Introduction
Known as a common malignancy in the digestive
system, pancreatic carcinoma is a deadly disease. The incidence
rates in males and females were 14.9 and 11.6 cases per 100,000
people annually in 2013–2017, and the mortality rates were 12.7 and
9.6 deaths per 100,000 people annually, respectively (1). Although great progress has been made
in surgical resection, radiotherapy and chemotherapy (2,3),
the five-year overall survival rate for patients with pancreatic
carcinoma is only 8.3% (4). There
is an urgent requirement to find new diagnostic markers and
therapeutic targets for pancreatic carcinoma.
In recent years, novel functional molecules in
cancer biology have been identified. Among them, long non-coding
RNAs (lncRNAs) have been demonstrated to be crucial regulators in
tumorigenesis and cancer progression. LncRNAs have >200
nucleotides but possess no protein-coding ability (5). LncRNAs have important biological
functions and they participate in regulating tumor cell growth,
migration, invasion and other malignant biological behaviors
(5). Reportedly, a number of
lncRNAs have been shown to serve a role in pancreatic carcinoma
development as cancer-promoting factors or tumor suppressors, such
as DNAH17-AS1 (6), CASC2
(7) and SNHG16 (8). However, the roles and mechanisms of
lncRNAs in pancreatic carcinoma remain to be elucidated.
CTBP1 antisense RNA 2 (CTBP1-AS2) is a newly
discovered oncogenic lncRNA that is abnormally expressed in
hepatocellular cancer (9) and
endometrial cell cancer (10);
however, its role in pancreatic carcinoma is unclear. Through the
Gene Expression Profiling Interactive Analysis (GEPIA) database it
was found that CTBP1-AS2 was significantly highly expressed in
pancreatic carcinoma tumor tissues, suggesting that it may have
important regulatory functions in the tumorigenesis of pancreatic
cancer. The present study aimed to analyze the effects of CTBP1-AS2
on pancreatic carcinoma cell proliferation, apoptosis, migration
and invasion and to decipher the molecular mechanism of CTBP1-AS2
in pancreatic carcinoma progression. The present study, for the
first time, to the best of our knowledge, elucidated the functions
of CTBP1-AS2 in pancreatic carcinoma and provided a novel
therapeutic target for this disease.
Materials and methods
Tissue sample collection
The present study followed the Declaration of
Helsinki and was endorsed by the Ethics Committee of the People's
Hospital of Ningxia Hui Autonomous Region (approval no.
2021-10-003). The human tissue samples were obtained from the Human
Tissue Bank of People's Hospital of Ningxia Hui Autonomous Region.
All patients signed the informed consent before surgery. Tumor
tissues and para-cancerous tissues (≥3 cm away from the tumor
margin) of 30 patients with pancreatic carcinoma (age range, 53–68
years; 14 male and 16 female) between October2021 and November 2012
were collected during surgery from the People's Hospital of Ningxia
Hui Autonomous Region and immediately stored in liquid nitrogen at
−196°C. None of the subjects had undergone chemotherapy or
radiotherapy prior to the surgery.
Cell culture
Normal human pancreatic duct epithelial cell line
(HPDE6-C7), human pancreatic carcinoma cell lines (Hs766T, SW1990,
CAPAN-1) and 293T were obtained from the American Type Culture
Collection; the JF305 pancreatic carcinoma cell line was purchased
from Yuchi (Shanghai) Biological Technology Co., Ltd. (cat. no.
SCO423). All the cells were cultured in RPMI-1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 10% fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.) in
5% CO2 at 37°C.
Cell transfection
Hs766T and JF305 cells were inoculated into 6-well
plates at a density of 2×105 cells/well. Small
interfering (si)RNA oligonucleotides targeting CTBP1-AS2
(si-CTBP1-AS2-1, 5′-GAGATCTAAGAAAAAATTCCAGA-3′; si-CTBP1-AS2-2,
5′-GCGCGTTATCATGACTTCTATTT-3′), scrambled siRNA [negative control
siRNA (si-NC); 5′-TTCTCCGAACGTGTCACGTTT-3′], miR-141-3p mimic
(5′-UAACACUGUCUGGUAAAGAUGG-3′), miR-141-3p inhibitor
(5′-CCAUCUUUACCAGACAGUGUUA-3′), mimics NC
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′), inhibitor NC
(5′-CAGUACUUUUGUGUAGUACAAA-3′), pcDNA3.1 vector overexpressing
ubiquitin-specific protease 22 (USP22) and empty plasmids were
provided by Guangzhou RiboBio Co., Ltd. The transfection was
performed by Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instruction at 37°C and 5% CO2 for 6 h, with final
concentrations of 50 nM siRNAs, miRNA mimics and inhibitor, and 2
µg overexpression plasmid. Finally, cells were incubated at 37°C
for 48 h and harvested for the following study.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
TRIzol® (Thermo Fisher Scientific, Inc.)
was used to extract lncRNA or mRNA from cells (5×105)
according to the manufacturer's instructions. A mirVana miRNA
isolation kit (Ambion; Thermo Fisher Scientific, Inc.) was used to
extract miRNA from tissues and cell lines according to the
manufacturer's instructions. cDNA synthesis was conducted using the
Mir-X miRNA First-Strand Synthesis kit (Takara Biotechnology Co.,
Ltd.) for miR-141-3p and a PrimeScript RT reagent kit (Promega
Corporation) was adopted for CTBP1-AS2 and USP22 according to the
manufacturer's instructions. An ABI 7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for
performing qPCR with a SYBR Green Premix Ex Taq kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The reaction conditions were as follows: 95°C for 10
min; followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and
72°C for 30 sec. GAPDH and U6 served as internal references, and
the 2−ΔΔCq method (11) was used for calculating the data.
Primer sequences are listed in Table
I.
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| Transcript | Primer sequences
(5′-3′) |
|---|
| CTBP1-AS2 | F:
CAAGGGCACTCAAAGGGCTA |
|
| R:
CAGGCAGGCAAACACAGAAC |
| miR-141-3p | F:
CGCAGTAACACTGTCTGGT |
|
| R:
GTCCAGTTTTTTTTTTTTTTTCCATCT |
| USP22 | F:
GGCGGAAGATCACCACGTAT |
|
| R:
TTGTTGAGACTGTCCGTGGG |
| GAPDH | F:
TGCACCACCAACTGCTTAGC |
|
| R:
GGCATGGACTGTGGTCATGAG |
| U6 | F:
GCTTCGGCAGCACATATACTAAAAT |
|
| R:
CGCTTCACGAATTTGCGTGTCAT |
Cell Counting Kit-8 (CCK-8) assay
Pancreatic carcinoma cells were seeded into 96-well
plates (2×103 cells/well) and cultured for 0, 24, 36 and
72 h. At each time point 10 µl of CCK-8 solution (5 mg/ml; Beyotime
Institute of Biotechnology) was added, and the cells were incubated
for an additional 4 h. A microplate reader (Thermo-Fisher
Scientific, Inc.) was used to measure the absorbance at 450 nm,
which indicated the viability of the cells.
