Introduction
Intervertebral disc degeneration (IDD) is a
prevalent complicated disease of the spine that reduces the quality
of life of patients (1). IDD is
usually associated with myelopathy, radiculopathy and lower back
pain, with an incidence rate of >90% (2). IDD is affected by several factors,
such as apoptosis, inflammatory factors, degradative enzymes,
mechanical load, cellular senescence and heredity (3). However, the exact mechanisms
responsible for IDD remain elusive.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs
that modulate their target genes by regulating mRNA stability and
translation, as well as controlling gene expression at the
post-transcriptional level by pairing with target mRNAs at the 3′
untranslated region (3′UTR) (4).
Increasing evidence has revealed that miRNAs are involved in the
progression of IDD. For instance, miR-140-5p overexpression reduces
lipopolysaccharide (LPS)-induced inflammation and degeneration of
intervertebral discs by downregulating Toll-like receptor 4 (TLR4)
(5). Moreover, miR-132
contributes to extracellular matrix (ECM) degradation by directly
targeting growth and differentiation factor 5 (GDF5) in NP cells
(6). miR-200c-3p is involved in
the pathogenesis of inflammatory diseases (7), and it has been shown that the
expression levels of miR-200c-3p in the synovial fluid of 150
patients with knee osteoarthritis were significantly lower compared
with those in 54 healthy controls (8). miR-200c-3p has also been reported to
inhibit apoptosis and inflammation in cigarette smoke
extract-stimulated 16HBE cells (9). However, the mechanism of action of
miR-200c-3p on IDD progression remains unknown.
Ras-related proteins (RAPs) organize various
biological processes, including metabolic turnover, cytoskeletal
organization, cell cycle, differentiation and cell adhesion
(10). RAP contains a broad
family of nucleotide-binding proteins, including five members:
RAP1A, RAP1B, RAP2A, RAP2B and RAP2C (11). Previous studies have revealed that
RAP2C was involved in multiple diseases, such as laryngeal squamous
cell carcinoma (12). However, to
the best of our knowledge, the role of RAP2C in the regulation of
IDD progression has not been previously reported. It has been shown
that RAP2C activates ERK signaling (13), which has been identified as
crucial in the modulation of IDD progression (14,15). Pro-inflammatory macrophages
promote the degeneration phenotypes of nucleus pulposus (NP) cells
partly via JNK and ERK signaling (16). However, the association of ERK
signaling with miR-200c-3p and RAP2C in IDD modulation is yet to be
fully elucidated.
Thus, the present study aimed to investigate the
role and underlying mechanism of miR-200c-3p in the modulation of
IDD development.
Materials and methods
IDD clinical samples
A total of 22 intervertebral disc tissues from
patients with IDD (age, 41–64 years; 10 male and 12 female) and 9
normal samples (age, 40–49 years; 4 male and 5 female) used in this
study were obtained from Qingdao No. 6 People's Hospital between
May 2018 and December 2019. All patients with chronic lower back
pain (lasting for >3 months) underwent MRI examinations
(17,18). The 22 patients with IDD underwent
fusion surgery and intervertebral disc excision. The 9 normal
tissues were obtained from patients who underwent traumatic lumbar
fracture. Patients who were treated with chemotherapy or
radiotherapy were excluded from this study. The samples were
immediately frozen in liquid nitrogen and stored at −80°C before
further analysis. The samples used in this study were obtained with
written approval from patients. This study conformed to the
experimental guidelines of the World Medical Association and the
Ethics Committee of Qingdao No.6 People's Hospital (approval no.
201907).
Cell culture and treatment
Human NP cells were obtained from ScienCell Research
Laboratories, Inc. The cells were cultured in DMEM (Gibco; Thermo
Fisher Scientific, Inc.) containing 15% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 0.1 mg/ml streptomycin (Beijing Solarbio Science
& Technology Co., Ltd.) and 100 U/ml penicillin (Beijing
Solarbio Science & Technology Co., Ltd.) at 37°C with 5%
CO2.
Lentiviral plasmids (GV248) carrying RAP2C short
hairpin RNA (shRNA) and the corresponding negative control (NC)
shRNA were synthesized and obtained from Shanghai GenePharma Co.,
Ltd. The sequence of RAP2C shRNA was 5′-GAAGCAAGAUCAGUGUUGU-3′ and
the sequence of the NC shRNA was 5′-TTCTCCGAACGTGTCACGT-3′. The
ratio of the lentiviral plasmid, packaging vector (Shanghai
GenePharma Co., Ltd.) and envelope vector (Shanghai GenePharma Co.,
Ltd.) was 4:3:3. A second-generation system was used. Briefly, 293T
cells (American Type Culture Collection) were plated into 6-well
plates (1×106/well), which were used as the interim cell
line, and they were transfected with 20 µg lentiviral plasmid using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 12 h. Lentiviral particles were
collected using 72,000 × g/min centrifugation for 120 min. NP cells
were plated into 24-well plates (2×105/well) and
subsequently infected with the lentiviral supernatant
(1×108 TU/ml) at a multiplicity of infection of 10 at
37°C for 12 h using TR-1003-G (Sigma-Aldrich; Merck KGaA). After 48
h of cell transduction, these cells were selected for in the
presence of puromycin (1 µg/ml; Beyotime Institute of
Biotechnology) for 3 days between transfection and experimentation
at 37°C to generate stable NC and RAP2C-knockdown cells. Puromycin
(0.5 µg/ml) was used for maintenance.
pcDNA3.1-RAP2C overexpression vector (OE-RAP2C) and
empty pcDNA3.1 plasmid were synthesized and obtained from
GenScript. miR-200c-3p mimic, miR-200c-3p inhibitor and
corresponding scrambled controls were synthesized and obtained from
Shanghai GenePharma Co., Ltd. NP cells (2×104/well) were
transfected with 100 pmol pcDNA-3.1 vectors or 50 nM miRNA
constructs using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions, at 37°C for 6 h. The sequences of each construct were
as follows: miR-200c-3p mimic sense, 5′-UAAUACUGCCGGGUAAUGAUGGA-3′
and antisense, 5′-CAUCAUUACCCGGCAGUAUUAUU-3′; mimic NC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-200c-3p inhibitor,
5′-UCCAUCAUUACCCGGCAGUAUUA-3′; and inhibitor NC,
5′-CAGUACUUUUGUGUAGUACAA-3′. NP cells were harvested 24 h after
transfection for use in subsequent experiments.
