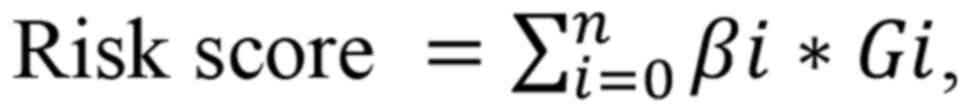Introduction
Gastric cancer (GC) has the fifth-highest incidence
among all cancer types and the third-highest cancer-associated
mortality rate globally (1). The
incidence of GC accounts for the second-highest cancer incidence in
China. In 2015, ~679,100 new cases and 498,000 deaths were
recorded, causing a considerable burden to society (2). As most individuals are already at
the advanced stage when they are diagnosed and based on the
incidences of chemotherapy resistance and recurrence, the overall
five-year OS of patients is <25% (3). Therefore, identifying its underlying
pathogenic mechanism and detecting novel and reliable potential
therapeutic targets is essential to enhance the prognosis of
individuals with GC.
The roles of ferroptosis in tumors have attracted
increasing attention recently. Ferroptosis is an iron-dependent
form of programmed cell death that is different from apoptosis,
cell necrosis and autophagy (4).
The primary mechanism of ferroptosis depends on the action of ester
oxygenase or divalent iron, which catalyzes lipid peroxidation of
unsaturated fatty acids, hence triggering cell death. Besides, it
also functions in the antioxidant system (the glutathione system)
to regulate the reduction of the core enzyme, phospholipid
hydroperoxide glutathione peroxidase 4 (GPX4) (5,6).
An increasing number of studies have indicated that long noncoding
RNAs (lncRNAs) are able to regulate ferroptosis and mediate
biological behavior in various tumors. Zhang et al (7) documented that lncRNA OIP5-antisense
1 (AS1) induced ferroptosis resistance and promoted prostate cancer
progression. Ma et al (8)
proved that the lncRNA MEG8 repressed proliferation and induced
ferroptosis in hemangioma endothelial cells. The lncRNA long
intergenic RNA (LINC)00618 was reported to accelerate ferroptosis
via increasing the contents of lipid reactive oxygen species and
iron in human leukemia (9).
However, there is still a lack of research that systematically
assesses ferroptosis-related lncRNA signatures and explains their
relationship with overall survival (OS) and progression-free
survival (PFS) in patients with stomach adenocarcinoma (STAD).
In the present study, two signatures of
differentially expressed ferroptosis-related lncRNAs were
established to evaluate OS and PFS prognosis based on The Cancer
Genome Atlas (TCGA) data. Furthermore, experiments were conducted
to validate the influence of a unique overlapping lncRNA, namely
lncRNA associated with spliceosome associated factor 3, U4/U6
recycling protein (SART3) regulation of splicing (LASTR) of the
signatures for PFS and OS in GC.
Materials and methods
Data collection
RNA-sequencing data of 407 patients (32
non-malignant and 375 tumors) were downloaded from the TCGA-STAD
data resource (https://portal.gdc.cancer.gov/). TCGA constitutes a
publicly funded project whose purpose includes cataloging and
discovering significant cancer-pathogenesis genome changes in large
datasets of >30 human cancer types via large-scale genome
sequencing along with integrated multidimensional analyses. The
matching TCGA clinical data were obtained from cBioPortal
(http://www.cbioportal.org/) (10). The matching ferroptosis-related
genes were abstracted from FerrDb (http://www.zhounan.org/ferrdb/) (11), an online consortium providing
comprehensive and up-to-date data resources for ferroptosis-related
biomarkers, their modulatory molecules and diseases.
Profiling differentially expressed
ferroptosis-related lncRNAs (DEFRLs)
To determine ferroptosis-related lncRNAs, to limma R
tool was employed to perform differential analyses for the STAD
samples from TCGA. Significant differences in expression were
determined using a false discovery rate <0.05 and |log2(fold
change)|≥1 as the threshold. Pearson correlation analysis was
adopted to assess the relationship of the lncRNAs with ferroptosis
markers. A correlation coefficient of |R2|>0.45 at
P<0.05 was considered to indicate a significant correlation.
Functional enrichment analysis of
DEFRLs
The clusterprofiler R tool was employed to perform
Gene Ontology (GO) coupled with Kyoto Encyclopedia of Genes and
Genomes (KEGG) functional analysis to elucidate the role of
unrecovered DEFRLs (12). An
adjusted P<0.05 denoted statistical significance.
Development of the ferroptosis-related
lncRNA prognostic signatures
OS and PFS were analyzed to gain insight into the
prognostic significance of DEFRLs in individuals with STAD. OS was
defined as the time from the first day of diagnosis until death
from any cause, while PFS included the time from the day of
diagnosis to cancer progression or death. First, a univariate Cox
analysis was adopted to explore OS- and PFS-related DEFRLs.
Furthermore, multivariate Cox regression was employed to determine
the potential OS- and PFS-related DEFRLs to create two predictive
signatures, the OS and PFS signatures, respectively. The DEFRLs'
coefficients in the final signatures were validated simultaneously
and utilized to compute the risk scores for each STAD patient. And
all subjects were stratified into either low-risk or high-risk
groups, as per the median score. The risk score was calculated as
follows:
where βi is the coefficient of lncRNA
i in the multivariate Cox analysis; Gi is the
expression value of lncRNA i; and n is the number of
lncRNAs in the signature.
To explore the efficiency of the signatures,
receiver operating characteristic (ROC) analysis was performed. The
‘survival ROC’ tool was employed to create ROC curves at 1, 3 and 5
years and the matching time-based areas under the curves (AUCs)
were computed. Furthermore, the Kaplan-Meier (K-M) survival plots
were generated and the log-rank test was used to assess the
differences in OS and PFS between the high- and low-risk
groups.
Predictive nomogram integrating DEFRL
signatures and clinical variables
Clinical characteristics, including sex, age and
grade, were abstracted from the cBioPortal data resource.
Univariate Cox regression integrating the signature with the
clinical information was performed for individuals with STAD.
Factors harboring P<0.05 were subjected to multivariate
regression to determine the independent predictive factors.
Subsequently, two predictive nomograms were created using the R
‘rms’ package on the basis of the independent predictive factors
for estimating OS and PFS of individuals with STAD. The concordance
index (C-index) was employed to explore the discrimination
efficiency of these two nomograms.
Cells and culture conditions
The AGS and MKN7 cell lines were acquired from the
cell bank of the Chinese Academy of Sciences and cultured in
RPMI-1640 medium (PM 150110; Procell Life Science & Technology
Co., Ltd) enriched with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) at 37°C in a humidified atmosphere containing 5%
CO2.
Cell line authentication
The appropriate amount of MKN-7 cells (cat. no.
PC244; 1×106) was processed with Chelex100 (Bio-Rad
Laboratories, Inc.) to extract DNA and then the 21CELLID System
(Promega Corporation) was used to amplify 20 short tandem repeat
sites and sex identification sites. The AB13130×1 genetic analyzer
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for
PCR product detection. GeneMapper IDX v4 software (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was employed to analyze
the test results and compare them with entries in databases such as
ATCC (American Type Culture Collection; http://www.atcc.org/), DSMZ (German Collection of
Microorganisms and Cell Cultures; http://www.dsmz.de/), JCRB (Japanese Cancer Research
Resources Bank; http://cellbank.nibiohn.go.jp/english/cellsearch_e/)
and Cellosaurus (https://web.expasy.org/cellosaurus/).
Transfection
The cells were transfected with small interfering
RNAs (siRNAs) (Table SI)
targeting LASTR (siLASTR; LncRNA-Pharma) and negative control
(siNC) with the Lipofectamine 2000 system (Invitrogen; Thermo
Fisher Scientific, Inc.) as described by the manufacturer. Cells
were incubated with LASTR siRNAs for 48 h and harvested for
subsequent experiments.
RNA isolation and reverse
transcription-quantitative (RT-q)PCR
The TRIzol reagent was used to extract and purify
total RNA from cells (Takara Bio, Inc.). cDNA was generated from
the RNA via RT with Prime Script RT Master Mix (Takara Bio, Inc.),
as per the manufacturer's instructions. Subsequently, qPCR was run
on the ABI 7500HT Fast Real-Time PCR Platform (Applied Biosystems;
Thermo Fisher Scientific, Inc.). Thermocycling conditions were:
95°C for 15 min; 40 cycles of 95°C for 1 min and 60°C for 1 min.
The 2−ΔΔCq method (13) was adopted to determine relative
lncRNA expression with GAPDH as the normalization control. The
oligonucleotide primers for RT-qPCR included were as follows: LASTR
forward, 5′-GAGAAGACAGTGGGTGAAGTCC-3′ and reverse,
5′-GACTCTAGGCACCAGCTGAC-3′; and GAPDH forward,
5′-GGAAGCTTGTCATCAATGGAAATC-3′ and reverse,
5′-TGATGACCCTTTTGGCTCCC-3′.
