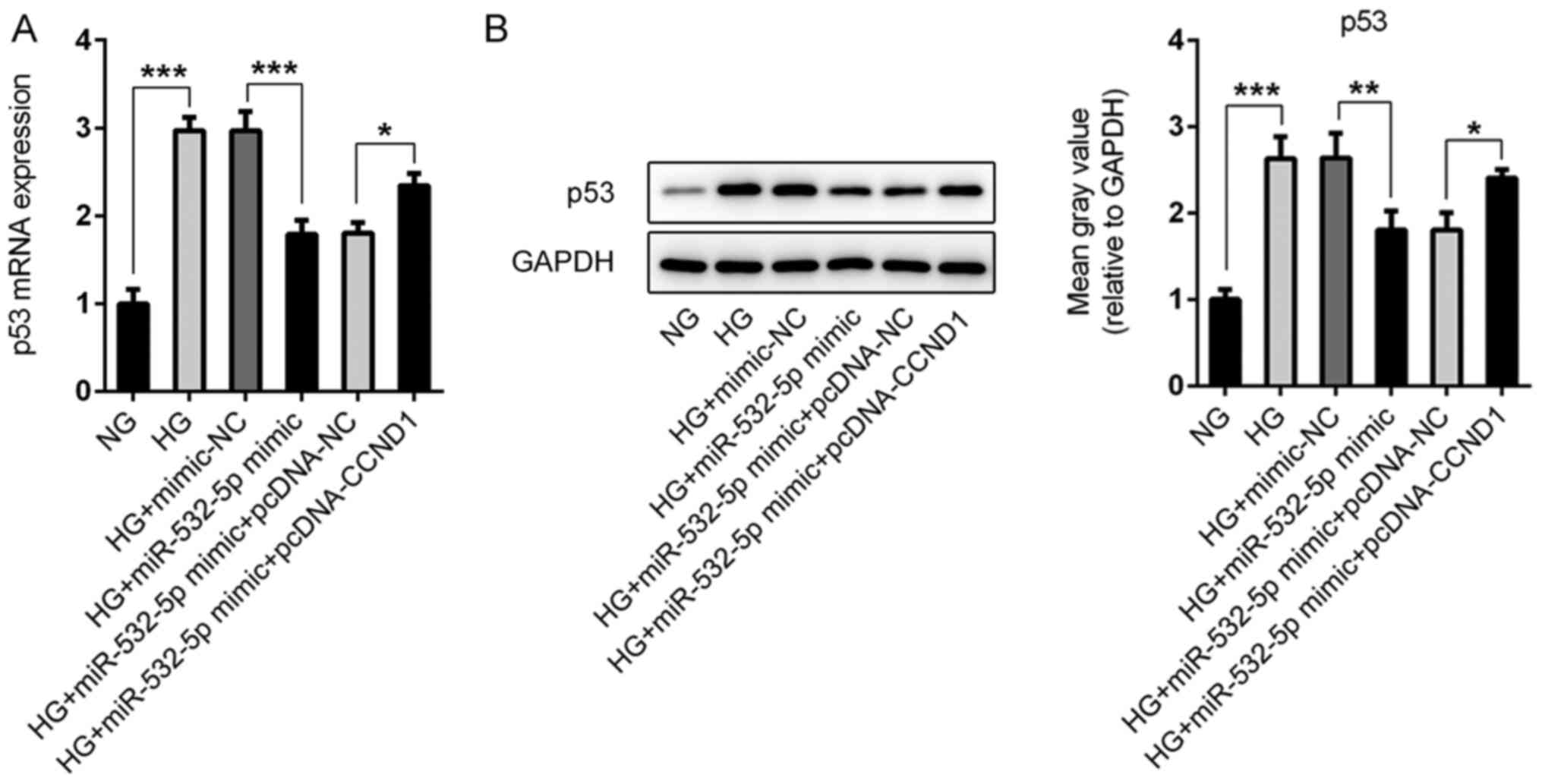
During the study, abnormal expression levels of p53
were observed in the cells (Fig. 8A and
B). Compared with those in the NG group, the expression levels
of p53 in the cells were increased following HG induction, whereas
the expression levels of p53 in the cells were decreased following
overexpression of miR-532-5p. Compared with that in the HG +
miR-532-5p mimic + pcDNA-NC group, p53 expression in the HG +
miR-532-5p mimic + pcDNA-CCND1 group was increased. Therefore, it
was preliminarily concluded that overexpression of miR-532-5p
regulated the expression levels of p53 in HG-induced cells by
downregulating CCND1 expression.