To construct the IDD cell model, NP cells were
plated into 6-well plates (3×105/well) and treated with
LPS (1 µg/ml; Sigma-Aldrich; Merck KGaA) for 24 h at 37°C after 24
h of transfection. The ERK inhibitor, SCH772984 (Selleck
Chemicals), was used at a dosage of 10 µmol/l for 24 h at 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNAs were extracted using TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.) and were treated with
RQ1 RNase-Free DNase (Promega Corporation) for 30 min at 37°C. The
quality of total RNA was detected at an absorbance 260/280 nm ratio
using NanoDrop life spectrophotometer (Thermo Fisher Scientific,
Inc.). cDNA was synthesized with the Maxima First-strand cDNA
Synthesis kit according to the manufacturer's instructions (Thermo
Fisher Scientific, Inc.). qPCR was performed using the SYBR
Real-time PCR I kit (Takara Bio, Inc.). The thermocycling
conditions used for the qPCR were as follows: Initial denaturation
for 3 min at 95°C; followed by 37 cycles of denaturation at 94°C
for 1 min, annealing at 60°C for 1 min and extension at 72°C for 1
min, followed by a final extension step at 72°C for 7 min. Relative
quantification was performed using the 2−ΔΔCq method
(19). The standard controls for
miRNA and mRNA were U6 and GAPDH, respectively (20). Quantitative determination of the
RNA levels was performed in three independent experiments. Data are
presented as the mean ± SD. The primer sequences were as follows:
miR-200c-3p NCBI Reference Sequences (RefSeq), NC_000012.12;
miR-200c-3p forward, 5′-GGGGTAGGGGAAGGTGGTTTA-3′ and reverse,
5′-CACCACCCCAATCCCTAAAAACACT-3′; RAP2C NCBI RefSeq, XM_035008753.1;
RAP2C forward, 5′-TGGCCATACCGAGCAGATAAAACTCA-3′ and reverse,
5′-ACAGGTTTACCAAGGCTCAGTTCTGC-3′; collagen II NCBI RefSeq,
XM_008951124.2; collagen II forward, 5′-CTGGTGATGATGGTGAAG-3′ and
reverse, 5′-CCTGGATAACCTCTGTGA-3′; AggrecanNCBI RefSeq,
NM_001369268.1; aggrecan forward, 5′-GCGAGCACTGTAACATAGACAT-3′ and
reverse, 5′-TCACACAGGTCCCCTTCGTA-3′; GAPDH NCBI RefSeq,
NM_001256799.3; GAPDH forward, 5′-CATGTTGCAACCGGGAAGGA-3′ and
reverse, 5′-GCCCAATACGACCAAATCAGAG-3′; and U6 NCBI RefSeq,
NC_015438.3; U6 forward, 5′-GCTTCGGCAGCACATATACTAA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
ELISA
The levels of TNF-α (cat. no. KHC3013), IL-6 (cat.
no. KAC1261) and IL-1β (cat. no. KAC1211) were analyzed using ELISA
kits (eBioscience; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. All standards and samples were tested
on a SpectraMax M5 microplate reader device (Molecular Devices,
LLC) at an absorbance of 450 nm. A standard curve was prepared, and
the concentration of the samples was calculated according to the
absorbance value. Data are presented as the mean ± SD of three
experiments.
Analysis of cell apoptosis
In total, ~2×105 NP cells were plated
into 6-well plates. Apoptosis was analyzed using the Annexin V-FITC
Apoptosis Detection kit (Cell Signaling Technology, Inc.) according
to the manufacturer's instructions. Briefly, ~2×105 NP
cells were collected and washed with binding buffer, followed by
flow cytometry analysis on a flow cytometer (BD Accuri™ C6 Plus; BD
Biosciences). The apoptotic rate was calculated as the percentage
of early + late apoptotic cells using FlowJo software (v10.7;
FlowJo, LLC). Data are presented as the mean ± SD of three
experiments.
Luciferase reporter gene assay
TargetScan (http://www.targetscan.org/vert_72) was used to verify
the potential interaction between RAP2C and miR-200c-3p. Luciferase
reporter gene assays were performed using a dual-luciferase
reporter assay system (Promega Corporation). The RAP2C wild type or
mutant 3′-untranslated region sequences containing the miR-200c-3p
binding site were constructed and subcloned into the psiCHECK2
vector (Promega Corporation). Briefly, the miR-200c-3p or control
mimic, and the vector containing RAP2C and RAP2C mutant fragments
were transfected into the cells (3×105/well) using
Lipofectamine 3000 for 48 h, followed by the analysis of luciferase
activities, in which Renilla was used as a normalized
control reporter. Data are presented as the mean ± SD of three
experiments.
Western blot analysis
Total proteins were extracted from cells using RIPA
buffer (Cell Signaling Technology, Inc.). Protein concentrations
were measured using the BCA Protein Quantification kit (Abbkine
Scientific Co., Ltd.). An equivalent concentration of protein (50
µg/lane) was separated via SDS-PAGE (12% polyacrylamide gels) and
transferred onto PVDF membranes (MilliporeSigma). The membranes
were blocked with 5% milk at 25°C for 60 min and incubated
overnight at 4°C with the primary antibodies for RAP2C (1:3,000;
cat. no. ab97805; Abcam), ERK (1:1,000; cat. no. ab32537; Abcam),
phosphorylated (p)-ERK (1:1,000; cat. no. ab194776; Abcam) and
β-actin (1:5,000; cat. no. ab8227; Abcam), in which β-actin served
as the control. Then, the goat anti-rabbit IgG H&L (HRP)
secondary antibodies (1:5,000; cat. no. ab7090; Abcam) were added
to membranes for 1 h at 25°C, followed by visualization using an
Odyssey CLx Infrared Imaging system (LI-COR Biosciences). Image
Studio™ Lite (v4.0; LI-COR Biosciences) was used for analysis. Data
are presented as the mean ± SD of three experiments.
Statistical analysis
Data are presented as the mean ± SD of three
experiments and statistical analysis was performed using GraphPad
Prism 7 (GraphPad Software, Inc.). An unpaired Student's t-test was
used to compare two groups. One-way ANOVA was used for multiple
group comparisons and Tukey's test was used for post hoc analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-200c-3p is downregulated in IDD
and inhibits LPS-induced NP cell apoptosis
To assess the potential association between
miR-200c-3p and IDD, the expression level of miR-200c-3p was
analyzed in patients with IDD and normal controls. The expression
level of miR-200c-3p was decreased in the intervertebral disc
tissues of patients with IDD (n=22) compared with that in normal
subjects (n=9) (Fig. 1A),
suggesting a potential association between miR-200c-3p and IDD
development. Next, the expression of miR-200c-3p was examined in
LPS-treated NP cells. LPS treatment reduced the expression of
miR-200c-3p in NP cells in a time-dependent manner (Fig. 1B), indicating that miR-200c-3p may
be involved in IDD progression. NP cell incubation with LPS for 24
h was selected for the next experiment.
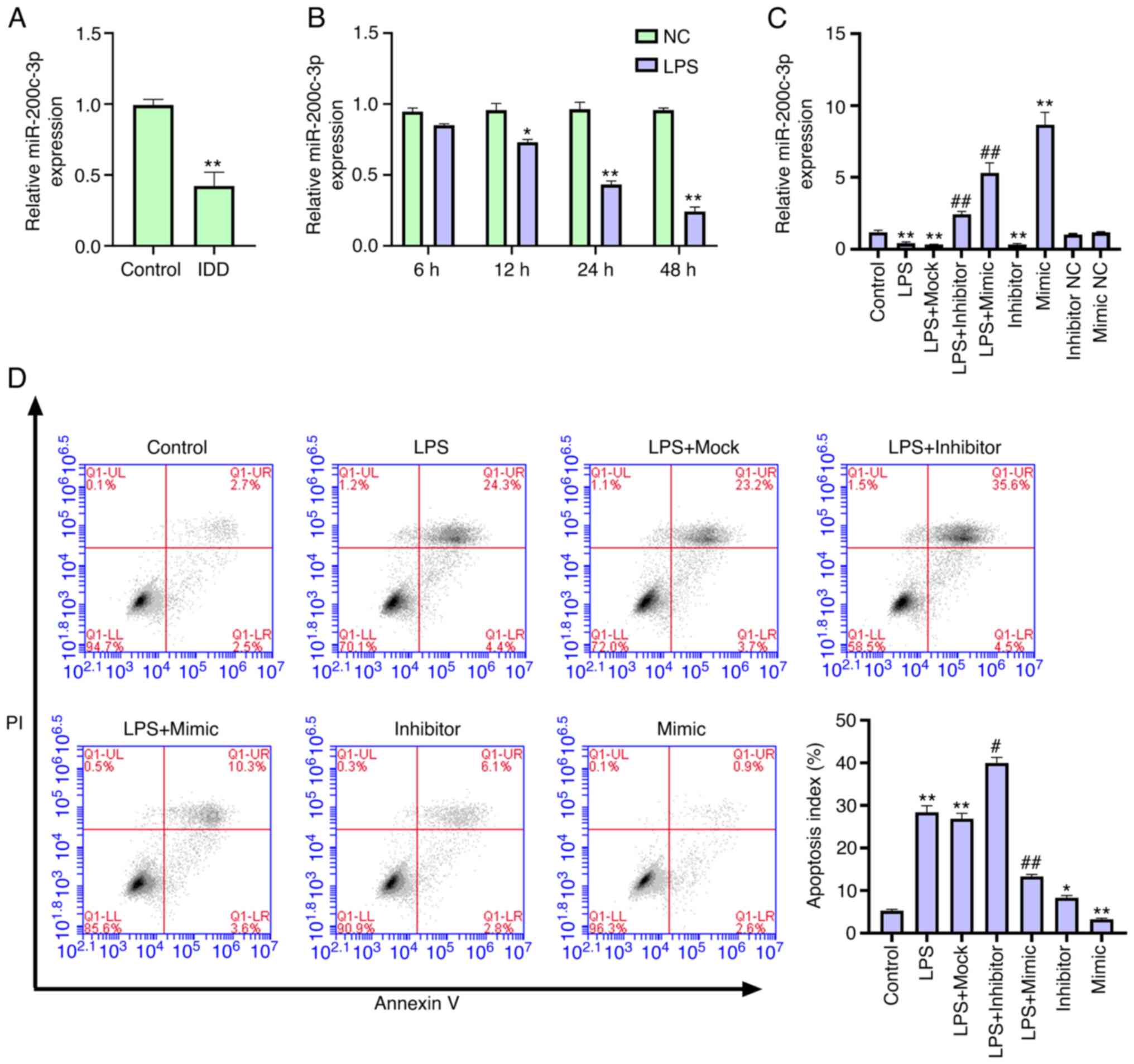 | Figure 1.miR-200c-3p inhibits LPS-induced NP
cell apoptosis. (A) Expression levels of miR-200c-3p were measured
via RT-qPCR in the intervertebral disc tissues of patients with IDD
(n=22) and normal cases (n=9). (B) Expression levels of miR-200c-3p
were assessed via RT-qPCR in NP cells treated with LPS (1 µg/ml).