Western blot analysis
The GC cells were inoculated onto 6-cm plates for 48
h and harvested via scraping. Lysis was performed by applying RIPA
lysis buffer enriched with protease and phosphatase inhibitors
(Beijing Solarbio) for 30 min. Subsequently, the cells were
centrifuged at 12,000 × g for 20 min at 4°C, and the protein was
quantitated with a BCA protein assay kit (Beyotime Institute of
Biotechnology, Inc.). Subsequently, 20 µg of the proteins were
fractionated by 12.5% SDS-PAGE and transfer-embedded onto PVDF
membranes (MilliporeSigma). Blocking of the membranes was performed
for 2 h using 5% skimmed milk (Nestlé S.A.) dispersed in
Tris-buffered saline containing Tween-20 (TBST). The membranes were
then inoculated overnight (4°C) with the indicated primary antibody
(1:1,000). Next, the membranes were rinsed in TBST for 10 min and
then inoculated at room temperature with the secondary
HRP-conjugated antibodies (1:8,000 dilution; abs20002; Absin
Bioscience Inc.) for 2 h at room temperature. Subsequently, the
membranes were rinsed with TBST and the bound antibodies were
visualized with a chemiluminescence (ECL) kit (cat. no. 34095,
Thermo Fisher Scientific, Inc.) on a ChemiDoc XRS+ gel imager
infrared imaging Platform (Bio-Rad Laboratories, Inc.). Antibodies
against β-actin (cat. no. 4970; Cell Signaling Technology, Inc.)
and anti-GPX4 (cat. no. ab18196) purchased from Abcam and secondary
antibodies (cat. no. abs20002) acquired from Absin Bioscience Inc.
were used for the western blotting.
5-Ethynyl-2′-deoxyuridine (EdU)
incorporation assay
GC cells (1×105/well) were plated onto
24-well plates, allowed to grow for 48 h and incubated with medium
enriched with 50 µM EdU (Beyotime Institute of Biotechnology) for 2
h. The cells were fixed in 4% paraformaldehyde (PFA; Beyotime
Institute of Biotechnology) followed by permeabilization and then a
click reaction mixture (cat. no. C0078S; Beyotime Institute of
Biotechnology) (200 µl/well) was added with subsequent incubation
for 30 min. Nuclear staining was performed with Hoechst 33342 (200
µl/well) for 30 min and a fluorescence microscope (Olympus IX 51;
Olympus Corporation) was employed to visualize the cells.
Colony-formation assay
Cells were transfected with siRNA for two days and
then inoculated into 6-well dishes at 300 cells/well and allowed to
grow for 10 days. Next, the cells were fixed in 4% PFA for 30–60
min, followed by staining with 0.5% crystal violet for 20 min at
room temperature. After numerous washes in ddH2O, images
of the colonies were acquired and their numbers determined. A
colony was considered to be >50 cells.
Transwell migration assay
Cell migration was evaluated using a Transwell
insert (cat. no. 354480; 8.0 µm pore; BD Biosciences) without
Matrigel. Following transfection, 2×105 cells in
serum-free medium were seeded into each upper compartment of the
Transwell insert. Medium enriched with 20% FBS was added to the
lower compartment and allowed to grow for one day. Subsequently,
the cells were fixed with 4% PFA for 30 min, followed by staining
with 0.5% crystal violet for 20 min all at room temperature) and
then rinsed with PBS. The cells were counted in five fields (top,
bottom, center, left and right) under a microscope (Olympus IX 51;
Olympus Corporation).
Wound-healing assay
The GC cells were seeded into 6-well plates to form
a confluent layer and a sterile pipette tip was employed to make a
linear scratch. Next, the cells were rinsed with PBS and then
further cultured in a medium enriched without FBS. Images were
acquired at 0 and 24 h with a phase-contrast microscope. The
fraction of wound healing was determined as follows: [1-(empty area
24 h/empty area 0 h)] ×100%.
Statistical analysis
All data analyses were implemented in Bioconductor
packages in R software, version 4.1.0, and GraphPad Prism 8.0
Software (GraphPad Software, Inc.). The unpaired Student's t-test,
the Wilcoxon rank-sum test, ANOVA and the Kruskal-Wallis test were
adopted to compare continuous variables. Pearson analysis was
implemented for the correlation analyses. P<0.05 was considered
to indicate statistical significance.
Results
Patient characteristics
Overall, 375 STAD tumor samples along with 32
non-malignant adjacent tissues were included and their expression
profiles were identified. The clinical features of the patients are
provided in Table I (male: female
ratio, 1.80:1; ≤65:>65 ratio, 0.79:1). Next, 382
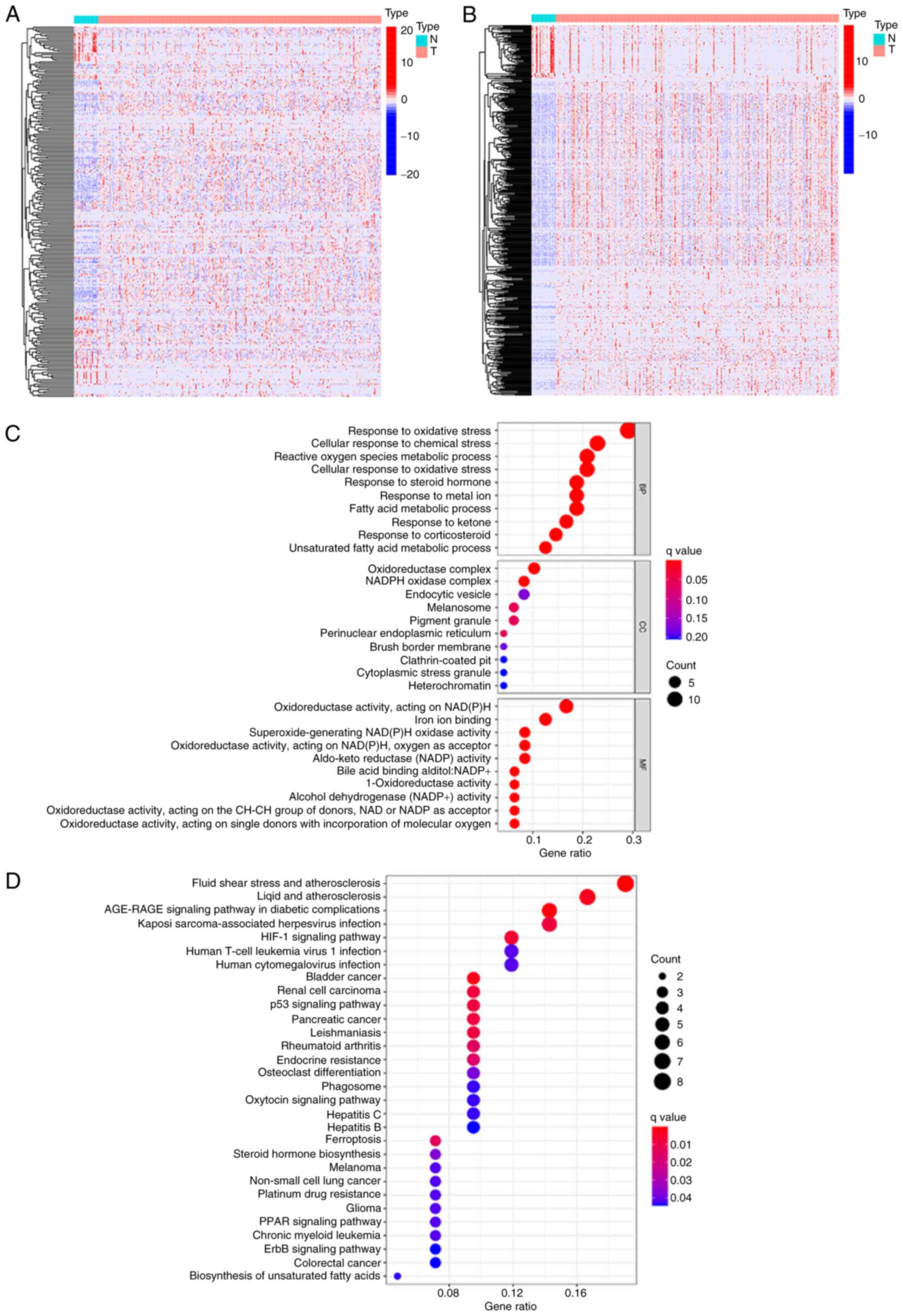
ferroptosis-related markers were obtained from FerrDb (Fig. 1A). Using Pearson's correlation
test, 503 differentially expressed ferroptosis-related lncRNAs were
obtained (Fig. 1B). To gain a
profound understanding of how these ferroptosis-related lncRNAs may
drive STAD development, GO along with KEGG enrichment analyses were
performed. In the category biological process, the terms were
related to the response to oxidative stress and the response to
metal ions. The cellular component terms were associated with the
production of oxidoreductase complex and NADPH oxidase complexes.