5-ethynyl-2-deoxyuridine (EdU)
incorporation assay
An EdU Labeling/Detection kit (Guangzhou RiboBio
Co., Ltd.) was used. The cells were inoculated into 96-well plates
(5×103 cells/well) for 24 h and then incubated with 50
mmol/l EdU solution in 5% CO2 at 37°C for 2 h. The cells
were then fixed for 30 min at room temperature with 4%
paraformaldehyde and incubated with glycine for 10 min at room
temperature. Next, 100 µl of PBS containing 0.5% TritonX-100 was
added into each well and the cells were incubated for 10 min at
room temperature. Then the cells were washed twice with PBS and
stained with 1X Apollo fluorochrome at room temperature for 30 min
in the dark. Next, the cell nuclei were stained by 1X DAPI staining
solution for 15 min at room temperature. A Nikon Eclipse Ti
fluorescence microscope (Nikon Corporation) was used to observe the
cells and to determine the percentage of EdU-positive cells, which
indicated the proliferative capacity.
Transwell assay
Transwell chambers (24-well; pore size of 8 µm;
Corning, Inc.) were used to evaluate cell invasion and migration.
Briefly, for the migration assay, pancreatic carcinoma cells in 200
µl FBS-free RPMI-1640 medium were transferred into the upper
compartment (1×105 cells/well) and the lower compartment
contained 600 µl of RPMI-1640 medium containing 10% FBS. After 24
h, the cells in the upper compartment were removed, the cells on
the lower surface of the filter were stained with 0.1% crystal
violet for 30 min at room temperature and the number of migrated
cells was counted under a microscope (Olympus Corporation). The
cell invasion assay was carried out following the same procedures
after the filter was precoated with 60 µl of Matrigel at 37°C for 2
h (BD Biosciences).
Flow cytometric analysis
Apoptosis was examined using an Annexin V-FITC
Apoptosis Detection kit (Beyotime Institute of Biotechnology,
Inc.). Briefly, 48 h after transfection, cells were harvested,
resuspended in binding buffer and incubated with 5 µl of Annexin
V-FITC and 10 µl of PI in the dark at room temperature for 15 min.
The apoptotic rate (early + late apoptotic cells) were analyzed by
flow cytometry using an Attune NxT flow cytometer (Thermo Fisher
Scientific, Inc.) and FlowJo software (version 10.0; BD
Biosciences) (12).
Luciferase reporter gene assay
Wild-type (WT) and mutant-type (MUT) CTBP1-AS2
luciferase reporter plasmids (CTBP1-AS2-WT and CTBP1-AS2-MUT) and
USP22 3′UTR luciferase reporter plasmids (USP22-3′UTR-WT and
USP22-3′UTR MUT) were obtained from Shanghai GenePharma Co., Ltd.
Subsequently, these luciferase reporters were co-transfected with
either miR-141-3p mimics or mimics NC into 293T cell line
(1×105 cells/well) by using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C. After 48
h, the luciferase activity of the cells was detected by the
Dual-Luciferase Reporter Assay System (Promega Corporation).
Firefly luciferase activity was normalized to Renilla
luciferase activity.
Western blot analysis
Proteins were extracted from pancreatic carcinoma
tissues and cell lines using RIPA lysis buffer (Sigma-Aldrich;
Merck KGaA), and a BCA Protein Assay kit (Thermo Fisher Scientific,
Inc.) was used for protein quantification. Proteins (30 µg/lane)
were separated using 10% SDS-PAGE (Sigma-Aldrich; Merck KGaA) and
then transferred to polyvinylidene difluoride membranes
(MilliporeSigma). After being blocked with 5% skimmed milk at 4°C
for 2 h, the membranes were incubated with primary antibodies
(anti-USP22; 1:2,000; cat. no. LS-C102769; LifeSpan BioSciences,
Inc.; anti-GAPDH; 1:1,000; ab9385; Abcam) at 4°C overnight and then
incubated with a HRP-conjugated secondary antibody (1:10,000;
ab6721; Abcam) for 1 h at 37°C. Amersham ECL Select Western
Blotting Detection Reagent (Cytiva) was used to visualize the
protein bands. ImageJ software 1.8.0 (National Institutes of
Health) was used to quantify the protein bands.
Bioinformatics analysis using GEPIA
and StarBase
The GEPIA database (http://gepia.cancer-pku.cn) is an interactive web
server for tumor gene expression analysis by using data from The
Cancer Genome Atlas Pan-Cancer project and Genotype-Tissue
Expression database. The GEPIA database was searched to analyze the
expression of CTBP1-AS2 in pancreatic carcinoma tissues and normal
pancreatic tissues. StarBase database (http://starbase.sysu.edu.cn) was used to search for
the miRNAs with potential binding sites for CTBP1-AS2 and mRNAs
that bind to miR-141-3p.
Statistical analysis
Each experiment was conducted at least three times,
and all data are expressed as mean ± standard deviation or median ±
interquartile range. SPSS 19.0 software (IBM Corp.) was used for
statistical analysis. Data were analyzed using unpaired or paired
Student's t-test, one-way ANOVA with Tukey's post hoc test,
Pearson's correlation analysis and χ2 test. P<0.05
was considered to indicate a statistically significant
difference.
Results
CTBP1-AS2 expression is significantly
elevated in pancreatic carcinoma tissues and is associated with
pathological characteristics
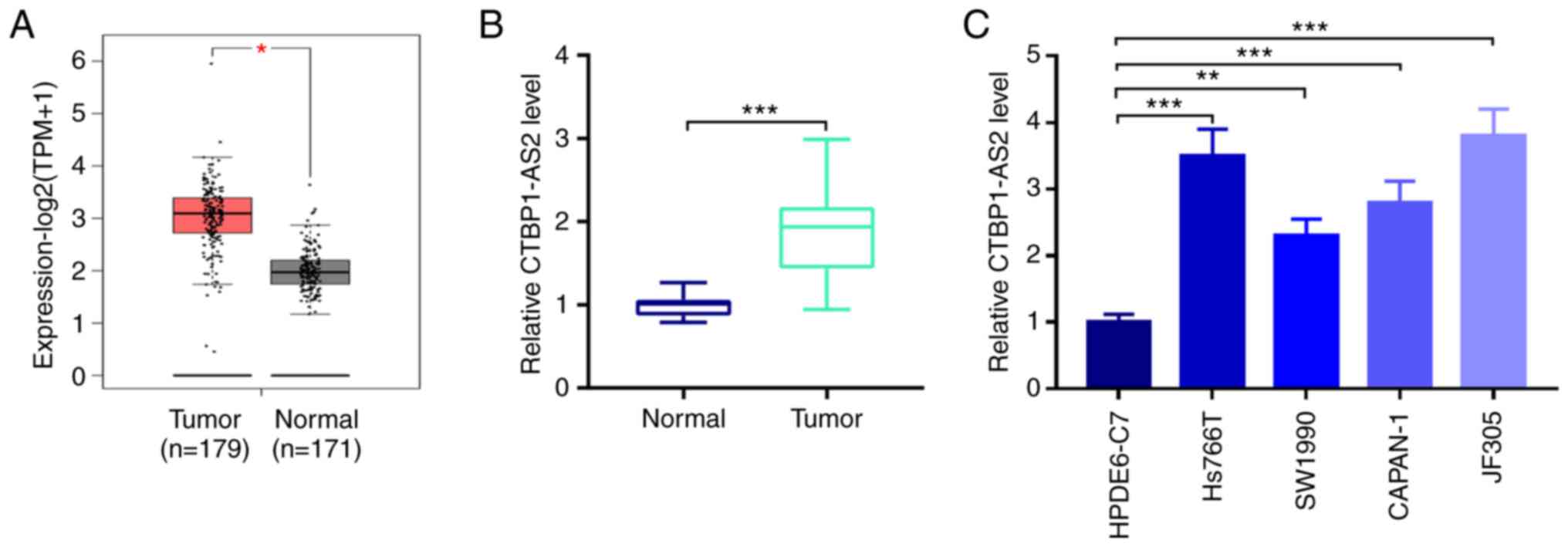
By searching the GEPIA database it was found that
CTBP1-AS2 expression in patients with pancreatic carcinoma tumor
tissues was significantly higher compared with that in normal
pancreatic tissues (Fig. 1A).