Discussion
miRNAs have been considered as potential biomarkers
of tissue-specific origin, which affect the occurrence and
development of diabetes by participating in processes such as
collective oxidative stress and the inflammatory response (19). miR-21-5p expression in extracellular
vesicles is increased during inflammatory responses and serves as a
biomarker for type 1 diabetes (20). Overexpression of miR-22 can reduce
oxidative stress injury in diabetic cardiomyopathy via sirtuin 1
(21). miR-365 promotes diabetic
retinopathy by inhibiting TIMP metallopeptidase inhibitor 3 and
increasing oxidative stress (22).
It has been reported that miR-532-5p expression is downregulated in
patients with type 2 diabetes (6).
In the present study, the expression levels of miR-532-5p were also
markedly decreased in HG-induced MIN6 cells.
At present, studies on miR-532-5p are concerned with
its role in other diseases. miR-532-5p expression is downregulated
in H9C2 cells exposed to hypoxia, and in the myocardium of rats
with acute myocardial infarction, to reduce the apoptosis of H9C2
cells (8). In addition, miR-532-5p
expression is downregulated in LPS-induced inflammation of RAW264.7
macrophages (23). It has been
revealed that miR-532-5p serves an important role in the
inflammatory response (24),
oxidative stress (25) and
apoptosis (8). However, to the best
of our knowledge, the role of miR-532-5p in diabetes has not yet
been reported. In the present study, it was revealed that
overexpression of miR-532-5p could increase the secretion of
insulin in HG-induced cells. Furthermore, miR-532-5p could inhibit
the levels of oxidative stress in cells, and could inhibit the
apoptosis of HG-induced cells.
The targeted binding of miR-532-5p to CCND1 was
identified using the StarBase website, and this targeting
relationship was further verified in the present study using a
luciferase reporter assay. Studies have reported that the
expression of CCND1 in diabetes is upregulated (9,26).
However, to the best of our knowledge, the specific role of CCND1
in diabetes has not yet been reported. In the present study, it was
revealed that the expression levels of CCND1 were upregulated in
HG-induced cells, and overexpression of miR-532-5p inhibited CCND1
expression, suggesting that miR-532-5p targeting CCND1 served a
regulatory role in diabetes. It has been reported that inhibition
of CCND1 expression can block the cell cycle and inhibit cell
proliferation (27). Furthermore,
in multiple myeloma, CCND1 can control redox metabolism by
producing ROS to disrupt the redox balance (11). In the present study, upregulation of
miR-532-5p was proposed to regulate insulin secretion, oxidative
stress and apoptosis in HG-induced cells by downregulating CCND1
expression. The experimental step of knocking down the expression
of CCND1 will be further explored in our future research.
During the experiment, it was revealed that
overexpression of miR-532-5p regulated the expression levels of p53
in HG-induced MIN6 cells by downregulating CCND1 expression. A
previous study reported that adaptive EGF expression sensitized
pancreatic cancer cells to ionizing radiation by activating the
CCND1/p53/PARP signaling pathway (15). Following glucose treatment, the
expression levels of p53 increase in a concentration-dependent
manner, and the upregulation of p53 induces apoptosis and reduces
insulin secretion (14). Therefore,
it was preliminarily concluded that miR-532-5p regulated oxidative
stress and insulin secretion of pancreatic β cells induced by HG by
downregulating CCND1 expression, which may be regulated via the
modulation of p53 expression levels. Further experimental
verification of this preliminary conclusion will be carried out in
subsequent research.