*P<0.05, **P<0.01 vs. NC. NP cells were treated with LPS (1
µg/ml) or co-treated with LPS (1 µg/ml) and miR-200c-3p mimic,
inhibitor or corresponding control. (C) Expression levels of
miR-200c-3p were analyzed via RT-qPCR. (D) Cell apoptosis was
tested using flow cytometry analysis. Data are presented as the
mean ± SD. *P<0.05, **P<0.01 vs. control;
#P<0.05, ##P<0.01 vs. LPS. LPS,
lipopolysaccharide; RT-qPCR, reverse transcription-quantitative
PCR; miR, microRNA; NC, negative control; IDD, intervertebral disc
degeneration; NP, nucleus pulposus. |
To evaluate the role of miR-200c-3p in the
development of IDD in vitro, LPS-treated NP cells were
transfected with miR-200c-3p mimic or miR-200c-3p inhibitor and the
efficiency was validated (Fig.
1C). As expected, LPS treatment increased the apoptosis of NP
cells, in which the miR-200c-3p inhibitor enhanced, while the
miR-200c-3p mimic suppressed this effect (Fig. 1D), suggesting that miR-200c-3p
inhibits LPS-induced NP cell apoptosis.
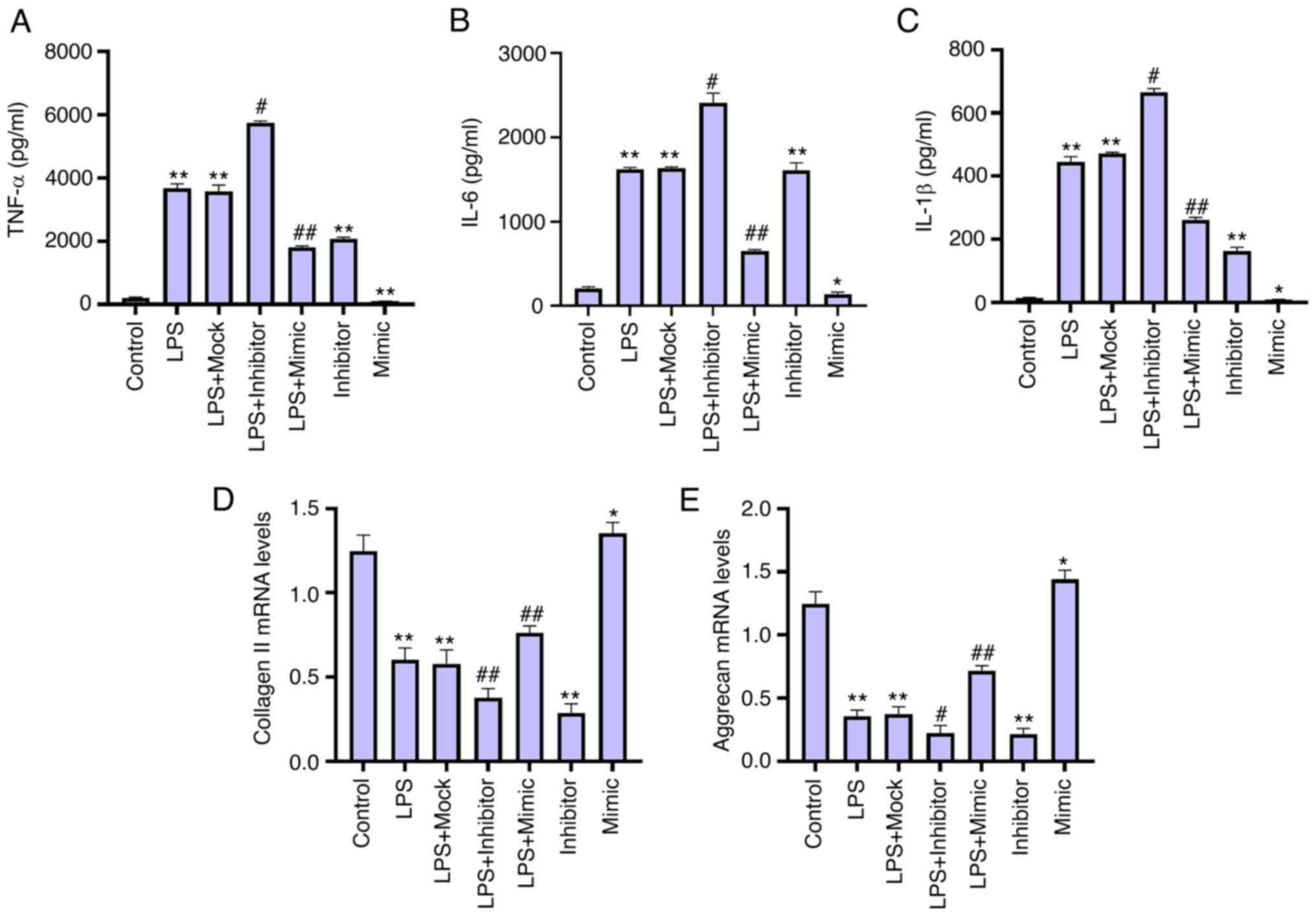
miR-200c-3p attenuates inflammatory
cytokine levels and ECM degradation in LPS-treated NP cells
Since inflammatory factors have been shown to be
involved in the pathological processes of IDD (21). The effect of miR-200c-3p on the
levels of inflammatory factors was determined in LPS-treated NP
cells. The levels of inflammatory cytokines in the culture medium
of NP cells, including TNF-α, IL-6 and IL-1β, were increased by LPS
stimulation, in which the miR-200c-3p inhibitor enhanced, but the
miR-200c-3p mimic reduced this phenotype in the cells (Fig. 2A-C). This suggests that
miR-200c-3p inhibits inflammatory cytokine levels in LPS-treated NP
cells. Moreover, the expression levels of collagen II and aggrecan
were significantly decreased by the miR-200c-3p inhibitor but were
upregulated by the miR-200c-3p mimic in the LPS-treated NP cells
(Fig. 2D and E), indicating that
miR-200c-3p attenuates ECM degradation in LPS-treated NP cells.