In the category molecular function, the terms were related to
oxidoreductase activity, iron ion binding, and
superoxide-generating NAD(P)H oxidase activity (Fig. 1C). KEGG pathway analysis indicated
that primary enrichment in ferroptosis, the HIF-1 signaling
pathway, the p53 signaling pathway, the PPAR signaling pathway and
the ErbB signaling pathway were significant (Fig. 1D). The enrichment analyses
indicated that ferroptosis-related lncRNAs are closely related to
iron metabolism and may mediate certain pivotal signaling pathways
in the tumorigenesis of STAD.
 | Table I.Clinical characteristics of patients
in The Cancer Genome Atlas-stomach adenocarcinoma dataset. |
Table I.
Clinical characteristics of patients
in The Cancer Genome Atlas-stomach adenocarcinoma dataset.
| Variable | Number of
samples |
|---|
| Sex
(male/female) | 241/134 |
| Age at diagnosis
(≤65/>65 years/NA) | 164/207/4 |
| Grade
(G1/G2/G3/NA) | 10/137/219/9 |
| TNM stage
(I/II/III/IV/NA) |
53/111/150/38/23 |
| TNM stage
(T0/T1/T2/T3/T4/NA) |
19/80/168/100/8 |
| N
(N0/N1/N2/N3/NA) |
111/97/75/74/18 |
| M (M0/M1/NA) | 330/25/20 |
Prognostic value of
ferroptosis-related lncRNAs in STAD
The lack of reliable markers for early tumor
diagnosis is still one of the critical factors in the dismal
prognosis of individuals with advanced STAD. Recent studies have
revealed that ferroptosis-related lncRNAs may act as prognostic
targets in diverse cancers (14,15). Thus, the possible predictive value
of ferroptosis-related lncRNAs in individuals with STAD was further
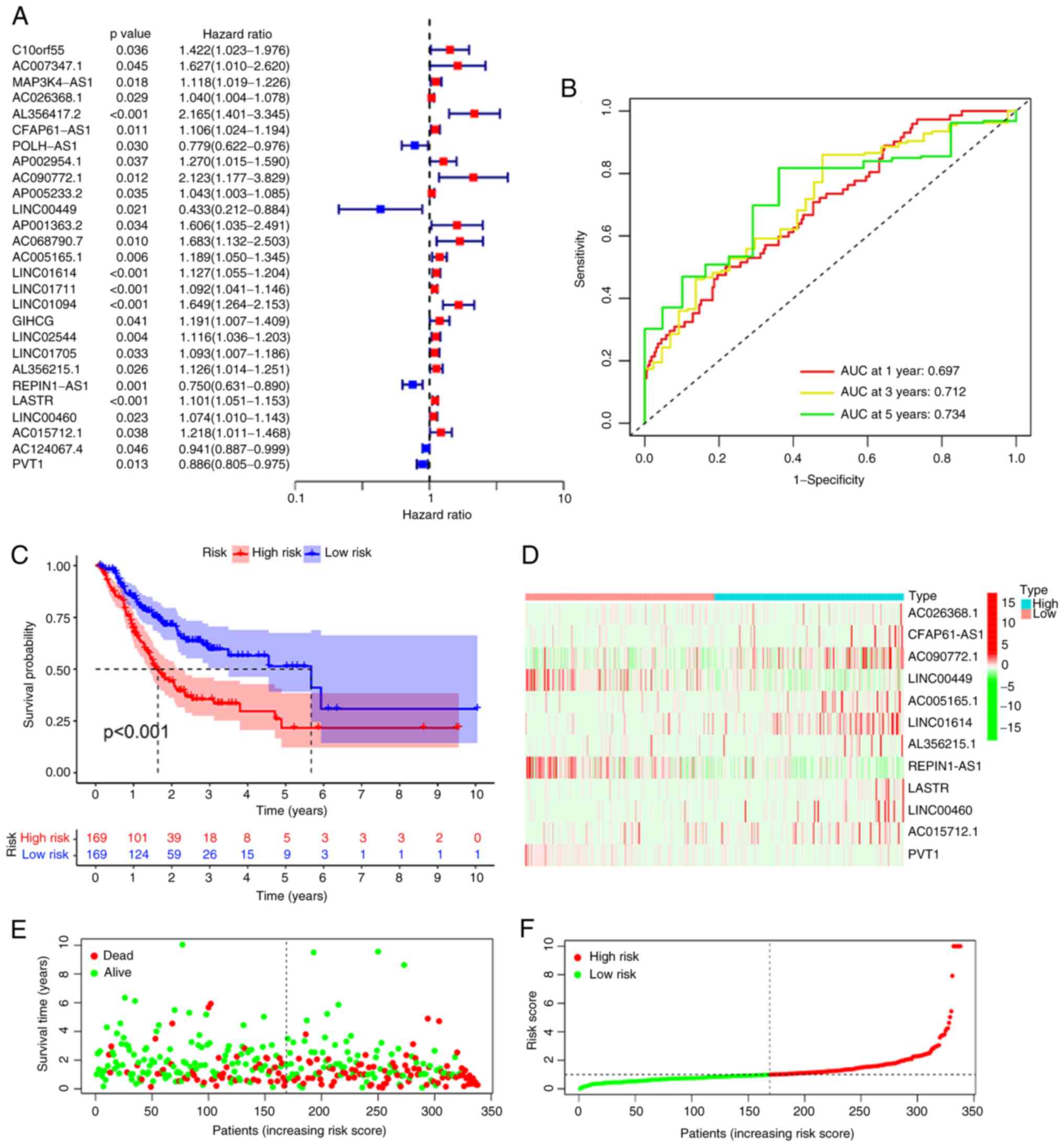
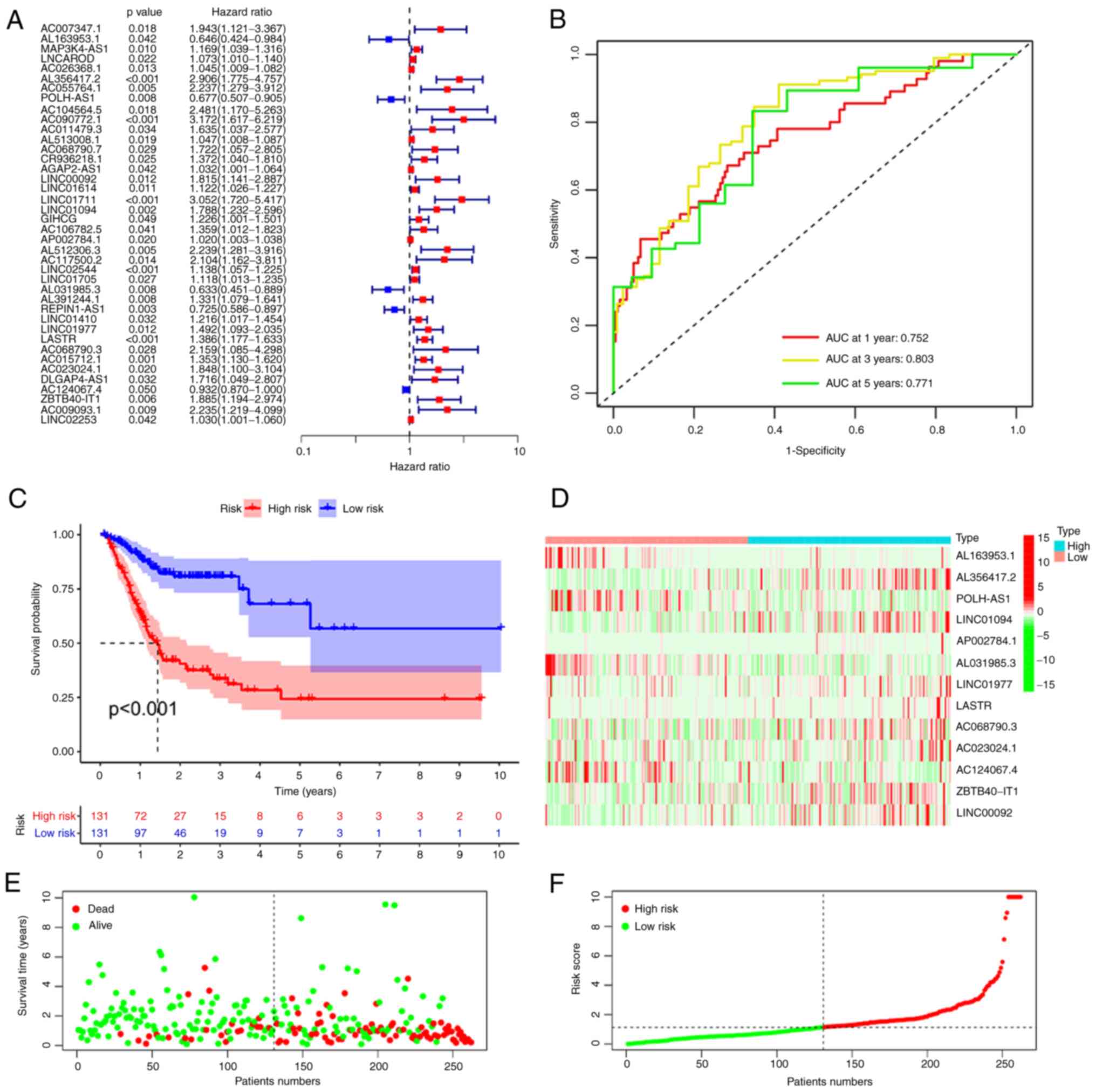
confirmed (Figs. 2 and 3). Univariate Cox analysis uncovered 27
and 40 ferroptosis-related lncRNAs that were significantly related
to OS and PFS, respectively (Figs.