Similarly, RT-qPCR showed that CTBP1-AS2 expression was
significantly higher in patient tumor tissues compared with
para-carcinoma tissues (Fig. 1B).
Additionally, compared with HPDE6-C7 normal pancreatic duct
epithelial cells, CTBP1-AS2 expression was significantly higher in
pancreatic carcinoma cell lines, Hs766T, SW1990, CAPAN-1 and JF305
(Fig. 1C). Since CTBP1-AS2
exhibited the highest expression level in Hs766T and JF305 cells,
these two cell lines were selected to perform the subsequent
loss-of-function experiments.
To analyze the clinical significance of high
CTBP1-AS2 expression in pancreatic carcinoma, the relationship
between patient clinicopathological characteristics and CTBP1-AS2
expression levels were evaluated (Table II). Based on the median value of
CTBP1-AS2 expression level, 30 patients were divided into high and
low CTBP1-AS2 expression groups (n=15 patients/group). The results
showed that high CTBP1-AS2 expression was associated with lymph
node metastasis and advanced clinical stage of patients with
pancreatic carcinoma. Together, these data indicated that CTBP1-AS2
may be a crucial regulatory factor for pancreatic carcinoma
development and may serve a cancer-promoting role.
 | Table II.Relationship between CTBP1-AS2
expression and clinicopathological characteristics of patients with
pancreatic carcinoma. |
Table II.
Relationship between CTBP1-AS2
expression and clinicopathological characteristics of patients with
pancreatic carcinoma.
|
|
| CTBP1-AS2 |
|
|---|
|
|
|
|
|
|---|
| Characteristic | Cases (n=30) | High (n=15) | Low (n=15) | P-value |
|---|
| Age, years |
|
|
|
|
|
≥60 | 17 | 10 | 7 | 0.269 |
|
<60 | 13 | 5 | 8 |
|
| Sex |
|
|
|
|
|
Male | 14 | 6 | 8 | 0.464 |
|
Female | 16 | 9 | 7 |
|
| Clinical stage |
|
|
|
|
|
III–IV | 14 | 10 | 4 | 0.028a |
|
I–II | 16 | 5 | 11 |
|
| Lymph node
metastasis |
|
|
|
|
|
Yes | 14 | 11 | 3 | 0.003a |
| No | 16 | 4 | 12 |
|
CTBP1-AS2 promotes pancreatic
carcinoma cell proliferation, migration and invasion and inhibited
cell apoptosis
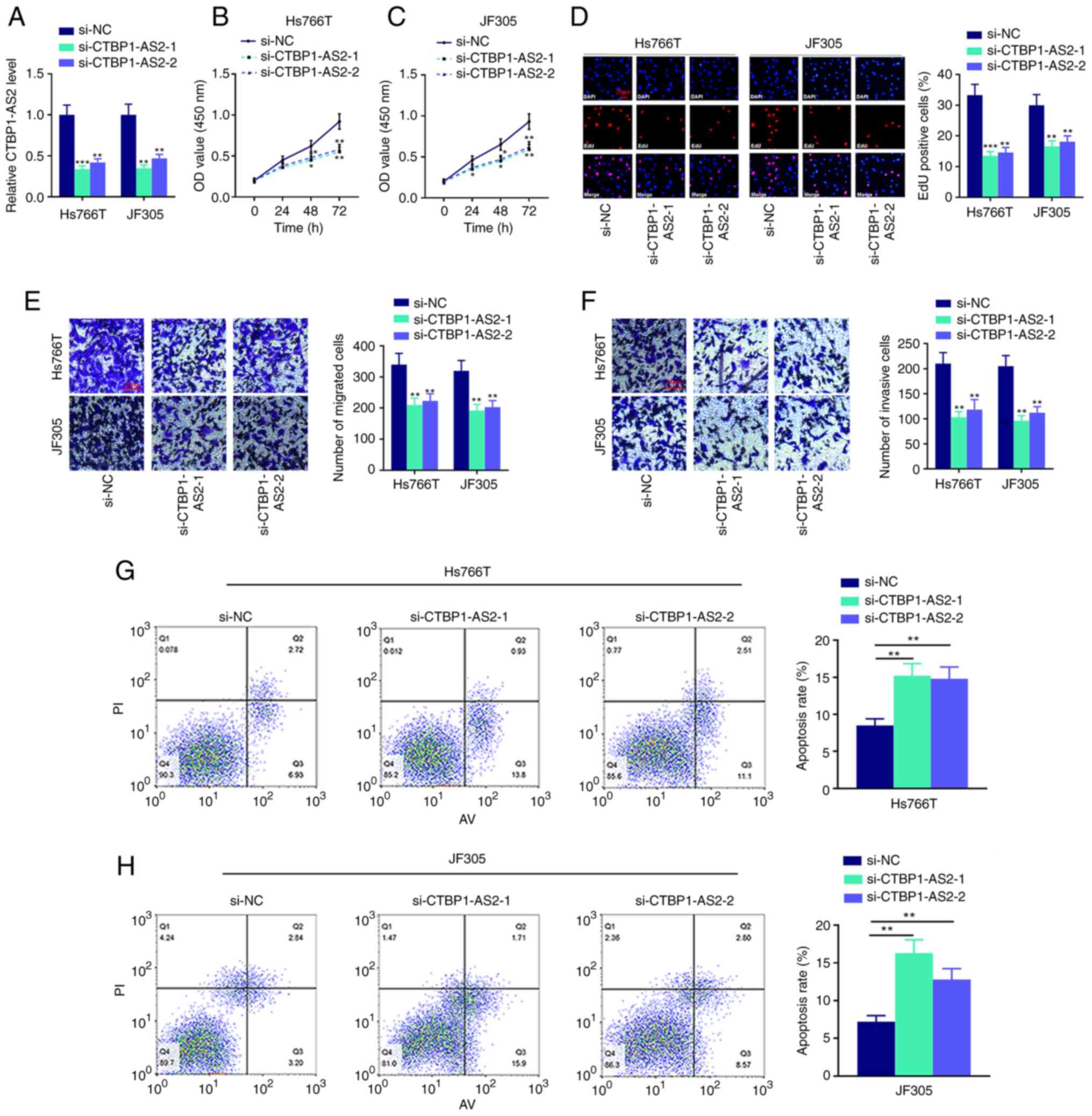
To examine the biological functions of CTBP1-AS2 in
pancreatic carcinoma cells, two CTBP1-AS2 siRNAs (si-CTBP1-AS2-1
and si-CTBP1-AS2-2) were used to knock down CTBP1-AS2 expression in
Hs766T and JF305 cell lines. It was demonstrated that CTBP1-AS2
expression levels in both CTBP1-AS2 knockdown groups were
significantly reduced compared with the si-NC group (Fig. 2A). CCK-8 and EdU assays confirmed
that the cell proliferation was significantly inhibited after
CTBP1-AS2 knockdown (Fig. 2B-D).