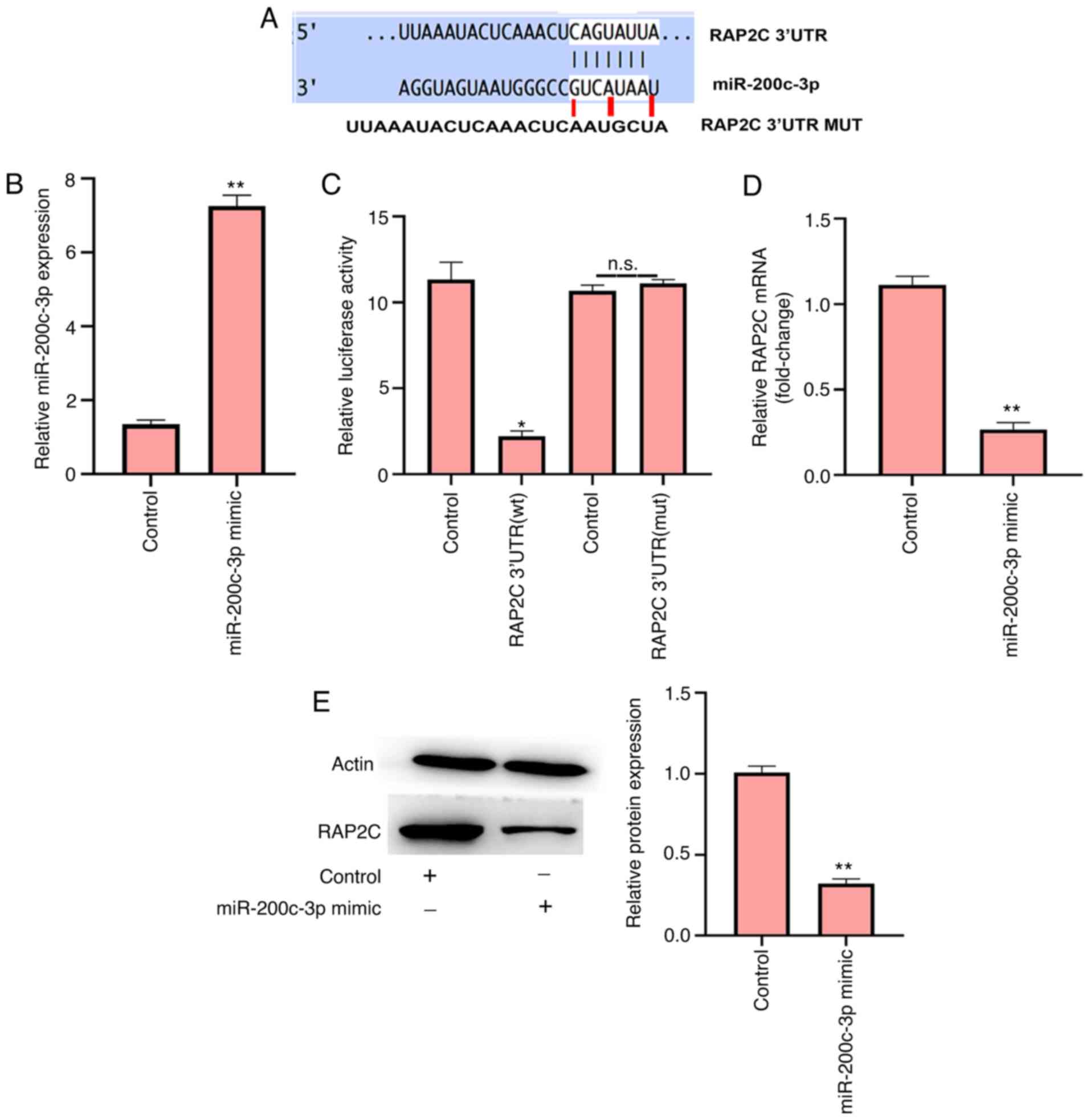
miR-200c-3p targets RAP2C in NP
cells
miR-200c-3p has a recognized target site in RAP2C
3′UTR, as determined using TargetScan (Fig. 3A). To examine the effect of
miR-200c-3p on RAP2C, the NP cells were transfected with
miR-200c-3p mimic (Fig. 3B).
Notably, the miR-200c-3p mimic inhibited luciferase activity of
wild-type RAP2C but failed to affect the RAP2C with the
miR-200c-3p-binding site mutant in the NP cells (Fig. 3C). Furthermore, the mRNA and
protein expression levels of RAP2C were significantly downregulated
by miR-200c-3p mimic transfection in NP cells (Fig. 3D and E), suggesting that
miR-200c-3p can target RAP2C in NP cells.
RAP2C promotes apoptosis, inflammatory
cytokine levels and ECM degradation in LPS-treated NP cells
LPS-treated NP cells were infected with lentiviral
plasmids carrying RAP2C shRNA or corresponding control shRNA or
transfected with the vector carrying the complete RAP2C coding
sequence for overexpression or the control vector. The efficiency
of RAP2C shRNA and RAP2C overexpression was validated in the cells
(Fig. 4A). It was demonstrated
that the LPS-induced apoptosis of NP cells was significantly
enhanced by RAP2C overexpression and reduced by RAP2C knockdown
(Fig. 4B), suggesting that RAP2C
promotes NP cell apoptosis. Moreover, the upregulation of TNF-α,
IL-6 and IL-1β by LPS treatment was increased by the overexpression
of RAP2C but was suppressed by RAP2C knockdown in the NP cells
(Fig. 4C-E), indicating that
RAP2C activates the inflammatory cytokine levels. Furthermore,
RAP2C overexpression inhibited and RAP2C knockdown increased the
expression levels of collagen II and aggrecan in NP cells (Fig. 4F and G), suggesting that RAP2C
contributes to ECM degradation in LPS-treated NP cells.