2A and 3A). Subsequently,
multivariate Cox analysis was performed and a total of 25
ferroptosis-related lncRNAs were employed to produce two predictive
signatures, consisting of 12 ferroptosis-related lncRNAs for the OS
signature and 13 ferroptosis-related lncRNAs for the PFS signature.
The formula for the OS signature was as follows: Risk score=0.03525
× AC026368.1 + 0.09945 × CFAP61-AS1 + 1.07809 × AC090772.1-0.62697
× LINC00449-0.12656 × AC005165.1 + 0.14984 × LINC01614 + 0.11706 ×
AL356215.1-0.17220 × REPIN1-AS1 + 0.05905 × LASTR + 0.07243 ×
LINC00460 + 0.21417 × AC015712.1-0.13778 × PVT1. The PFS signature
was as follows: Risk score=−0.47559 × AL163953.1 + 0.972225 ×
AL356417.2-0.28607 × POLH-AS1 + 0.377634 × LINC01094 + 0.034465 ×
AP002784.1-0.75762 × AL031985.3 + 0.532136 × LINC01977 + 0.300628 ×
LASTR + 0.838305 × AC068790.3 + 0.70416 × AC023024.1-0.0644 ×
AC124067.4 + 0.578635 × ZBTB40-IT1 + 0.434186 × LINC00092. The
time-based ROC curves illustrated that the AUCs of the OS signature
for estimating 1-, 3- and 5-year OS were 0.697, 0.712 and 0.734,
respectively (Fig. 2B).
Subsequently, based on the median risk score, patients were divided
into high- and low-risk groups. The K-M plots illustrated that the
high-risk group had a dismal OS in comparison with the low-risk
group (Fig. 2C). The AUCs of the
signature for estimating 1-, 3- and 5-year PFS were 0.752, 0.803
and 0.771, respectively (Fig.
3B). Similar to the situation for the OS signature, those
patients with STAD with higher risk scores had unfavorable PFS
(Fig. 3C). These data illustrated
that the OS and PFS signature were valuable tools for estimating
the prognosis of individuals with STAD. To more clearly illustrate
differences in the prognosis and expression trends of lncRNAs,
heatmaps were then constructed (Figs.
2D and 3D), as well as
survival status plots (Figs. 2E
and 3E) and risk score plots
(Figs. 2F and 3F). All of the above results indicated
that the signatures based on DEFRLs are able to effectively assess
the prognosis of patients with STAD.
Creation of nomograms on the basis of
the DEFRL signatures and clinical parameters
To enhance the clinical utility of the prognostic
signatures determined in the present study, two comprehensive
signatures were constructed based on independent clinical
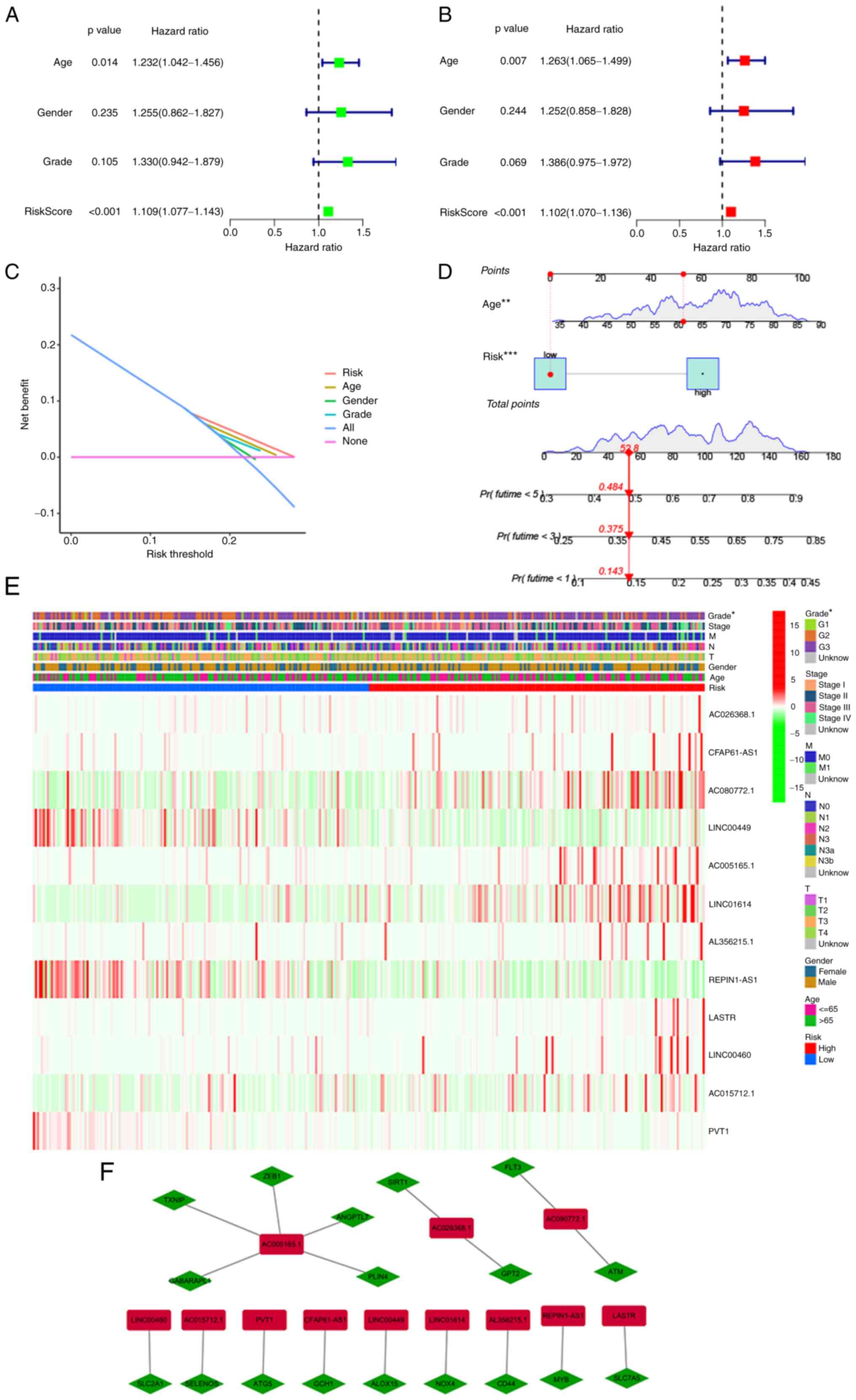
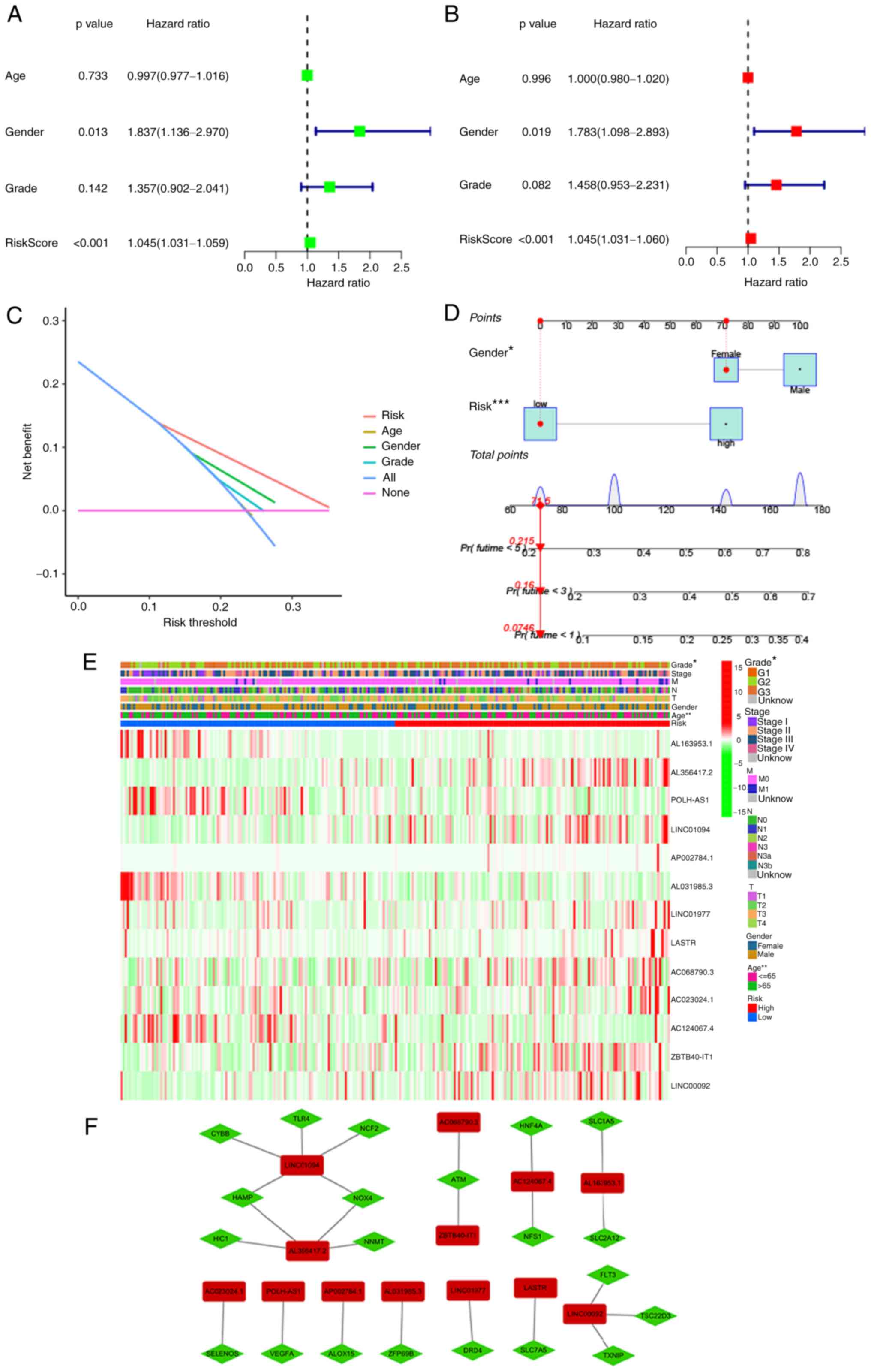