Transwell and Matrigel assays showed that cell migration and
invasion, respectively, were notably suppressed after CTBP1-AS2
knockdown (Fig. 2E and F). It was
also revealed that Hs766T and JF305 cells transfected with
si-CTBP1-AS2 had higher apoptosis rates compared with cells
transfected with si-NC (Fig. 2G and
H). These results suggested that knocking down CTBP1-AS2 could
inhibit the malignant phenotypes of pancreatic carcinoma cells.
CTBP1-AS2 targets miR-141-3p
LncRNAs can take part in tumor progression as
competing endogenous RNAs (ceRNAs), which means that lncRNAs can
adsorb miRNAs like molecular sponges to regulate the expression
levels of mRNAs (13). To further
pinpoint the mechanism by which CTBP1-AS2 serves a role in
pancreatic carcinoma, StarBase database was searched and it was
found that CTBP1-AS2 likely binds with miR-141-3p (Fig. 3A). RT-qPCR results demonstrated
that miR-141-3p expression was significantly reduced in tumor
tissues and pancreatic carcinoma cell lines compared with that in
the respective para-cancerous tissues and normal human pancreatic
duct epithelial cell line (Fig. 3B
and C). Furthermore, CTBP1-AS2 expression in pancreatic
carcinoma tumor tissues was inversely correlated with miR-141-3p
expression (Fig. 3D), and
CTBP1-AS2 knockdown markedly promoted miR-141-3p expression in
Hs766T and JF305 cell lines compared with si-NC (Fig. 3E). Subsequently, miR-141-3p
expression was overexpressed or knocked down in Hs766T and JF305
cells by transfecting with miR-141-3p mimics or miR-141-3p
inhibitor, respectively. The results showed that miR-141-3p mimics
significantly increased the expression of miR-141-3p compared with
mimics NC and miR-141-3p inhibitor decrease the miR-141-3p
expression compared with the inhibitor NC (Fig. 3F). No significant difference in
miR-141-3p expression was observed between mimic NC and inhibitor
NC, so mimic NC was selected as the control of miR-141-3p mimics or
miR-141-3p inhibitor in the following study. Dual-luciferase
reporter assay results showed that miR-141-3p mimics could
significantly reduce CTBP1-AS2-WT luciferase activity but failed to
affect the luciferase activity of CTBP1-AS2-MUT, which verified the
targeting relationship between CTBP1-AS2 and miR-141-3p (Fig. 3G). To determine whether CTBP1-AS2
could serve a role in pancreatic carcinoma through miR-141-3p,
rescue assays were performed. The results showed that the effects
of CTBP1-AS2 knockdown on pancreatic carcinoma cell proliferation,
migration, invasion and apoptosis could be partially counteracted
by inhibiting miR-141-3p (Fig.
3H-M and Fig. S1). These
results displayed that CTBP1-AS2 exerted a cancer-promoting effect
by repressing miR-141-3p expression in pancreatic carcinoma.
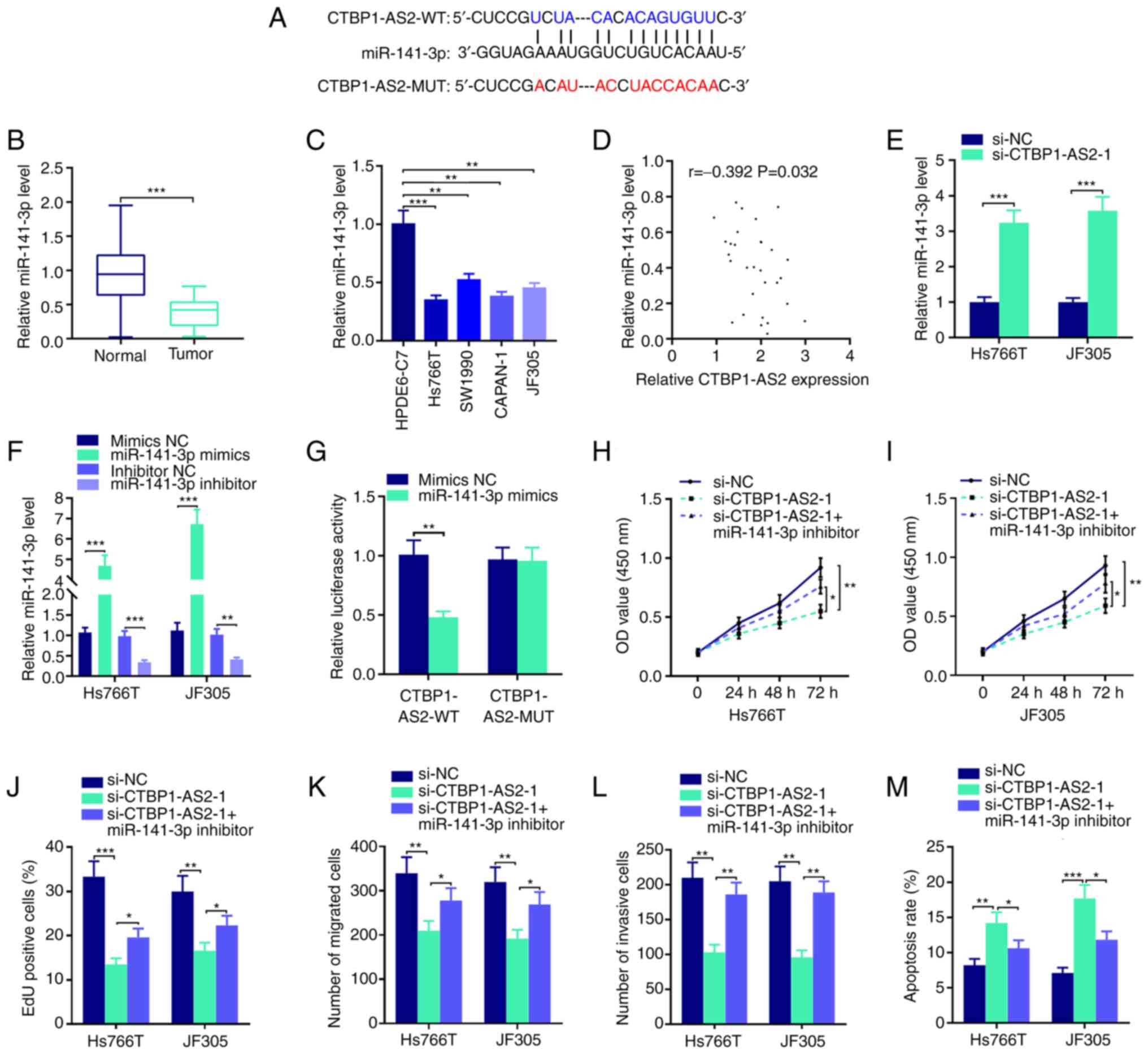 | Figure 3.CTBP1-AS2 targets miR-141-3p to
promote pancreatic carcinoma progression. (A) StarBase database
showed that CTBP1-AS2 sequence contained the binding site
complementary to miR-141-3p. RT-qPCR was performed to detect the
expression level of miR-141-3p in pancreatic carcinoma (B) tissues
and (C) cell lines. (D) Pearson's correlation analysis of CTBP1-AS2
and miR-141-3p expression levels in pancreatic carcinoma tissues.