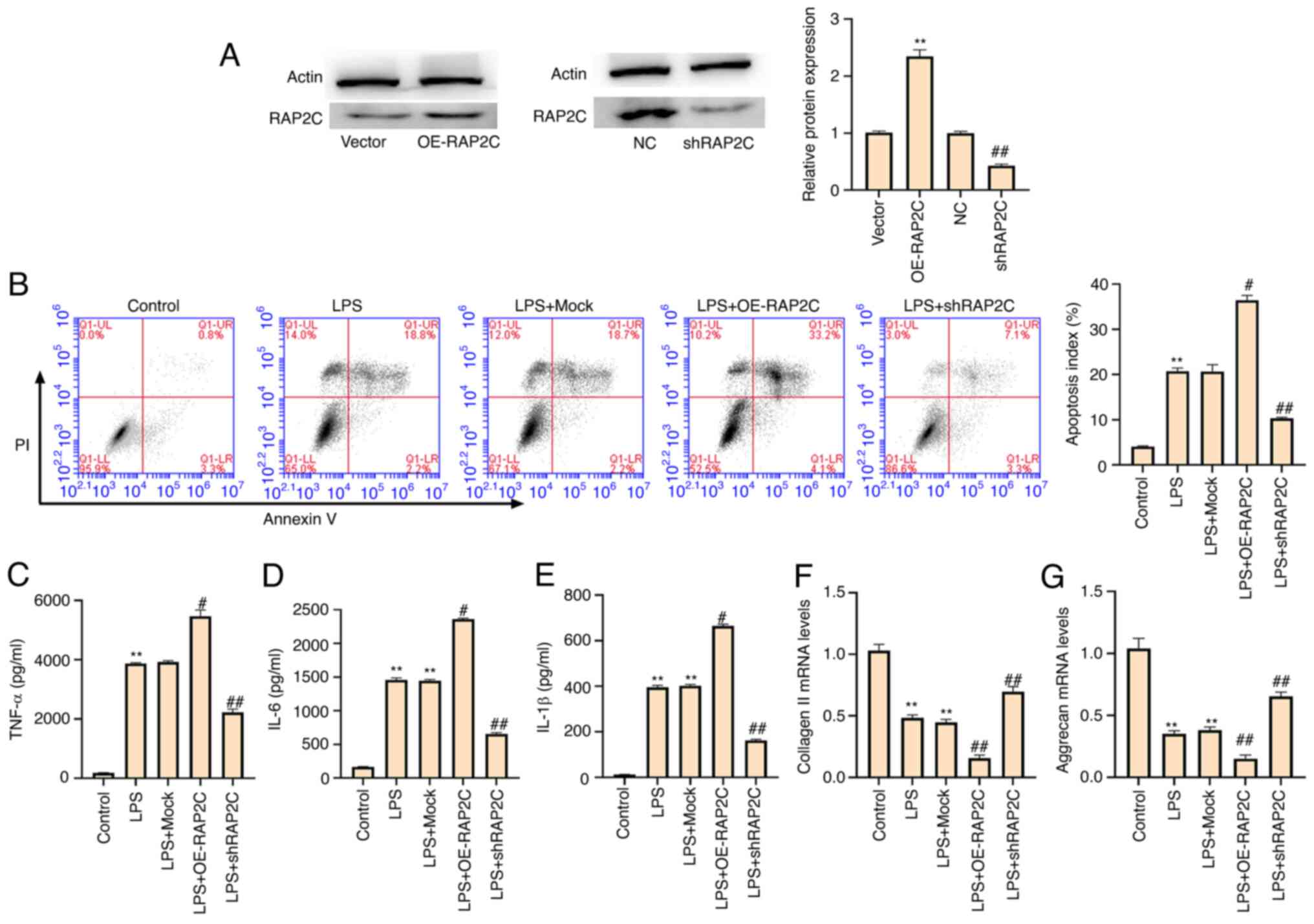 | Figure 4.RAP2C promotes apoptosis,
inflammatory cytokine levels and ECM degradation in LPS-treated NP
cells. LPS (1 µg/ml)-treated NP cells were infected with lentiviral
plasmids carrying RAP2C shRNA or the corresponding control shRNA,
or transfected with the control vector or the vector carrying the
complete RAP2C coding sequence for overexpression. (A) Expression
levels of RAP2C were measured via western blotting. **P<0.01 vs.
vector; ##P<0.01 vs. NC. (B) Cell apoptosis was
measured using flow cytometry analysis. Levels of (C) TNF-α, (D)
IL-6 and (E) IL-1β in the culture medium of the cells were analyzed
using ELISAs. Expression levels of (F) collagen II and (G) aggrecan
were examined via reverse transcription-quantitative PCR. Data are
presented as the mean ± SD. **P<0.01 vs. control;
#P<0.05, ##P<0.01 vs. LPS. LPS,
lipopolysaccharide; shRNA/sh, short hairpin RNA; NC, negative
control; RAP2C, Ras-related protein 2C; NP, nucleus pulposus; OE,
overexpression vector. |
RAP2C enhances apoptosis, inflammatory
cytokine levels and ECM degradation by activating ERK
signaling
It has been reported that the activation of ERK
signaling contributes to the development of IDD (14,22). The phosphorylation of ERK is
required for activation of ERK signaling (23,24). The expression level of RAP2C was
significantly upregulated by the overexpression of RAP2C, whereas
the ERK inhibitor SCH772984 partially reduced this upregulation in
NP cells. Moreover, the knockdown of RAP2C significantly decreased
RAP2C expression (Fig. 5A). The
knockdown of RAP2C increased the ratio of p-/total ERK protein
expression (Fig. 5A). Moreover,
RAP2C induced LPS-treated NP cell apoptosis, and SCH772984
partially restored this phenotype (Fig. 5B). In addition, SCH772984
partially blocked the elevated levels of TNF-α, IL-6 and IL-1β
caused by the overexpression of RAP2C in LPS-treated NP cells
(Fig. 5C-E). In addition, RAP2C
overexpression-inhibited expression of collagen II and aggrecan was
rescued by SCH772984 treatment in LPS-treated NP cells (Fig. 5F and G). Taken together, these
data suggest that RAP2C enhances apoptosis, inflammatory cytokine
levels and ECM degradation by activating ERK signaling.
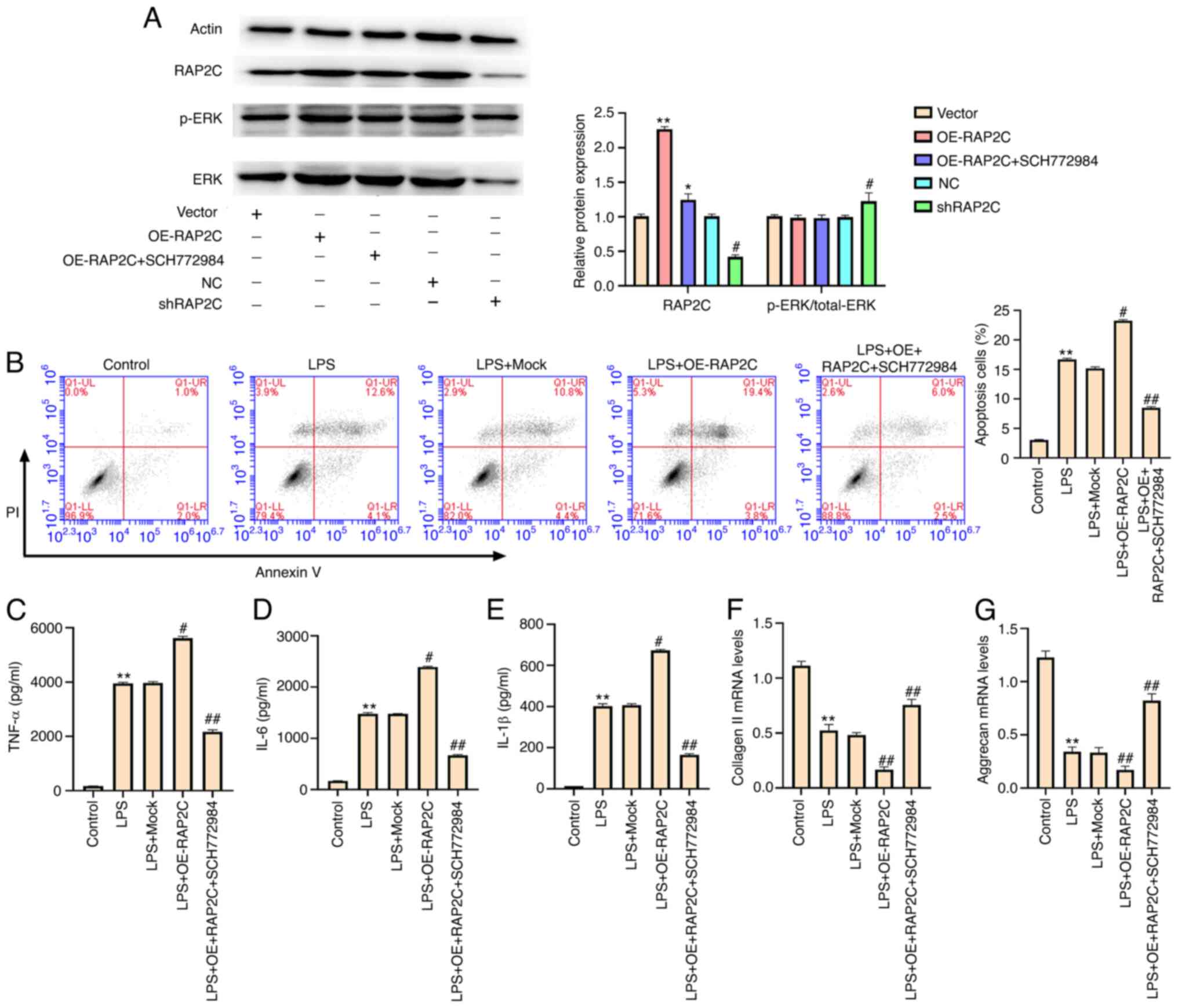 | Figure 5.RAP2C enhances apoptosis,
inflammatory cytokine levels and extracellular matrix degradation
by activating ERK signaling. (A) NP cells were infected with the
lentiviral plasmids carrying RAP2C shRNA or the corresponding
control shRNA, transfected with the control vector or the vector
carrying the complete RAP2C coding sequence for overexpression or
co-treated with the RAP2C overexpression vector and the ERK
inhibitor SCH772984 (10 µmol/l). The expression levels of RAP2C,
ERK and β-actin, and the phosphorylation of ERK (p-ERK) were
examined via western blotting. *P<0.05, **P<0.01 vs. vector;
#P<0.05 vs. NC. LPS (1 µg/ml)-treated NP cells were
transfected with the control vector, or the vector carrying the
complete RAP2C coding sequence for overexpression or co-treated
with RAP2C overexpression vector and the ERK inhibitor SCH772984
(10 µmol/l). (B) Cell apoptosis was examined via flow cytometry