parameters (Figs. 4 and 5). First, univariate and multivariate
Cox analyses were performed to explore independent predictive
variables of OS along with PFS. Risk score and age were identified
as independent OS-related variables (Fig. 4A and B). Two independent
variables, namely the risk score and sex, were identified as
PFS-related variables (Fig. 5A and
B). Clinical factors of patients with STAD that correlated with
Decision Curve Analysis parameters were screened (Figs. 4C and 5C). Furthermore, on the basis of the
independent predictive variables, two new nomograms were created to
estimate OS and PFS (Figs. 4D and
5D). The C-indices were 0.674
(95% CI, 0.625–0.723) and 0.664 (95% CI, 0.6052–0.7228) for the OS
and PFS nomograms. These data illustrated that two nomograms may be
adapted to precisely estimate the prognosis of individuals with
STAD. Heatmaps for the associations of ferroptosis-related lncRNA
prognostic signatures with clinicopathological manifestations were
also generated (Figs. 4E and
5E). Furthermore, the association
between lncRNAs and mRNAs for the OS and PFS signatures is
displayed in Figs. 4F and
5F, revealing a complex
regulatory relationship between them.
Roles of LASTR in GC
One overlapping ferroptosis-related lncRNA among
those identified by multivariate regression of OS and PFS
signatures, namely LASTR, was indicated to be the most critical
factor in the prognostic models for STAD. It was reported that
LASTR was able to foster the fitness of cancer cells via modulating
the activity of the U4/U6 recycling factor SART3 (16). The specific roles of LASTR in GC
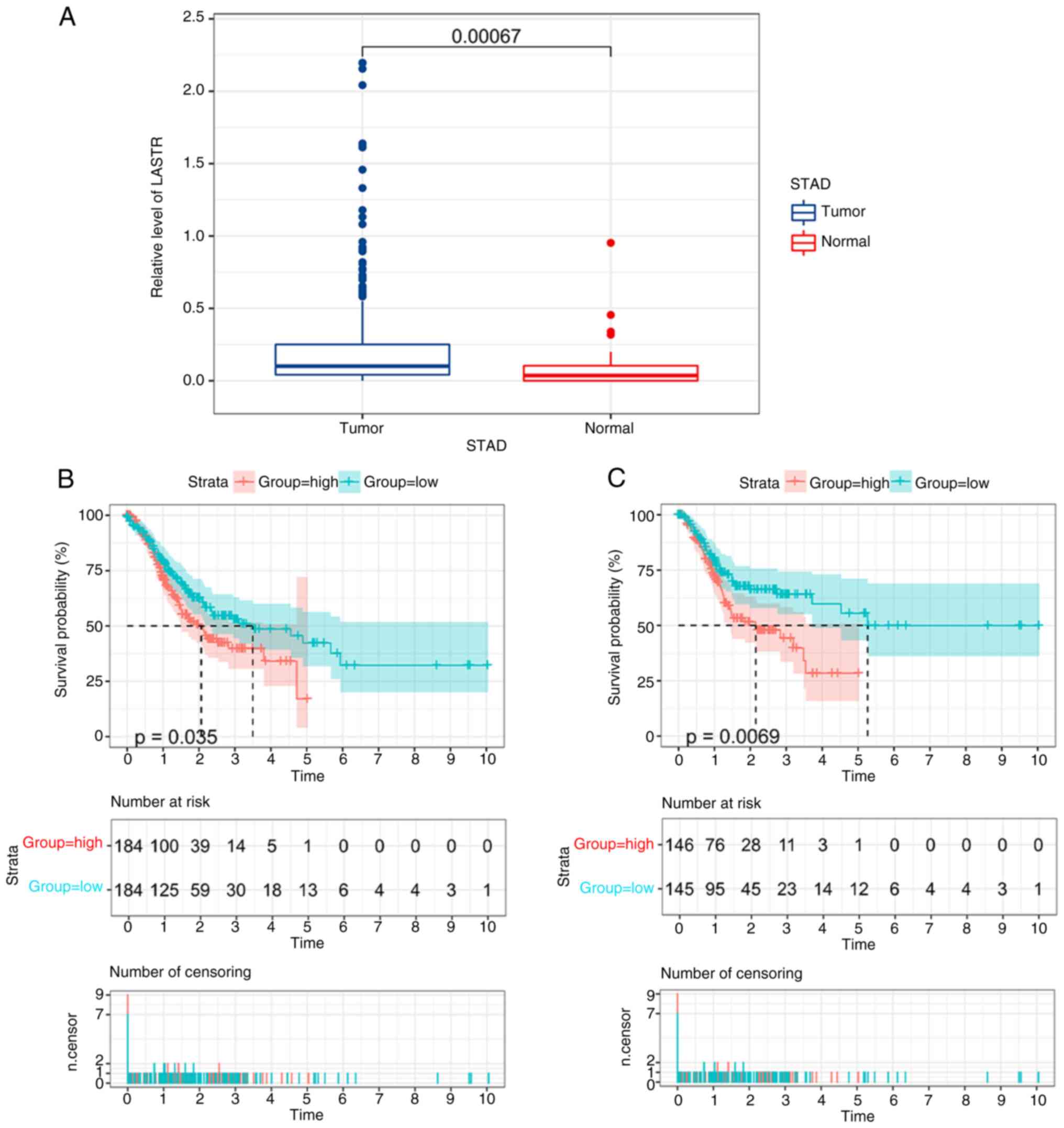
cells were then explored. The expression of LASTR in GC tissues and
neighboring non-malignant tissues from the TCGA data resource is
provided in Fig. 6A. K-M survival
curves illustrated that a low level of LASTR was significantly
associated with favorable OS and PFS (Fig. 6B and C). Next, siLASTR was used to
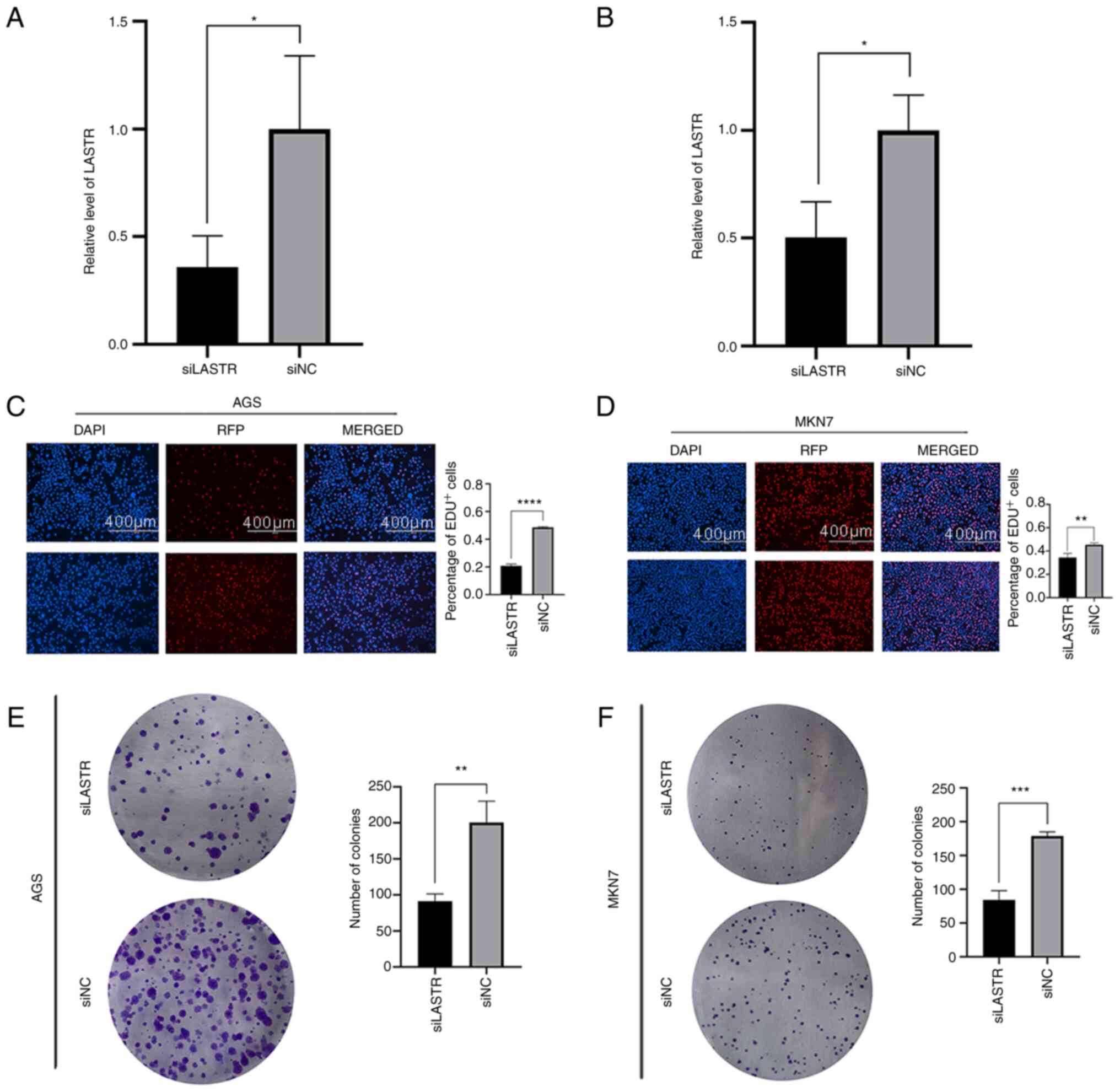
infect two cell lines (AGS and MKN7) and the knockdown efficiency
was confirmed via RT-qPCR (Fig. 7A
and B). Subsequently, an EdU incorporation assay (Fig. 7C and D) and colony formation assay
(Fig. 7E and F) were performed on
the two GC cell lines to measure changes in proliferative capacity,
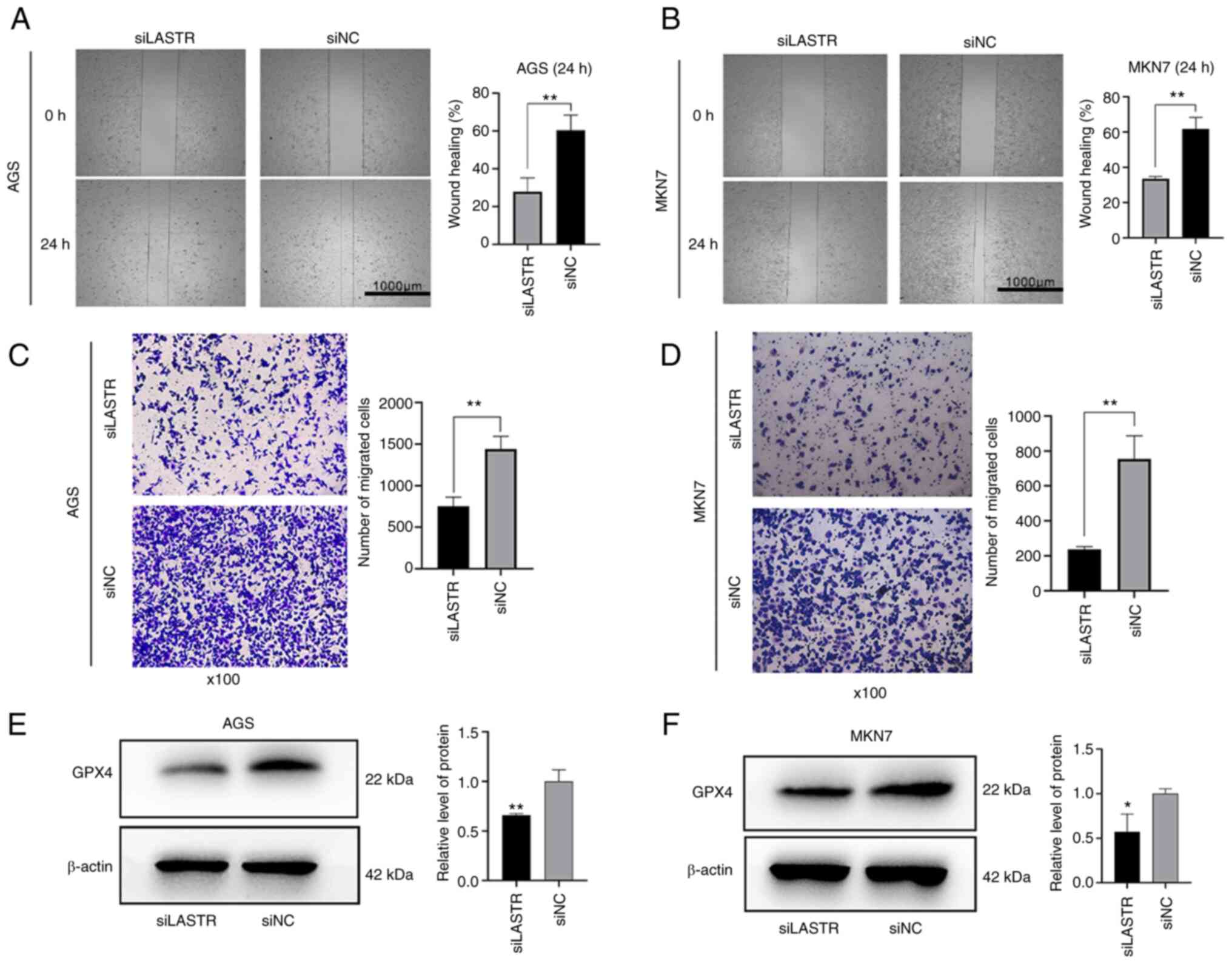
and a wound-healing assay (Fig. 8A