(E) RT-qPCR was used to examine miR-141-3p expression levels in
Hs766T and JF305 cell lines following the transfection of
siCTBP1-AS2-1. (F) RT-qPCR was used to examine miR-141-3p
expression in Hs766T and JF305 cell lines following the
transfection of miR-141-3p mimics or miR-141-3p inhibitor. (G)
Dual-luciferase reporter assay was used to verify the inaction
between miR-141-3p and CTBP1-AS2. CCK-8 assay was conducted to
examine proliferation of (H) Hs766T and (I) JF305 cell lines
following the co-transfection of si-CTBP1-AS2-1 and miR-141-3p
inhibitor. (J) EdU assay was also used to examine proliferation in
Hs766T and JF305 cell line following the co-transfection of
si-CTBP1-AS2-1 and miR-141-3p inhibitor. Transwell and Matrigel
assays were performed to detect (K) migratory and (L) invasive
abilities, respectively, of Hs766T and JF305 cell lines following
the co-transfection of si-CTBP1-AS2-1 and miR-141-3p inhibitor. (M)
Flow cytometry was performed to detect the apoptosis of Hs766T and
JF305 cells following the co-transfection of si-CTBP1-AS2-1 and
miR-141-3p inhibitor. *P<0.05 vs. si-CTBP1-AS2-1, **P<0.01
vs. HPDE6-C7, inhibitor NC, mimics NC, si-NC or si-CTBP1-AS2-1.
***P<0.001 vs. Normal, HPDE6-C7, si-NC or mimics NC.
CTBP1-AS2-1, CTBP1 antisense RNA 2 siRNA-1; EdU,
5-ethynyl-2-deoxyuridine; miR, microRNA; MUT, mutant; NC, negative
control; RT-qPCR, reverse transcription-quantitative PCR; si, short
interfering RNA; WT, wild-type. |
miR-141-3p directly targets USP22
Putative downstream mRNAs interacting with
miR-141-3p were also investigated. StarBase database analysis
identified a number of potential targets of miR-141-3p, such as E2F
transcription factor 3, cyclin G1 and ring finger protein 38. Among
them, USP22 was selected in a previous study which demonstrated
that USP22 served as a cancer-promoting gene in the progression of
pancreatic cancer (14) (Fig. 4A). Results from the
dual-luciferase reporter assay showed that miR-141-3p mimics
inhibited USP22-WT luciferase activity but did not change USP22-MUT
luciferase activity (Fig. 4A). In
addition, miR-141-3p inhibitor was transfected into Hs766T and
JF305 cell lines and it was found that USP22 mRNA and protein
expression levels were significantly decreased following the
transfection compared with the control group (Fig. 4B). Notably, USP22 expression