analysis. Levels of (C) TNF-α, (D) IL-6 and (E) IL-1β in the
culture medium of the cells were tested using ELISAs. Expression
levels of (F) collagen II and (G) aggrecan were determined using
reverse transcription-quantitative PCR. Data are presented as the
mean ± SD. *P<0.05, **P<0.01 vs. control;
#P<0.05, ##P<0.01 vs. LPS. LPS,
lipopolysaccharide; shRNA/sh, short hairpin RNA; NC, negative
control; RAP2C, Ras-related protein 2C; NP, nucleus pulposus; OE,
overexpression vector; p-, phosphorylated. |
miR-200c-3p suppresses IDD by
targeting RAP2C/ERK signaling
Knockdown of miR-200c-3p with a miR-200c-3p
inhibitor promoted the expression of RAP2C. However, SCH772984
treatment blocked miR-200c-3p inhibitor-induced RAP2C expression in
NP cells. The miR-200c-3p mimic inhibited the expression of RAP2C
(Fig. 6A). The miR-200c-3p mimic
also increased the ratio of p-/total ERK protein expression
(Fig. 6A). Moreover, the
miR-200c-3p inhibitor-induced apoptosis of LPS-treated NP cells was
partially inhibited by RAP2C shRNA or SCH772984 (Fig. 6B). In addition, the levels of
TNF-α, IL-6 and IL-1β enhanced by the miR-200c-3p inhibitor were
partially decreased by RAP2C knockdown or SCH772984 treatment in NP
cells (Fig. 6C-E). RAP2C
knockdown and SCH772984 treatment also partially restored the
expression levels of collagen II and aggrecan reduced by
miR-200c-3p inhibitor (Fig. 6F and
G). Collectively, these results suggest that miR-200c-3p
suppresses IDD by targeting RAP2C/ERK signaling.
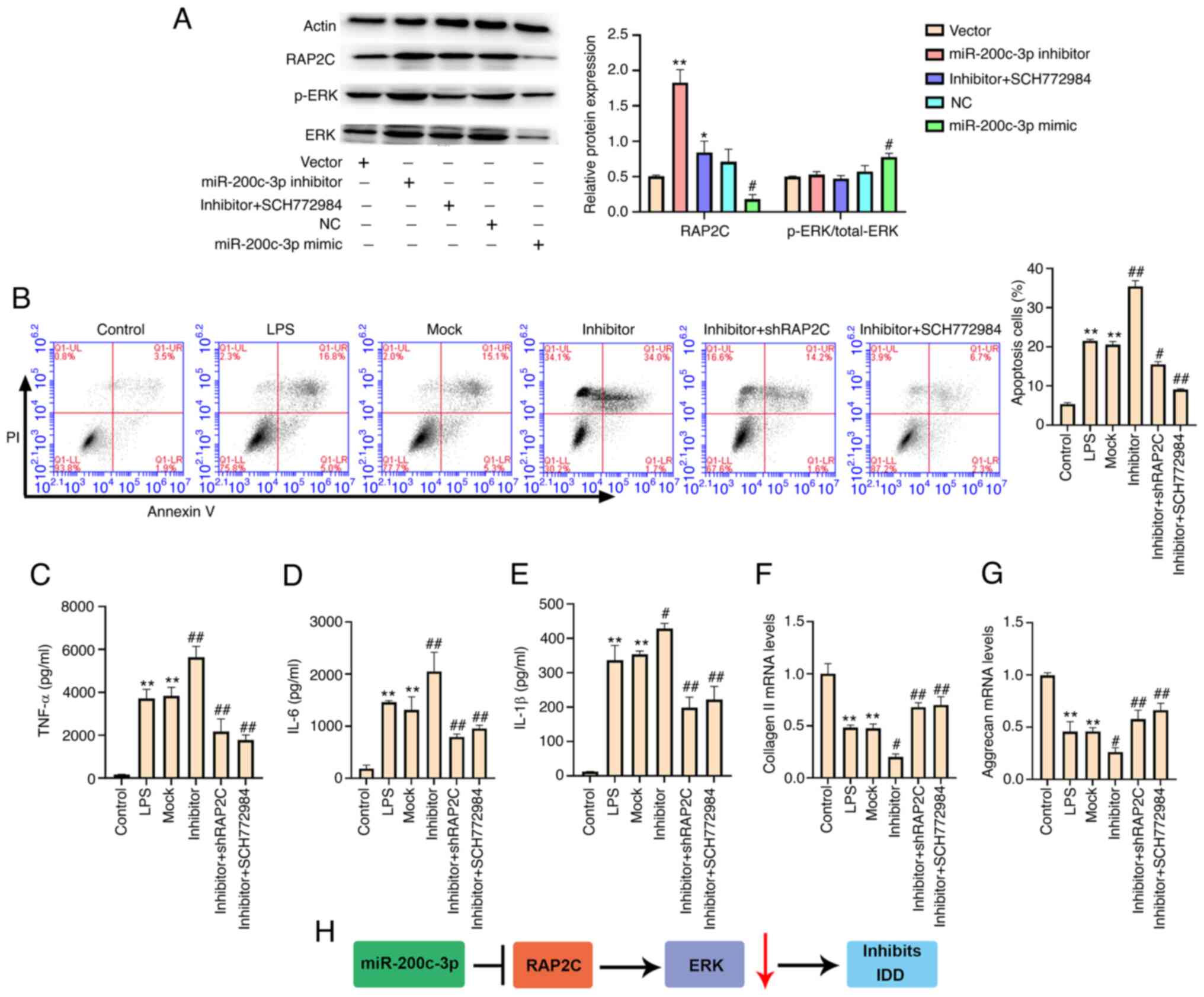 | Figure 6.miR-200c-3p suppresses IDD by
targeting RAP2C/ERK signaling. (A) NP cells were transfected with
miR-200c-3p mimic, inhibitor or corresponding control, or
co-treated with miR-200c-3p inhibitor and the ERK inhibitor
SCH772984 (10 µmol/l). The expression levels of RAP2C, ERK and
β-actin, and the phosphorylation of ERK (p-ERK) were examined via
western blotting. LPS (1 µg/ml)-treated NP cells were transfected
with miRNA control inhibitor or miR-200c-3p inhibitor, or
co-transfected with miR-200c-3p inhibitor and the lentiviral
plasmids carrying RAP2C shRNA, or co-transfected with miR-200c-3p
inhibitor and the ERK inhibitor SCH772984 (10 µmol/l). *P<0.05,
**P<0.01 vs. vector; #P<0.05 vs. NC. (B) Cell
apoptosis was analyzed using flow cytometry. The levels of (C)
TNF-α, (D) IL-6 and (E) IL-1β in the culture medium of the cells
were assessed using ELISA. The expression levels of (F) collagen II
and (G) aggrecan were examined using reverse
transcription-quantitative PCR. (H) The regulatory pathway of
miR-200c-3p in LPS-treated NP cells. Data are presented as the mean
± SD. **P<0.01 vs. control; #P<0.05,
##P<0.01 vs. LPS. IDD, intervertebral disc
degeneration; LPS, lipopolysaccharide; shRNA/sh, short hairpin RNA;
NC, negative control; RAP2C, Ras-related protein 2C; NP, nucleus
pulposus; p-, phosphorylated; miRNA/miR, microRNA. |
Discussion
IDD is a complex disorder characterized by genotypic
and phenotypic alterations, such as ECM degradation, apoptosis and
inflammation (25). miRNAs are
essential regulators of multiple physiological and pathological
processes, and are involved in the modulation of IDD progression.