and B) and Transwell migration assay (Fig. 8C and D) were carried out to assess
changes in migration capacity. The results suggested that the cell
proliferation and migration abilities were decreased following
LASTR knockdown. Western blot analysis was used to compare the
expression of the ferroptosis marker GPX4 between the knockdown
group and control group. Compared with the control group, the level
of GPX4 was significantly decreased in the knockout group (Fig. 8E and F). The results indicated
that knockdown of LASTR repressed cell growth and migration and may
trigger ferroptosis in GC.
Discussion
Despite advances in detection approaches and medical
standards, the five-year survival rate of patients with STAD
remains low (17). One of the
main reasons is the lack of sufficiently specific and sensitive
biomarkers for early diagnosis (18). It has been indicated that
ferroptosis is involved in proliferation, migration, drug
resistance and other biological behaviors of STAD (19–23). LncRNAs are pivotal regulators of
ferroptosis, having different biological roles in various cancer
types (7–9,24–27). However, an effective predictive
tool featuring ferroptosis-related lncRNAs for patients with STAD
is still lacking. In the present study, 12 and 13
ferroptosis-related lncRNAs were identified and used to produce
signatures to predict OS and PFS, respectively, in patients with
STAD. First, 382 ferroptosis-related markers were determined from
the TCGA dataset and 503 lncRNAs were identified as candidate
predictive biomarkers. Furthermore, and GO and KEGG analyses
uncovered the prospective mechanisms of these lncRNAs. Of note, an
OS predictive signature was constructed based on 12 key lncRNAs and
a PFS predictive signature consisting of 13 key lncRNAs. According
to the risk scores, STAD subjects were categorized into high-risk
and low-risk groups. The differences in OS and PFS between the
high-risk and low-risk groups were statistically significant. The
reliability of these two signatures was further supported by the
prediction ability of the ROC curves of the two signatures. In
addition, two comprehensive nomograms integrating the
lncRNA-related prognostic signatures and clinical features were
established to enhance clinical utility. These nomograms allow
clinicians to evaluate the OS and PFS for each patient with STAD by
inputting the score for each parameter.