levels were significantly enhanced in pancreatic carcinoma cell
lines and tumor tissues (Fig. 4C and
D, respectively). miR-141-3p expression was negatively
correlated with USP22 mRNA expression (Fig. 4E), and CTBP1-AS2 expression was
positively correlated with USP22 mRNA expression in pancreatic
carcinoma tissues (Fig. 4F).
These findings revealed that miR-141-3p targeted and downregulate
USP22 expression in pancreatic carcinoma.
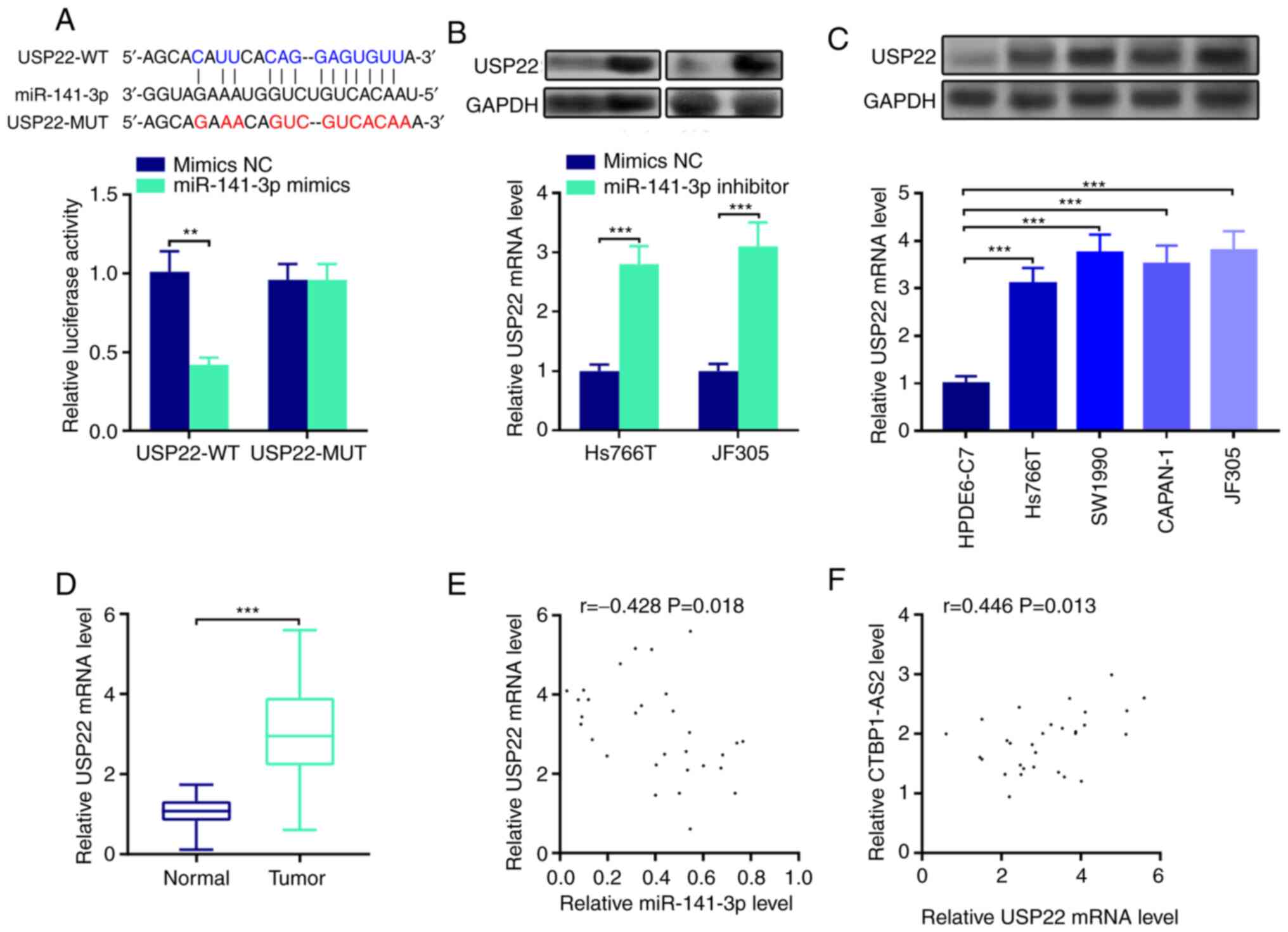 | Figure 4.miR-141-3p directly targets USP22. (A)
StarBase database search indicated that the 3′UTR of USP22 mRNA
contained a target site for miR-141-3p, which was confirmed by
luciferase reporter assay. (B) Western blotting (top) and RT-qPCR
(bottom) were conducted to detect the expression levels of USP22
protein and mRNA, respectively, in pancreatic carcinoma cells
transfected with miR-141-3p inhibitor. (C) Western blotting (top)
and RT-qPCR (bottom) were used to detect USP22 protein and mRNA
expression, respectively, in pancreatic carcinoma cell lines. (D)
RT-qPCR was used to detect USP22 expression in pancreatic tumor
tissues. Pearson's correlation analysis of (E) USP22 mRNA and
miR-141-3p expression levels and (F) USP22 mRNA and CTBP1-AS2
expression levels in pancreatic carcinoma tissues. **P<0.01 vs.
mimics NC and ***P<0.001 vs. mimics NC, HPDE6-C7 or Normal.
CTBP1-AS2, CTBP1 antisense RNA 2; miR, microRNA; MUT, mutant;
RT-qPCR, reverse transcription-quantitative PCR; NC, negative
control; USP22, ubiquitin-specific protease 22; WT, wild-type. |
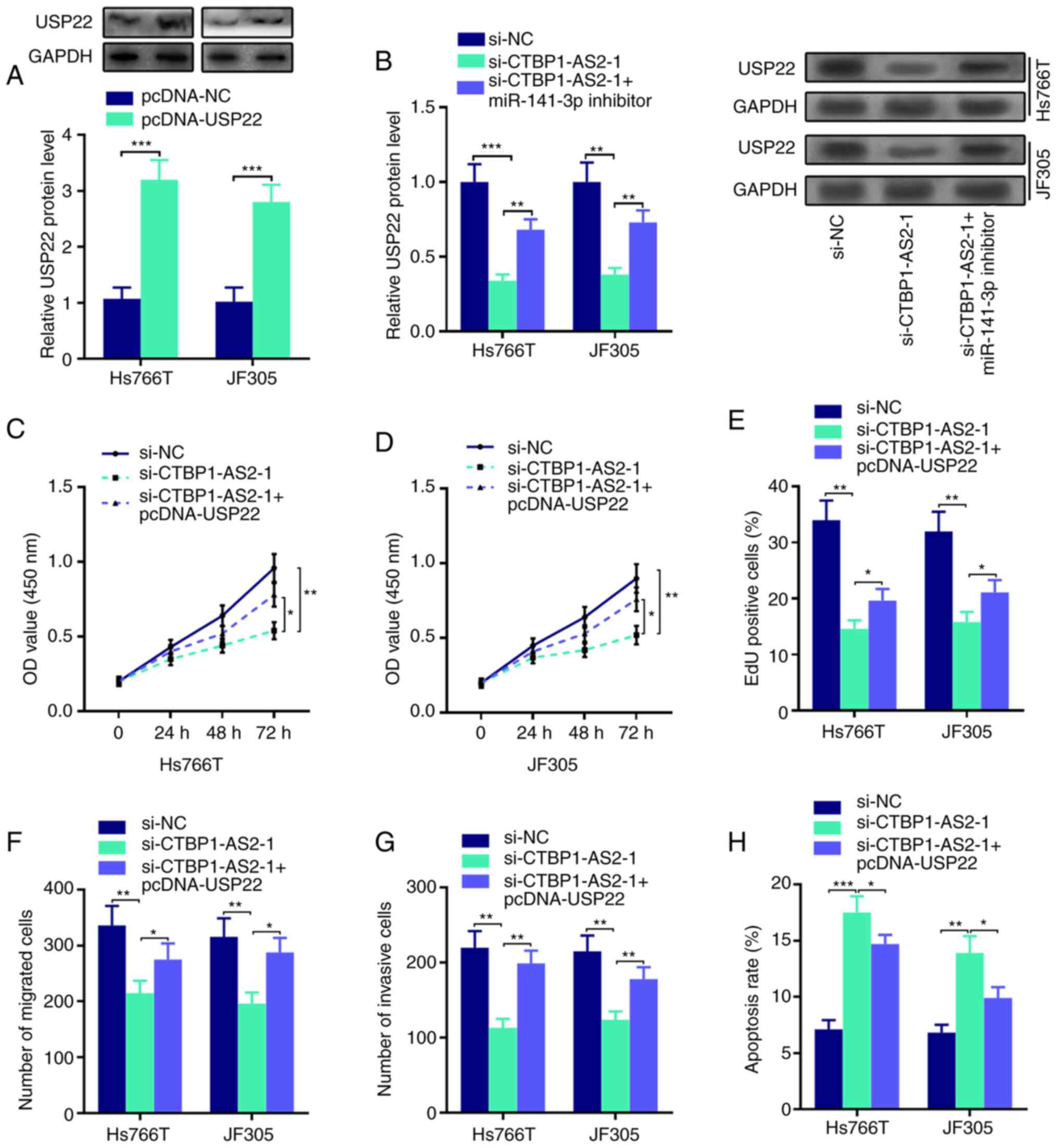
CTBP1-AS2 upregulates USP22 expression
through miR-141-3p inhibition
Western blot assay results showed that USP22 protein
expression was upregulated in Hs766T and JF305 cells transfected
with USP22 overexpression plasmid compared with the control
(Fig. 5A). In addition, the
inhibitory effects of CTBP1-AS2 knockdown on USP22 protein
expression could be abolished by co-transfection with miR-141-3p
inhibitor (Fig. 5B). In addition,
the transfection of USP22 overexpression plasmid could reverse
CTBP1-AS2 knockdown-mediated inhibitory effects on the
proliferation, migration and invasion of Hs766T and JF305 cells
(Fig. 5C-H; Fig. S2). These results suggested that
CTBP1-AS2 may serve a cancer-promoting role through regulating the
miR-141-3p/USP22 axis.