Due to the length of the article, only in
vitro experiments were performed and described in the present
study. In addition, our research group is conducting the relevant
in vivo experiments to further verify the conclusion
obtained from these in vitro experiments.
In conclusion, the present study revealed that
miR-532-5p regulated oxidative stress and insulin secretion in
HG-induced pancreatic β cells by downregulating the expression
levels of CCND1, which was involved in the regulation of p53
expression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ and WS made substantial contributions to the
conception and design of the study, and the acquisition of data. HC
made substantial contributions to analysis and interpretation of
data. ZZ and WS confirm the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patients consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yang X, Zhang Y, Xu W, Deng R, Liu Y, Li
F, Wang Y, Ji X, Bai M, Zhou F, et al: Potential role of Hsp90 in
rat islet function under the condition of high glucose. Acta
Diabetol. 53:621–628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao YC, Zhu J, Song GY and Li XS:
Relationship between thioredoxin-interacting protein (TXNIP) and
islet β-cell dysfunction in patients with impaired glucose
tolerance and hypertriglyceridemia. Int J Clin Exp Med.
8:4363–4368. 2015.PubMed/NCBI
|
|
3
|
Nelson P, Smith N, Ciupe S, Zou W, Omenn
GS and Pietropaolo M: Modeling dynamic changes in type 1 diabetes
progression: Quantifying beta-cell variation after the appearance
of islet-specific autoimmune responses. Math Biosci Eng. 6:753–778.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dahan T, Ziv O, Horwitz E, Zemmour H, Lavi
J, Swisa A, Leibowitz G, Ashcroft FM, In't Veld P, Glaser B and Dor
Y: Pancreatic beta-cells express the fetal islet hormone gastrin in
rodent and human diabetes. Diabetes. 66:426–436. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jones A, Danielson KM, Benton MC, Ziegler
O, Shah R, Stubbs RS, Das S and Macartney-Coxson D: MiRNA
signatures of insulin resistance in obesity. Obesity (Silver
Spring). 25:1734–1744. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ortega FJ, Mercader JM, Moreno-Navarrete
JM, Rovira O, Guerra E, Esteve E, Xifra G, Martínez C, Ricart W,
Rieusset J, et al: Profiling of circulating microRNAs reveals
common microRNAs linked to type 2 diabetes that change with insulin
sensitization. Diabetes Care. 37:1375–1383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma J, Zhang J, Wang Y, Long K, Wang X, Jin
L, Tang Q, Zhu L, Tang G, Li X and Li M: MiR-532-5p alleviates
hypoxia-induced cardiomyocyte apoptosis by targeting PDCD4. Gene.
675:36–43. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taneera J, Fadista J, Ahlqvist E, Zhang M,
Wierup N, Renström E and Groop L: Expression profiling of cell
cycle genes in human pancreatic islets with and without type 2
diabetes. Mol Cell Endocrinol. 375:35–42. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shen J and Zhu B: Integrated analysis of
the gene expression profile and DNA methylation profile of obese
patients with type 2 diabetes. Mol Med Rep. 17:7636–7644.
2018.PubMed/NCBI
|
|
11
|
Bustany S, Bourgeais J, Tchakarska G, Body
S, Hérault O, Gouilleux F and Sola B: Cyclin D1 unbalances the
redox status controlling cell adhesion, migration, and drug
resistance in myeloma cells. Oncotarget. 7:45214–45224. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhong Q, Hu Z, Li Q, Yi T, Li J and Yang
H: Cyclin D1 silencing impairs DNA double strand break repair,
sensitizes BRCA1 wildtype ovarian cancer cells to olaparib. Gynecol
Oncol. 152:157–165. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pistritto G, Trisciuoglio D, Ceci C,
Garufi A and D'Orazi G: Apoptosis as anticancer mechanism: Function
and dysfunction of its modulators and targeted therapeutic
strategies. Aging (Albany NY). 8:603–619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu RX, Ma Y, Hu XL, Liao YP, Hu XN, He BC
and Sun WJ: Pioglitazone/metformin adduct regulates insulin
secretion and inhibits high glucose-induced apoptosis via
p21-p53-MDM2 signaling in INS-1 cells. J Cell Biochem.
