Introduction
Thyroid cancer is the most widespread form of
endocrine cancers worldwide. Its incidence has been reported to
have increased in recent years (1). According to histological features,
thyroid cancer can be divided into four subgroups: Papillary,
follicular, anaplastic and medullary. Among these subgroups, the
most common type (~80%) is papillary thyroid cancer (PTC)
originating from follicular cells (2,3).
Even though current forms of treatment (e.g., surgical intervention
and radioactive iodine) can keep this disease under control for a
number of patients, it nevertheless has a high morbidity rate;
likewise, in some cases, the tumors can be aggressive (4,5).
In addition, while it is known that environmental and genetic
factors serve a role in development of this disease (6), its pathogenesis is not fully
clarified. Therefore, there is a need to elucidate the molecular
mechanisms involved in the development of the disease and to
discover effective treatment targets, such as biomarkers that can
be used in early diagnosis and prognosis prediction.
Long non-coding RNAs (lncRNAs) are molecules that
are >200 base pairs long and do not encode a protein. However,
they perform important roles in regulating gene expression. LncRNAs
can regulate the expression of genes involved in processes such as
cell cycle, proliferation, differentiation, stem cell
differentiation, apoptosis, invasion, migration and autophagy
through different mechanisms such as regulation of chromatin
structure, transcription, splicing process, post-transcriptional
events and interaction with microRNAs (2,7).
The changes in the expression of lncRNAs and impairment of their
regulatory functions have been associated with a number of
diseases, including numerous types of cancer. It has been reported
that several lncRNAs (e.g. NEAT1, MALAT1, H19, HOTAIR, BANCR,
PTCSC3) are associated with the development and progression of
thyroid cancer (3,8).
The SOX2 overlapping transcript (SOX2OT) gene
is located in the 3q26.3 region and contains the SOX2 gene
in its intronic region. This gene serves a key role in maintaining
both pluripotency and self-renewal properties of embryonic stem
cells (9,10). Many studies have revealed that the
increase in SOX2OT expression in cancer cells induces
SOX2 expression and this in turn contributes to
tumorigenesis (11–14). The changes in SOX2OT
expression have been associated with a number of cancers such as
lung, breast, esophageal, gastric, hepatocellular and ovarian
(10). However, no study has been
published so far investigating the relationship between the
expression level of SOX2OT and thyroid cancer, to the best
of the authors' knowledge. Therefore, this association required
investigation.
Differentiation antagonizing non-protein coding RNA
(DANCR), localized on chromosome 4, is 855 base pairs long
and helps suppress progenitor cell differentiation (15). The relationship between
DANCR and tumorigenesis and clinicopathological features of
the tumors have been investigated in relation to various types of
cancer such as hepatocellular, gastric, colorectal, breast and lung
cancer (16–20). It has been reported that
DANCR is an oncogenic lncRNA that is overexpressed in tumor
tissues and is associated with poor prognostic factors (16). In molecular studies, it has been
demonstrated that DANCR induces proliferation, migration,
invasion, metastasis, angiogenesis and drug resistance and inhibits
apoptosis (16,21). There are a few studies reporting
that its expression decreases in some tumors and even acts as a
tumor suppressor (22,23). In the literature, only one study
examined the relationship between DANCR expression and
thyroid cancer and reported that the expression of DANCR
decreases in thyroid cancer (23). Therefore, the relationship between
DANCR and thyroid cancer and its clinicopathological
features requires investigation.
Tissue differentiation-induced non-coding RNA
(TINCR), which is ~3.7 kb long, is localized on chromosome
19. TINCR regulates the expression of genes that serve a
role in epidermal differentiation. The changes in TINCR
expression have been found in esophageal, breast, colon, lung,
prostate and bladder cancers and have associated with tumorigenesis
(24,25). Only one study has been published
that evaluated TINCR expression in thyroid cancer, to the
best of the authors' knowledge. In this study, the Cancer Genome
Atlas (TCGA) RNA sequencing data for PTC was examined and it was
reported that TINCR was one of a number of lncRNAs whose
expression increased in thyroid tumor tissue (26). Therefore, the relationship between
development of thyroid cancer, clinicopathological features and
TINCR expression needs to be supported by other data.
The aim of the present study was to investigate the
relationship between expressions of DANCR, TINCR, SOX2OT
lncRNAs and development and clinopathological features of PTC.