Discussion
The morbidity and mortality rates of pancreatic
carcinoma both increased by an average of 0.3% per year during the
past decade (1), so it is
necessary to clarify the mechanism of pancreatic carcinoma
tumorigenesis to find new therapeutic targets (15). The present study demonstrated that
CTBP1-AS2 expression was upregulated in pancreatic carcinoma
tissues and cell lines, and high CTBP1-AS2 expression was
associated with lymph node metastasis and the advanced clinical
stage of patients with pancreatic carcinoma. CTBP1-AS2 expression
increased pancreatic carcinoma cell proliferation, migration and
invasion and repressed apoptosis by regulating miR-141-3p/USP22
axis.
LncRNAs participate in the pathogenesis of a number
of cancers, including pancreatic carcinoma (16). Previous studies have reported the
role of CTBP1-AS2 in cancers and other diseases. Specifically,
CTBP1-AS2 can facilitate hepatocellular carcinoma cell
proliferation by regulating the miR-623/Cyclin D1 axis (9). In gastric cancer, CTBP1-AS2 promotes
cells proliferation and metastasis and suppresses apoptosis by
regulating the miR-139-3p/MMP11 axis (17). CTBP1-AS2 expression is upregulated
in osteoarthritis and increases the methylation of miR-130a gene to
suppress the proliferation of chondrocytes (18). CTBP1-AS2 regulates the
epithelial-mesenchymal transition (EMT) of glioma by modulating the
miR-370-3p/Wnt7a axis (19).
Additionally, in ovarian cancer, CTBP1-AS2 overexpression results
in the reduced proliferation rate of cancer cells through the
miR-216a/PTEN axis (20). In
cervical cancer, CTBP1-AS2 facilitates cervical cancer progression
by sponging miR-3163 to upregulate ZNF217 expression (21). The present study demonstrated that
CTBP1-AS2 expression was significantly higher in pancreatic
carcinoma tumor tissues compared with para-cancerous tissues, and
high CTBP1-AS2 expression was associated with the unfavorable
pathological indices of the patients. Functional experiments
verified that knocking down CTBP1-AS2 could significantly inhibit
pancreatic carcinoma cell proliferation, migration and invasion and
induce cell apoptosis. These results displayed that CTBP1-AS2 may
serve a tumor-promoting function in pancreatic carcinoma
progression.
LncRNAs can adsorb miRNAs like sponges and therefore
indirectly regulate downstream mRNA expression, by which they serve
a role in regulating biological processes (13). There are also a number of
lncRNA-miRNA-mRNA regulatory networks in pancreatic carcinoma. For
instance, lncRNA DNAH17-AS1 upregulates PPME1 expression through
decoying miR-432-5p to promote pancreatic carcinoma cell
proliferation, migration and invasion (6). Another study reported that CASC2
induces the expression of PTEN to suppress pancreatic carcinoma
cell metastasis by sponging miR-21 (7). The present study found that
miR-141-3p was the target of CTBP1-AS2, whose expression could be
negatively regulated by CTBP1-AS2. miR-141-3p expression is
reported to be decreased and act as a tumor suppressor in several
types of tumors. For example, miR-141-3p expression is
downregulated in osteosarcoma; miR-141-3p targets FUS to degrade
L-lactate dehydrogenase B chain to reduce the degree of malignancy
of osteosarcoma (22). In
addition, miR-141-3p suppresses colorectal cancer cell
proliferation, migration and invasion by targeting TNF
receptor-associated factor 5 (23). A recent study reported that in
pancreatic carcinoma, miR-141-3p expression is significantly
reduced (24), which is
consistent with the findings of the present study, and that XIST
upregulates TGF-β2 expression by targeting miR-141-3p, therefore
promoting pancreatic carcinoma cell proliferation, migration and
invasion. Another study demonstrated that miR-141-3p can directly
target MAP4K4 to repress pancreatic carcinoma cell invasion
(25). The present study showed
that miR-141-3p expression in pancreatic carcinoma tissues was
negatively correlated with CTBP1-AS2 expression and that inhibition
of miR-141-3p counteracted the effects of CTBP1-AS2 knockdown on
regulating pancreatic carcinoma cell proliferation, migration,
invasion and apoptosis. It was suggested that CTBP1-AS2 may serve a
role in pancreatic carcinoma by sponging miR-141-3p.
In the present study, USP22 was identified as one of
the targets of miR-141-3p. USP22 is a member of the
deubiquitinating enzyme family; it can regulate cell metabolism and
cell cycle, and it is also involved in the pathogenesis of a number
of human diseases (26). In
gastric cancer, USP22 expression is significantly upregulated; it
can promote cancer cell proliferation and inhibit apoptosis through
regulating son of sevenless 1/RAS protein axis (27). In retinoblastoma, USP22 knockdown
promotes the senescence and apoptosis of retinoblastoma cells by
regulating telomerase reverse transcriptase/p53 pathway (28). Importantly, in pancreatic
carcinoma, USP22 is overexpressed and promotes migration, invasion
and EMT of pancreatic carcinoma cells through the focal adhesion
kinase pathway (14).
Furthermore, USP22 knockdown in pancreatic carcinoma cells can
promote the infiltration of T cells and natural killer cells in the
tumor microenvironment (29). The
present study revealed that USP22 expression was markedly elevated
in pancreatic carcinoma cell lines and tumor tissues. Notably,
CTBP1-AS2 could upregulate USP22 expression by repressing
miR-141-3p expression. The rescue assays showed that the inhibiting
effects of CTBP1-AS2 knockdown on pancreatic carcinoma cell
proliferation, migration, invasion and apoptosis could be reversed
by USP22 overexpression. Collectively, these findings validated the
hypothesis that CTBP1-AS2 could function as a ceRNA to facilitate
pancreatic carcinoma cell proliferation, migration and invasion by
adsorbing miR-141-3p and upregulating USP22 expression.
Collectively, the present study revealed that
CTBP1-AS2 was an oncogenic lncRNA by regulating miR-141-3p/USP22
axis in pancreatic cancer. The current study may provide an
improved understanding of the lncRNA-miRNA-mRNA ceRNA network in
pancreatic cancer development. However, there were some limitations
to the study. For example, the function of
CTBP1-AS2/miR-141-3p/USP22 in pancreatic cancer needs to be
verified with in vivo experiment. In addition, the
downstream signaling pathway of USP22 in pancreatic cancer also
needs to be investigated in the future.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
MZ, SM and JM made substantial contributions to the
design of the study and wrote the manuscript. MZ, SM and XL
collected samples and analyzed the data. MZ, SM, XL, HY, YT, JH and
XW performed the experiments. MZ and SM confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study followed the Declaration of
Helsinki and was endorsed by the Ethics Committee of the People's
Hospital of Ningxia Hui Autonomous Region (approval no.