The present study also indicated that oxidative
stress and tumor-associated signaling pathways, such as the HIF-1
signaling pathway, the p53 signaling pathway, the PPAR signaling
pathway and the ErbB signaling pathway, were significantly enriched
through GO and KEGG functional enrichment analyses. It was
previously reported that ferroptosis induced by oxidative stress is
associated with various diseases, such as Alzheimer's disease
(28), intervertebral disc
degeneration (29) and cancer
(30). Ni et al (31) demonstrated that targeting HIF-1α
is able to induce osteoclast ferroptosis to treat osteoporosis.
Certain studies have indicated that p53 is able to regulate
ferroptosis and mediate certain diseases (32,33). The above studies have proved the
regulatory relationship between signaling pathways and ferroptosis
in different disease types, confirming the reliability of the
signatures constructed in the present study.
A total of 12 lncRNAs (AC026368.1, CFAP61-AS1,
AC090772.1, LINC00449, AC005165.1, LINC01614, AL356215.1,
REPIN1-AS1, LASTR, LINC00460, AC015712.1 and PVT1) were included in
the OS signature and 13 lncRNAs (AL163953.1, AL356417.2, POLH-AS1,
LINC01094, AP002784.1, AL031985.3, LINC01977, LASTR, AC068790.3,
AC023024.1, AC124067.4, ZBTB40-IT1 and LINC00092) in the PFS
signature. LINC00449 (34),
LINC01614 (35–39), LINC00460 (40–44), PVT1 (24,26,45–49), LINC01094 (50–55), LINC01977 (56), ZBTB40-IT1 (57) and LINC00092 (58) were previously reported to regulate
the biological behavior or serve as prognostic tumor biomarkers in
various cancer types. Except for these lncRNAs mentioned above, the
other lncRNAs included in the signatures have remained largely
unexplored, which will be the focus of future studies by our group.
Among the OS and PFS signatures, LASTR was the only lncRNA included
in both signatures, which may thus have a relatively greater
prognostic value in patients with STAD. LASTR (LINC02657) was
originally named as it was indicated to be associated with SART3
regulation of splicing. It was previously reported that LASTR was
upregulated in triple-negative breast cancer with hypoxia and
affected the adaptability of tumor cells (16). Apart from that, there is virtually
no information about it in the literature. Thus, the biological
roles of LASTR in GC were further explored in the present study.
Cell experiments, omics experiments and bioinformatics analysis
were performed to confirm that LASTR has a role in enhancing GC
progression. When verifying the association between LASTR and
ferroptosis, due to limitations of experimental technology and the
experimental environment, only the changes in the ferroptosis
marker GPX4 protein after LASTR was knocked down were examined to
determine their regulatory relationship. As this analysis was not
very rigorous, this matter will be explored in advanced research
settings in the future.
Ferroptosis has become a hot topic in research in
recent years and provides a novel mechanism for cancer treatment.
There are still numerous unknown areas in the relation between
ferroptosis and lncRNAs worth exploring. In the present study, 12
and 13 ferroptosis-related lncRNAs were identified and included in
an OS signature and PFS signature for STAD, respectively, and
experimental verification of the roles of LASTR was performed.
In conclusion, in the present study, novel
ferroptosis-related biomarkers were identified for STAD prognosis.
One of the markers, LASTR, was experimentally verified as a
cancer-promoting factor and to be associated with ferroptosis. This
study provided a novel approach for the treatment of cancer and
predict the survival of patients with STAD.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the Beijing Xisike
Clinical Oncology Research Foundation (grant no. Y-BMS2019-038), Wu
Jieping Medical Foundation (grant no. 320.6750.19088-29), Shandong
Medical and Health Technology Development Foundation (grant no.
202003030451) and Qingdao Municipal People's Livelihood Science and
Technology Foundation (grant no. 17-3-3-34-nsh).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GW, LS and SW analyzed the data. GW and JG wrote and
reviewed the manuscript. WQiu, WQi and JG contributed to the design
of the study. GW, RX and WL performed the experiments. All authors
contributed to the article and read and approved the final version.
All authors confirm the authenticity of the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang D, Kang R, Berghe TV, Vandenabeele P
and Kroemer G: The molecular machinery of regulated cell death.
Cell Res. 29:347–364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hassannia B, Vandenabeele P and Vanden
Berghe T: Targeting ferroptosis to Iron out cancer. Cancer Cell.
35:830–849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Guo S, Wang S, Li X, Hou D, Li H,
Wang L, Xu Y, Ma B, Wang H and Jiang X: LncRNA OIP5-AS1 inhibits
ferroptosis in prostate cancer with long-term cadmium exposure
through miR-128-3p/SLC7A11 signaling. Ecotoxicol Environ Saf.
220:1123762021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma Q, Dai X, Lu W, Qu X, Liu N and Zhu C:
Silencing long non-coding RNA MEG8 inhibits the proliferation and
induces the ferroptosis of hemangioma endothelial cells by
regulating miR-497-5p/NOTCH2 axis. Biochem Biophys Res Commun.
556:72–78. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Z, Chen X, Liu N, Shi Y, Liu Y,
Ouyang L, Tam S, Xiao D, Liu S, Wen F and Tao Y: A nuclear long
non-coding RNA LINC00618 accelerates ferroptosis in a manner
dependent upon apoptosis. Mol Ther. 29:263–274. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:pl12013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou N and Bao J: FerrDb: A manually
curated resource for regulators and markers of ferroptosis and
ferroptosis-disease associations. Database (Oxford).
2020:baaa0212020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chew WL, Tabebordbar M, Cheng JK, Mali P,
Wu EY, Ng AH, Zhu K, Wagers AJ and Church GM: A multifunctional
AAV-CRISPR-Cas9 and its host response. Nat Methods. 13:868–874.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai HJ, Zhuang ZC, Wu Y, Zhang YY, Liu X,
Zhuang JF, Yang YF, Gao Y, Chen B and Guan GX: Development and
validation of a ferroptosis-related lncRNAs prognosis signature in
colon cancer. Bosn J Basic Med Sci. 21:569–576. 2021.PubMed/NCBI
|
|
15
|
Tang Y, Li C, Zhang YJ and Wu ZH:
Ferroptosis-related long non-coding RNA signature predicts the
prognosis of Head and neck squamous cell carcinoma. Int J Biol Sci.
17:702–711. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De Troyer L, Zhao P, Pastor T, Baietti MF,
Barra J, Vendramin R, Dok R, Lechat B, Najm P, Van Haver D, et al:
Stress-induced lncRNA LASTR fosters cancer cell fitness by
regulating the activity of the U4/U6 recycling factor SART3.
Nucleic Acids Res. 48:2502–2517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang F and Shen X: Current prevalence
status of gastric cancer and recent studies on the roles of
circular RNAs and methods used to investigate circular RNAs. Cell
Mol Biol Lett. 24:532019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dang Y, Ouyang X, Zhang F, Wang K, Lin Y,
Sun B, Wang Y, Wang L and Huang Q: Circular RNAs expression
profiles in human gastric cancer. Sci Rep. 7:90602017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu D, Wang C, Yu L and Yu R: Induction of
ferroptosis by ATF3 elevation alleviates cisplatin resistance in
gastric cancer by restraining Nrf2/Keap1/xCT signaling. Cell Mol
Biol Lett. 26:262021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao Z, Deng G, Li Y, Huang H, Sun X, Shi
H, Yao X, Gao L, Ju Y and Luo M: Actinidia chinensis Planch
prevents proliferation and migration of gastric cancer associated
with apoptosis, ferroptosis activation and mesenchymal phenotype
suppression. Biomed Pharmacother. 126:1100922020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Tian Y, Liang Y and Li Q:
Circ_0008035 contributes to cell proliferation and inhibits
apoptosis and ferroptosis in gastric cancer via miR-599/EIF4A1
axis. Cancer Cell Int. 20:842020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao D, Zhang C, Jiang M, Wang Y, Liang Y,
Wang L, Qin K, Rehman FU and Zhang X: Survival-associated
alternative splicing signatures in non-small cell lung cancer.
Aging (Albany NY). 12:5878–5893. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang H, Deng T, Liu R, Ning T, Yang H,
Liu D, Zhang Q, Lin D, Ge S, Bai M, et al: CAF secreted miR-522
suppresses ferroptosis and promotes acquired chemo-resistance in
gastric cancer. Mol Cancer. 19:432020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu J, Xu F and Lu H: LncRNA PVT1 regulates
ferroptosis through miR-214-mediated TFR1 and p53. Life Sci.