119:5449–5459. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Chen H, Hou Y, Ma X, Ye M, Huang R,
Hu B, Cao H, Xu L, Liu M, et al: Adaptive EGF expression sensitizes
pancreatic cancer cells to ionizing radiation through activation of
the cyclin D1/P53/PARP pathway. Int J Oncol. 54:1466–1480.
2019.PubMed/NCBI
|
|
16
|
Ruan D, Liu Y, Wang X, Yang D and Sun Y:
MiR-149-5p protects against high glucose-induced pancreatic beta
cell apoptosis via targeting the BH3-only protein BIM. Exp Mol
Pathol. 110:1042792019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Abouzaripour M, Pasbakhsh P, Atlasi N,
Shahverdi AH, Mahmoudi R and Kashani IR: In vitro differentiation
of insulin secreting cells from mouse bone marrow derived
stage-specific embryonic antigen 1 positive stem cells. Cell J.
17:701–710. 2016.PubMed/NCBI
|
|
19
|
Ye D, Zhang T, Lou G, Xu W, Dong F, Chen G
and Liu Y: Plasma miR-17, miR-20a, miR-20b and miR-122 as potential
biomarkers for diagnosis of NAFLD in type 2 diabetes mellitus
patients. Life Sci. 208:201–207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lakhter AJ, Pratt RE, Moore RE, Doucette
KK, Maier BF, DiMeglio LA and Sims EK: Beta cell extracellular
vesicle miR-21-5p cargo is increased in response to inflammatory
cytokines and serves as a biomarker of type 1 diabetes.
Diabetologia. 61:1124–1134. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Q, Len Q, Liu Z and Wang W:
Overexpression of miR-22 attenuates oxidative stress injury in
diabetic cardiomyopathy via Sirt 1. Cardiovasc Ther. 36:2018.
View Article : Google Scholar
|
|
22
|
Wang J, Zhang J, Chen X, Yang Y, Wang F,
Li W, Awuti M, Sun Y, Lian C, Li Z, et al: MiR-365 promotes
diabetic retinopathy through inhibiting Timp3 and increasing
oxidative stress. Exp Eye Res. 168:89–99. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng Y, Kuang W, Hao Y, Zhang D, Lei M,
Du L, Jiao H, Zhang X and Wang F: Downregulation of miR-27a* and
miR-532-5p and upregulation of miR-146a and miR-155 in LPS-induced
RAW264.7 macrophage cells. Inflammation. 35:1308–1313. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan X, Zeng D, Zhu H, Zhang Y, Shi Y, Wu
Y, Tang H and Li D: MiRNA-532-5p regulates CUMS-induced
depression-like behaviors and modulates LPS-induced proinflammatory
cytokine signaling by targeting STAT3. Neuropsychiatr Dis Treat.
16:2753–2764. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cai X, Wang S, Hong L, Yu S, Li B, Zeng H,
Yang X, Zhang P and Shao L: Long noncoding RNA taurine-upregulated
gene 1 knockdown protects cardiomyocytes against
hypoxia/reoxygenation-induced injury through regulating
miR-532-5p/Sox8 axis. J Cardiovasc Pharmacol. 76:556–563. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gurke J, Schindler M, Pendzialek SM,
Thieme R, Grybel KJ, Heller R, Spengler K, Fleming TP, Fischer B
and Navarrete Santos A: Maternal diabetes promotes mTORC1
downstream signalling in rabbit preimplantation embryos.
Reproduction. 151:465–476. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li N, Zeng J, Sun F, Tong X, Meng G, Wu C,
Ding X, Liu L, Han M, Lu C and Dai F: p27 inhibits CDK6/CCND1
complex formation resulting in cell cycle arrest and inhibition of
cell proliferation. Cell Cycle. 17:2335–2348. 2018. View Article : Google Scholar : PubMed/NCBI
|