2021-10-003). The human tissue samples were obtained from the Human
Tissue Bank of People's Hospital of Ningxia Hui Autonomous Region.
All patients signed the informed consent before surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Grossberg AJ, Chu LC, Deig CR, Fishman EK,
Hwang WL, Maitra A, Marks DL, Mehta A, Nabavizadeh N, Simeone DM,
et al: Multidisciplinary standards of care and recent progress in
pancreatic ductal adenocarcinoma. CA Cancer J Clin. 70:375–403.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang P, Wang J, Tan H, Weng S, Cheng L,
Zhou Z and Wen S: Acid- and reduction-sensitive micelles for
improving the drug delivery efficacy for pancreatic cancer therapy.
Biomater Sci. 6:1262–1270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sheng J, Wang L, Han Y, Chen W, Liu H,
Zhang M, Deng L and Liu YN: Dual roles of protein as a template and
a sulfur provider: A general approach to metal sulfides for
efficient photothermal therapy of cancer. Small. 14:Nov
17–2017.doi: 10.1002/smll.201702529. PubMed/NCBI
|
|
4
|
Batista IA and Melo SA: Exosomes and the
future of immunotherapy in pancreatic cancer. Int J Mol Sci.
20:5672019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan JJ and Tay Y: Noncoding RNA: RNA
regulatory networks in cancer. Int J Mol Sci. 19:13102018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu T, Lei T, Li SQ, Mai EH, Ding FH and
Niu B: DNAH17-AS1 promotes pancreatic carcinoma by increasing PPME1
expression via inhibition of miR-432-5p. World J Gastroenterol.
26:1745–1757. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang H, Feng X, Zhang M, Liu A, Tian L,
Bo W, Wang H and Hu Y: Long non-coding RNA CASC2 upregulates PTEN
to suppress pancreatic carcinoma cell metastasis by downregulating
miR-21. Cancer Cell Int. 19:182019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo JQ, Yang ZJ, Wang S, Wu ZZ, Yin LL and
Wang DC: LncRNA SNHG16 functions as an oncogene by sponging
miR-200a-3p in pancreatic cancer. Eur Rev Med Pharmacol Sci.
24:72182020.PubMed/NCBI
|
|
9
|
Wang M and Zhao H: LncRNA CTBP1-AS2
promotes cell proliferation in hepatocellular carcinoma by
regulating miR-623/Cyclin d1 axis. Cancer Biother Radiopharm.
35:765–770. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang QA, Yang Y and Liang X: LncRNA
CTBP1-AS2 sponges miR-216a to upregulate PTEN and suppress
endometrial cancer cell invasion and migration. J Ovarian Res.
13:372020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tang M, Xie X, Shi M, Xin W, Zheng G,
Zhang Y, Zhang Z and Lian X: Antileukemic effect of caffeic acid
3,4-dihydroxyphenetyl ester. Evidences for its mechanisms of
action. Phytomedicine. 80:1533832021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ning Z, Wang A, Liang J, Xie Y, Liu J, Yan
Q and Wang Z: USP22 promotes epithelial-mesenchymal transition via
the FAK pathway in pancreatic cancer cells. Oncol Rep.
32:1451–1458. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mattiuzzi C and Lippi G: Cancer
statistics: A comparison between World Health Organization (WHO)
and global burden of disease (GBD). Eur J Public Health.
30:1026–1027. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Weng W, Zhang Z, Huang W, Xu X, Wu B, Ye
T, Shan Y, Shi K and Lin Z: Identification of a competing
endogenous RNA network associated with prognosis of pancreatic
adenocarcinoma. Cancer Cell Int. 20:2312020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y, Gao M, Li Y, Li M and Ma Q: LncRNA
CTBP1-AS2 facilitates gastric cancer progression via regulating the
miR-139-3p/MMP11 axis. Onco Targets Ther. 13:11537–11547. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang H, Li J, Shao W and Shen N: LncRNA
CTBP1-AS2 is upregulated in osteoarthritis and increases the
methylation of miR-130a gene to inhibit chondrocyte proliferation.
Clin Rheumatol. 39:3473–3478. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Zong J and Zhao C: lncRNA CTBP1-AS2
promotes proliferation and migration of glioma by modulating
miR-370-3p-Wnt7a-mediated epithelial-mesenchymal transition.
Biochem Cell Biol. 98:661–668. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui K and Zhu G: LncRNA CTBP1-AS2
regulates miR-216a/PTEN to suppress ovarian cancer cell
proliferation. J Ovarian Res. 13:842020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang S, Shi F, Du Y, Wang Z, Feng Y, Song
J, Liu Y and Xiao M: Long non-coding RNA CTBP1-AS2 enhances
cervical cancer progression via up-regulation of ZNF217 through
sponging miR-3163. Cancer Cell Int. 20:3432020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L: MiR-141-3p overexpression
suppresses the malignancy of osteosarcoma by targeting FUS to
degrade LDHB. Biosci Rep. 40:BSR201934042020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang Z, Li X, Liu S, Li C, Wang X and
Xing J: MiR-141-3p inhibits cell proliferation, migration and
invasion by targeting TRAF5 in colorectal cancer. Biochem Biophys
Res Commun. 514:699–705. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun J and Zhang Y: LncRNA XIST enhanced
TGF-β2 expression by targeting miR-141-3p to promote pancreatic
cancer cells invasion. Biosci Rep. 39:BSR201903322019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao G, Wang B, Liu Y, Zhang JG, Deng SC,
Qin Q, Tian K, Li X, Zhu S, Niu Y, et al: MiRNA-141, downregulated
in pancreatic cancer, inhibits cell proliferation and invasion by
directly targeting MAP4K4. Mol Cancer Ther. 12:2569–2580. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Melo-Cardenas J, Zhang Y, Zhang DD and
Fang D: Ubiquitin-specific peptidase 22 functions and its
involvement in disease. Oncotarget. 7:44848–44856. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lim C, Xu JC, Chen TY, Xu JX, Chen WF, Hu
JW, Li QL and Zhang YQ: Ubiquitin-specific peptide 22 acts as an
oncogene in gastric cancer in a son of sevenless 1-dependent
manner. Cancer Cell Int. 20:452020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou D, Liu P, Sun DW, Chen ZJ, Hu J, Peng
SM and Liu YL: USP22 down-regulation facilitates human
retinoblastoma cell aging and apoptosis via inhibiting TERT/P53
pathway. Eur Rev Med Pharmacol Sci. 21:2785–2792. 2017.PubMed/NCBI
|
|
29
|
Li J, Yuan S, Norgard RJ, Yan F, Yamazoe
T, Blanco A and Stanger BZ: Tumor Cell-Intrinsic USP22 suppresses
antitumor immunity in pancreatic cancer. Cancer Immunol Res.
8:282–291. 2020. View Article : Google Scholar : PubMed/NCBI
|