It has been reported that TLR4/NF-κB signaling increases the
expression of miR-625-5p, thereby contributing to the pathological
process of IDD by modulating collagen type I α 1 (26). miR-665 contributes to the
proliferation and matrix degradation of NP cells by regulating the
expression of GDF5 in IDD (27).
Moreover, miR-24-3p upregulation is able to increase IDD by
targeting insulin-like growth factor-binding protein 5 and ERK
signaling (28), while miR-640
induces IDD by modulating Wnt and NF-κB signaling (29). Contrary to the aforementioned
results, miR-200c-3p expression was significantly decreased in the
intervertebral disc tissues of patients with IDD and it inhibited
apoptosis, inflammatory cytokine levels and ECM degradation in IDD
by targeting RAP2C/ERK signaling.
RAP2C is an evolutionarily conserved Ras-like GTPase
that serves as a molecular switch that pairs receptor signaling to
remodel the actin cytoskeleton, cell adhesion and cell polarity
(30,31). Previous studies have identified a
correlation between miRNAs and RAP2C in multiple pathological
processes (10,32,33). Spinal cord ischemia/reperfusion
injury promotes neurocyte apoptosis by reducing RAP2C via sponging
miR-204 (34). miR-188-5p
increases apoptosis and represses proliferation in breast cancer
cells by regulating RAP2C/MAPK signaling (35). In addition, it has been shown that
RAP2C can activate ERK signaling (12). Similarly, in the present study,
RAP2C was targeted by miR-200c-3p in NP cells, whereby RAP2C
promoted apoptosis, inflammatory cytokine levels and ECM
degradation in LPS-treated NP cells by activating ERK
signaling.
ECM degradation plays a critical role in the
development of IDD, in which collagen II and aggrecan serve as
markers of ECM degradation (36).
In the present study, the expression levels of collagen II and
aggrecan were decreased by miR-200c-3p inhibitors but upregulated
by miR-200c-3p mimics in LPS-treated NP cells. This finding was in
agreement with a previous study showing that long non-coding RNA
nuclear paraspeckle assembly transcript 1 downregulated aggrecan,
collagen II, thrombospondin-type motifs 5 and MMP13 by modifying
ERK/MAPK signaling in NP cells (37). Consistently, TGF-β1 inhibits the
expression of C-C motif chemokine ligand 3/4 via ERK signaling and
restricts IDD and inflammation-induced pain (38). Moreover, autophagy alleviates
compression-related NP cell apoptosis in IDD via MEK/ERK/nuclear
respiratory factor 1/autophagy related 7 signaling (39). The present study demonstrated that
ERK signaling was involved in the miR-200c-3p/RAP2C-mediated
apoptosis, inflammatory cytokine levels and ECM degradation in NP
cells.
The current study has some limitations. The
expression levels of aggrecan and collagen II were determined only
via RT-qPCR, while levels of inflammatory factors were assessed
using ELISA alone. In future studies, the results of the present
study should be verified using in vivo models and the
regulation of IDD by miR-200c-3p requires further
investigation.
In conclusion, the present study demonstrated that
miR-200c-3p inhibited NP cell apoptosis, inflammatory cytokine
levels and ECM degradation in IDD by targeting RAP2C/ERK signaling.
Moreover, miR-200c-3p and RAP2C may serve as potential targets for
IDD therapy.
Acknowledgements
Not applicable.
Funding
This study was supported in part by the Qingdao
Medical Research Guidance Program in 2018: Three-dimensional
Decompression Comprehensive Therapy for the Treatment of Lumbar
Disc Herniation and Degeneration (grant no. 2018-WJZD113).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC and KX designed the study. MJ performed the
experiments. HR analyzed data. JC and KX confirm the authenticity
of all the raw data. KX wrote the paper. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The protocol of this research has been approved by
the Ethics Committee of Qingdao No.6 People's Hospital. All
patients have signed written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have competing
interests.
Glossary
Abbreviations
Abbreviations:
|
IDD
|
intervertebral disc degeneration
|
|
NP
|
nucleus pulposus
|
|
ECM
|
extracellular matrix
|
|
LPS
|
lipopolysaccharide
|
|
miRNAs/miRs
|
microRNAs
|
|
RAP
|
Ras-related proteins
|
References
|
1
|
Wang SZ, Chang Q, Lu J and Wang C: Growth
factors and platelet-rich plasma: Promising biological strategies
for early intervertebral disc degeneration. Int Orthop. 39:927–934.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tisherman R, Coelho P, Phillibert D, Wang
D, Dong Q, Vo N, Kang J and Sowa G: NF-κB signaling pathway in
controlling intervertebral disk cell response to inflammatory and
mechanical stressors. Phys Ther. 96:704–711. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Yang J, Zhou X, Wang N, Li Z,
Zhou Y, Feng J, Shen D and Zhao W: Knockdown of miR-222 inhibits
inflammation and the apoptosis of LPS-stimulated human
intervertebral disc nucleus pulposus cells. Int J Mol Med.
44:1357–1365. 2019.PubMed/NCBI
|
|
4
|
Zhou Q, Huang SX, Zhang F, Li SJ, Liu C,
Xi YY, Wang L, Wang X, He QQ, Sun CC and Li DJ: MicroRNAs: A novel
potential biomarker for diagnosis and therapy in patients with
non-small cell lung cancer. Cell Prolif. 50:e123942017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Q, Weng Y, Jiang Y, Zhao S, Zhou D
and Xu N: Overexpression of miR-140-5p inhibits
lipopolysaccharide-induced human intervertebral disc inflammation
and degeneration by downregulating toll-like receptor 4. Oncol Rep.
40:793–802. 2018.PubMed/NCBI
|
|
6
|
Liu W, Xia P, Feng J, Kang L, Huang M,
Wang K, Song Y, Li S, Wu X, Yang S and Yang C: MicroRNA-132
upregulation promotes matrix degradation in intervertebral disc
degeneration. Exp Cell Res. 359:39–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu G, Jiao Y, Huang JJ, Fan MD, Hao YC,
Han JZ and Qu L: Acidic preconditioning reduces
lipopolysaccharide-induced acute lung injury by upregulating the
expression of angiotensin-converting enzyme 2. Exp Ther Med.
21:4412021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lai Z and Cao Y: Plasma miR-200c-3p,
miR-100-5p, and miR-1826 serve as potential diagnostic biomarkers
for knee osteoarthritis: Randomized controlled trials. Medicine
(Baltimore). 98:e181102019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen P, Jiang P, Chen J, Yang Y and Guo X:
XIST promotes apoptosis and the inflammatory response in
CSE-stimulated cells via the miR-200c-3p/EGR3 axis. BMC Pulm Med.