260:1183052020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C
and Xu S: LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress
during erastin-induced ferroptosis in HepG2 hepatocellular
carcinoma cells. Sci Rep. 9:161852019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu
N, Shi Y, Chen L, Xiao D, Yu F, et al: Long noncoding RNA LINC00336
inhibits ferroptosis in lung cancer by functioning as a competing
endogenous RNA. Cell Death Differ. 26:2329–2343. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang Y, Tai W, Lu N, Li T, Liu Y, Wu W, Li
Z, Pu L, Zhao X, Zhang T and Dong Z: lncRNA ZFAS1 promotes lung
fibroblast-to-myofibroblast transition and ferroptosis via
functioning as a ceRNA through miR-150-5p/SLC38A1 axis. Aging
(Albany NY). 12:9085–1102. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park MW, Cha HW, Kim J, Kim JH, Yang H,
Yoon S, Boonpraman N, Yi SS, Yoo ID and Moon JS: NOX4 promotes
ferroptosis of astrocytes by oxidative stress-induced lipid
peroxidation via the impairment of mitochondrial metabolism in
Alzheimer's diseases. Redox Biol. 41:1019472021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang RZ, Xu WN, Zheng HL, Zheng XF, Li B,
Jiang LS and Jiang SD: Involvement of oxidative stress-induced
annulus fibrosus cell and nucleus pulposus cell ferroptosis in
intervertebral disc degeneration pathogenesis. J Cell Physiol.
236:2725–2739. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kou L, Sun R, Jiang X, Lin X, Huang H, Bao
S, Zhang Y, Li C, Chen R and Yao Q: Tumor
Microenvironment-responsive, multistaged liposome induces apoptosis
and ferroptosis by amplifying oxidative stress for enhanced cancer
therapy. ACS Appl Mater Interfaces. 12:30031–30043. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ni S, Yuan Y, Qian Z, Zhong Z, Lv T, Kuang
Y and Yu B: Hypoxia inhibits RANKL-induced ferritinophagy and
protects osteoclasts from ferroptosis. Free Radic Biol Med.
169:271–282. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tang LJ, Zhou YJ, Xiong XM, Li NS, Zhang
JJ, Luo XJ and Peng J: Ubiquitin-specific protease 7 promotes
ferroptosis via activation of the p53/TfR1 pathway in the rat
hearts after ischemia/reperfusion. Free Radic Biol Med.
162:339–352. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Z, Guo M, Shen M, Kong D, Zhang F,
Shao J, Tan S, Wang S, Chen A, Cao P and Zheng S: The
BRD7-P53-SLC25A28 axis regulates ferroptosis in hepatic stellate
cells. Redox Biol. 36:1016192020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi Y, Zhu Y, Zheng X and Zheng Z:
LINC00449 regulates the proliferation and invasion of acute
monocytic leukemia and predicts favorable prognosis. J Cell
Physiol. 235:6536–6547. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cai Q, Zhao X, Wang Y, Li S, Wang J, Xin Z
and Li F: LINC01614 promotes osteosarcoma progression via
miR-520a-3p/SNX3 axis. Cell Signal. 83:1099852021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen Y, Cheng WY, Shi H, Huang S, Chen H,
Liu D, Xu W, Yu J and Wang J: Classifying gastric cancer using
FLORA reveals clinically relevant molecular subtypes and highlights
LINC01614 as a biomarker for patient prognosis. Oncogene.
40:2898–2909. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu AN, Qu HJ, Yu CY and Sun P: Knockdown
of LINC01614 inhibits lung adenocarcinoma cell progression by
up-regulating miR-217 and down-regulating FOXP1. J Cell Mol Med.
22:4034–4044. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tang L, Chen Y, Peng X, Zhou Y, Jiang H,
Wang G and Zhuang W: Identification and validation of potential
pathogenic genes and prognostic markers in ESCC by integrated
bioinformatics analysis. Front Genet. 11:5210042020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang Y, Song B, Zhu L and Zhang X: Long
non-coding RNA, LINC01614 as a potential biomarker for prognostic
prediction in breast cancer. PeerJ. 7:e79762019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cheng J, Lou Y and Jiang K: Downregulation
of long non-coding RNA LINC00460 inhibits the proliferation,
migration and invasion, and promotes apoptosis of pancreatic cancer
cells via modulation of the miR-320b/ARF1 axis. Bioengineered.
12:96–107. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hong H, Sui C, Qian T, Xu X, Zhu X, Fei Q,
Yang J and Xu M: Long noncoding RNA LINC00460 conduces to tumor
growth and metastasis of hepatocellular carcinoma through
miR-342-3p-dependent AGR2 up-regulation. Aging (Albany NY).
12:10544–10555. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hou P, Meng S, Li M, Lin T, Chu S, Li Z,
Zheng J, Gu Y and Bai J: LINC00460/DHX9/IGF2BP2 complex promotes
colorectal cancer proliferation and metastasis by mediating HMGA1
mRNA stability depending on m6A modification. J Exp Clin Cancer
Res. 40:522021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jiang Y, Cao W, Wu K, Qin X, Wang X, Li Y,
Yu B, Zhang Z, Wang X, Yan M, et al: LncRNA LINC00460 promotes EMT
in head and neck squamous cell carcinoma by facilitating
peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res.
38:3652019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li F, Zhu W and Wang Z: Long noncoding RNA
LINC00460 promotes the progression of cervical cancer via
regulation of the miR-361-3p/Gli1 axis. Hum Cell. 34:229–327. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cho SW, Xu J, Sun R, Mumbach MR, Carter
AC, Chen YG, Yost KE, Kim J, He J, Nevins SA, et al: Promoter of
lncRNA Gene PVT1 Is a tumor-suppressor DNA boundary element. Cell.
173:1398–1412.e22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ghetti M, Vannini I, Storlazzi CT,
Martinelli G and Simonetti G: Linear and circular PVT1 in
hematological malignancies and immune response: Two faces of the
same coin. Mol Cancer. 19:692020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shigeyasu K, Toden S, Ozawa T, Matsuyama
T, Nagasaka T, Ishikawa T, Sahoo D, Ghosh P, Uetake H, Fujiwara T
and Goel A: The PVT1 lncRNA is a novel epigenetic enhancer of MYC,
and a promising risk-stratification biomarker in colorectal cancer.
Mol Cancer. 19:1552020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu Y, Li Y, Jin J, Han G, Sun C, Pizzi MP,
Huo L, Scott A, Wang Y, Ma L, et al: LncRNA PVT1 up-regulation is a
poor prognosticator and serves as a therapeutic target in
esophageal adenocarcinoma. Mol Cancer. 18:1412019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou C, Yi C, Yi Y, Qin W, Yan Y, Dong X,
Zhang X, Huang Y, Zhang R, Wei J, et al: LncRNA PVT1 promotes
gemcitabine resistance of pancreatic cancer via activating
Wnt/β-catenin and autophagy pathway through modulating the
miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 19:1182020.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jiang Y, Li W, Yan Y, Yao X, Gu W and
Zhang H: LINC01094 triggers radio-resistance in clear cell renal
cell carcinoma via miR-577/CHEK2/FOXM1 axis. Cancer Cell Int.
20:2742020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jiang Y, Zhang H, Li W, Yan Y, Yao X and
Gu W: FOXM1-activated LINC01094 promotes clear cell renal cell
carcinoma development via MicroRNA 224-5p/CHSY1. Mol Cell Biol.
40:e00357–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li XX and Yu Q: Linc01094 accelerates the
growth and metastatic-related traits of glioblastoma by sponging
miR-126-5p. Onco Targets Ther. 13:9917–9928. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Xu H, Wang X, Wu J, Ji H, Chen Z, Guo H
and Hou J: Long Non-coding RNA LINC01094 promotes the development
of clear cell renal cell carcinoma by upregulating SLC2A3 via
MicroRNA-184. Front Genet. 11:5629672020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu J, Zhang P, Sun H and Liu Y:
LINC01094/miR-577 axis regulates the progression of ovarian cancer.
J Ovarian Res. 13:1222020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhu B, Liu W, Liu H, Xu Q and Xu W:
LINC01094 Down-regulates miR-330-3p and enhances the expression of
MSI1 to promote the progression of glioma. Cancer Manag Res.
12:6511–6521. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li Z, Li Y, Wang X, Liang Y, Luo D, Han D,
Li C, Chen T, Zhang H, Liu Y, et al: LINC01977 promotes breast
cancer progression and chemoresistance to doxorubicin by targeting
miR-212-3p/GOLM1 Axis. Front Oncol. 11:6570942021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mei B, Wang Y, Ye W, Huang H, Zhou Q, Chen
Y, Niu Y, Zhang M and Huang Q: LncRNA ZBTB40-IT1 modulated by
osteoporosis GWAS risk SNPs suppresses osteogenesis. Hum Genet.
138:151–166. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao L, Ji G, Le X, Wang C, Xu L, Feng M,
Zhang Y, Yang H, Xuan Y, Yang Y, et al: Long noncoding RNA
LINC00092 acts in cancer-associated fibroblasts to drive glycolysis
and progression of ovarian cancer. Cancer Res. 77:1369–1382. 2017.
View Article : Google Scholar : PubMed/NCBI
|