21:021–01582. 2021. View Article : Google Scholar
|
|
10
|
Wang Z, Huang C, Zhang A, Lu C and Liu L:
Overexpression of circRNA_100290 promotes the progression of
laryngeal squamous cell carcinoma through the miR-136-5p/RAP2C
axis. Biomed Pharmacother. 125:1098742020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu J, Du W, Wang X, Wei L, Pan Y, Wu X,
Zhang J and Pei D: Ras-related protein Rap2c promotes the migration
and invasion of human osteosarcoma cells. Oncol Lett. 15:5352–5358.
2018.PubMed/NCBI
|
|
12
|
Wang Z, Huang C, Zhang A, Lu C and Liu L:
Overexpression of circRNA_100290 promotes the progression of
laryngeal squamous cell carcinoma through the miR-136-5p/RAP2C
axis. Biomed Pharmacother. 125:109872020. View Article : Google Scholar
|
|
13
|
Wang X, Wang C, Xi L and Yu Z: Rap2c as a
novel biomarker for predicting poor prognosis in glioma. Onco
Targets Ther. 13:3073–3083. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mi D, Cai C, Zhou B, Liu X, Ma P, Shen S,
Lu W and Huang W: Long noncoding RNA FAF1 promotes intervertebral
disc degeneration by targeting the Erk signaling pathway. Mol Med
Rep. 17:3158–3163. 2018.PubMed/NCBI
|
|
15
|
Li L, Wei K, Ding Y, Ahati P, Xu H, Fang H
and Wang H: M2a macrophage-secreted CHI3L1 promotes extracellular
matrix metabolic imbalances via activation of IL-13Rα2/MAPK pathway
in rat intervertebral disc degeneration. Front Immunol.
12:6663612021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ni L, Zheng Y, Gong T, Xiu C, Li K,
Saijilafu, Li B, Yang H and Chen J: Proinflammatory macrophages
promote degenerative phenotypes in rat nucleus pulpous cells partly
through ERK and JNK signaling. J Cell Physiol. 234:5362–5371. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pfirrmann CW, Metzdorf A, Zanetti M,
Hodler J and Boos N: Magnetic resonance classification of lumbar
intervertebral disc degeneration. Spine (Phila Pa 1976).
26:1873–1878. 1976. View Article : Google Scholar
|
|
18
|
Ji Y, Hong W, Liu M, Liang Y, Deng Y and
Ma L: Intervertebral disc degeneration associated with vertebral
marrow fat, assessed using quantitative magnetic resonance imaging.
Skeletal Radiol. 49:1753–1763. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Y, Zhong Z, Zhao Y, Ren K and Li N:
LincRNA-SLC20A1 (SLC20A1) promotes extracellular matrix degradation
in nucleus pulposus cells in human intervertebral disc degeneration
by targeting the miR-31-5p/MMP3 axis. Int J Clin Exp Pathol.
12:3632–3643. 2019.PubMed/NCBI
|
|
21
|
Navone SE, Marfia G, Giannoni A, Beretta
M, Guarnaccia L, Gualtierotti R, Nicoli D, Rampini P and Campanella
R: Inflammatory mediators and signalling pathways controlling
intervertebral disc degeneration. Histol Histopathol. 32:523–542.
2017.PubMed/NCBI
|
|
22
|
Han YC, Ma B, Guo S, Yang M, Li LJ, Wang
SJ and Tan J: Leptin regulates disc cartilage endplate degeneration
and ossification through activation of the MAPK-ERK signalling
pathway in vivo and in vitro. J Cell Mol Med. 22:2098–2109. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Roskoski R Jr: ERK1/2 MAP kinases:
Structure, function, and regulation. Pharmacol Res. 66:105–143.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roskoski R Jr: Targeting ERK1/2
protein-serine/threonine kinases in human cancers. Pharmacol Res.
142:151–168. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sampara P, Banala RR, Vemuri SK, Av GR and
Gpv S: Understanding the molecular biology of intervertebral disc
degeneration and potential gene therapy strategies for
regeneration: A review. Gene Ther. 25:67–82. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen L, Xiao Y, Wu Q, Liu L, Zhang C and
Pan X: TLR4/NF-κB axis signaling pathway-dependent up-regulation of
miR-625-5p contributes to human intervertebral disc degeneration by
targeting COL1A1. Am J Transl Res. 11:1374–1388. 2019.PubMed/NCBI
|
|
27
|
Tan H, Zhao L, Song R, Liu Y and Wang L:
microRNA-665 promotes the proliferation and matrix degradation of
nucleus pulposus through targeting GDF5 in intervertebral disc
degeneration. J Cell Biochem. 119:7218–7225. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen Z, Liu M, Zhang W, Deng M, Zhou Y and
Li Y: miR-24-3p induces human intervertebral disc degeneration by
targeting insulin-like growth factor binding protein 5 and the ERK
signaling pathway. Life Sci. 243:1172882020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dong W, Liu J, Lv Y, Wang F, Liu T, Sun S,
Liao B, Shu Z and Qian J: miR-640 aggravates intervertebral disc
degeneration via NF-κB and WNT signalling pathway. Cell Prolif.
52:e126642019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lagarrigue F, Kim C and Ginsberg MH: The
Rap1-RIAM-talin axis of integrin activation and blood cell
function. Blood. 128:479–487. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Paganini S, Guidetti GF, Catricalà S,
Trionfini P, Panelli S, Balduini C and Torti M: Identification and
biochemical characterization of Rap2C, a new member of the Rap
family of small GTP-binding proteins. Biochimie. 88:285–295. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liao X, Zhan W, Tian B, Luo Y, Gu F and Li
R: Circular RNA ZNF609 promoted hepatocellular carcinoma
progression by upregulating PAP2C expression via sponging
miR-342-3p. Onco Targets Ther. 13:7773–7783. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shen Z, Zhou R, Liu C, Wang Y, Zhan W,
Shao Z, Liu J, Zhang F, Xu L, Zhou X, et al: MicroRNA-105 is
involved in TNF-α-related tumor microenvironment enhanced
colorectal cancer progression. Cell Death Dis. 8:32132017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qiao Y, Peng C, Li J, Wu D and Wang X:
Spinal cord ischemia-reperfusion causes damage of neurocyte by
inhibiting RAP2C. Neurol Res. 39:877–884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu X, Qiu J, Zhang T, Yang Y, Guo S, Li
T, Jiang K, Zahoor A, Deng G and Qiu C: MicroRNA-188-5p promotes
apoptosis and inhibits cell proliferation of breast cancer cells
via the MAPK signaling pathway by targeting Rap2c. J Cell Physiol.
235:2389–2402. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen Y, Zheng Z, Wang J, Tang C, Khor S,
Chen J, Chen X, Zhang Z, Tang Q, Wang C, et al: Berberine
suppresses apoptosis and extracellular matrix (ECM) degradation in
nucleus pulposus cells and ameliorates disc degeneration in a
rodent model. Int J Biol Sci. 14:682–692. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ruan Z, Ma H, Li J, Liu H, Jia H and Li F:
The long non-coding RNA NEAT1 contributes to extracellular matrix
degradation in degenerative human nucleus pulposus cells. Exp Biol
Med (Maywood). 243:595–600. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang J, Li Z, Chen F, Liu H, Wang H, Li
X, Liu X, Wang J and Zheng Z: TGF-β1 suppresses CCL3/4 expression
through the ERK signaling pathway and inhibits intervertebral disc
degeneration and inflammation-related pain in a rat model. Exp Mol
Med. 49:e3792017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li S, Hua W, Wang K, Gao Y, Chen S, Liu W,
Song Y, Wu X, Tu J, Kang L, et al: Autophagy attenuates
compression-induced apoptosis of human nucleus pulposus cells via
MEK/ERK/NRF1/Atg7 signaling pathways during intervertebral disc
degeneration. Exp Cell Res. 370:87–97. 2018. View Article : Google Scholar : PubMed/NCBI
|