Materials and methods
Patients
A total of 112 PTC patients who had undergone a
thyroidectomy between January 2015 and December 2020 at Giresun
University's Faculty of Medicine were included in this study. Of
the 102 patients included in this study, 21 were male and 81 were
female. Their age range was 23–85 (median 51). The pathology
department histopathologically examined the tumor samples and
confirmed the diagnosis of PTC. Those samples (tumor and adjacent
non-cancerous thyroid tissue) were obtained from formalin-fixed
paraffin-embedded (FFPE) tissue blocks that were archived in the
pathology department. The patients included in the study upon
examination of the patient records were selected from the subjects
who had not undergone chemotherapy, radiotherapy, or other cancer
treatments before surgery. As a result of spectrophotometric
measurements performed after RNA isolation, 10 patients were
excluded from the study because RNA quality was not at optimum
level (A260/A280 ratio of 1.8-2.1) and the expression analysis was
performed on samples taken from the 102 patients. The present study
was approved by Giresun University's Faculty of Medicine Clinical
Trials Ethics Committee (approval no. 2018-06-10).
RNA isolation from FFPE tissue samples
and cDNA synthesis
After removing the excess paraffin, 4–5 5 µm-thick
sections were taken from the FFPE tissue blocks of the patients and
then transferred into 1.5 ml sterile micro centrifuge tubes. Total
RNA isolation was conducted using an RNeasy FFPE kit (Qiagen GmbH),
in accordance with the manufacturer's protocol. The quality and the
concentration of the RNA samples were assessed by measuring the
ratio of the absorbances at 260/280 nm on a NanoDrop One/OneC
Microvolume UV–Vis Spectrophotometer (Thermo Fisher Scientific,
Inc.). Then the integrity of the RNA samples was analyzed using 1%
agarose gel electrophoresis. Then, the samples that had poor RNA
quantity and quality were excluded from the study. All samples were
kept at −80°C for further research. The cDNA synthesis was carried
out using the RevertAid RT Reverse Transcription kit (Thermo Fisher
Scientific, Inc.), in accordance with the manufacturer's
protocol.
Reverse transcription-quantitative
(RT-q) PCR analysis
Expression analysis of lncRNAs was performed using a
LightCycler 480 SYBR-Green I Master (Roche Diagnostics GmbH) and
specific primer pairs according to the manufacturer's protocol on
the LightCycler 480 Real-Time PCR system (Roche Diagnostics GmbH).
β-actin was used as an internal control to normalize the expression
of lncRNAs. The primer pairs for RT-qPCR were: SOX2OT,
Forward 5′-GTAAGGCGATGTGGGTGAAG-3′; Reverse
5′-AGTTGAAGGAGCTTGCAGTT-3′; DANCR, Forward
5′-CTGCATTCCTGAACCGTTATCT-3′; Reverse 5′-GGGTGTAATCCACGTTTCTCAT-3′;
TINCR, Forward 5′-AGATGACAGTGGCTGGAGTTGTCA-3′; Reverse
5′-TGTGGCCCAAACTCAGGGATACAT-3′; β-actin, Forward
5′-TCTACAATGAGCTGCGTGTG-3′; Reverse 5′-GGTCTCAAACATGATCTGGGT-3′ The
thermocycling conditions were: 95°C for 5 min, 6 cycles of 95°C for
10 sec and 57°C for 25 sec, 45 cycles of 95°C for 10 sec, 57°C for
10 sec and 72°C for 10 sec. All samples were tested in triplicate.
Relative gene expression levels in lncRNAs were calculated using
the 2−ΔΔCq method (27).
Statistical analysis
Statistical analyses were performed using SPSS 15.0
(SPSS, Inc.). Continuous variables were expressed as mean ±
standard deviation. The expression levels of SOX2OT, DANCR
and TINCR in tumor and adjacent normal thyroid tissues were
compared using the Wilcoxon signed-rank test. The patient group was
divided into two subgroups (low expression and high expression)
according to median lncRNA expression values to determine the
relationship between the lncRNA expressions and clinicopathological
features in more detail. Categorical data were compared using the
chi-square test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression levels of SOX2OT, DANCR and
TINCR in PTC tissues
RT-qPCR was performed to determine the expression of
SOX2OT, DANCR and TINCR lncRNAs in tumor and adjacent
noncancerous thyroid tissue samples. The data revealed that
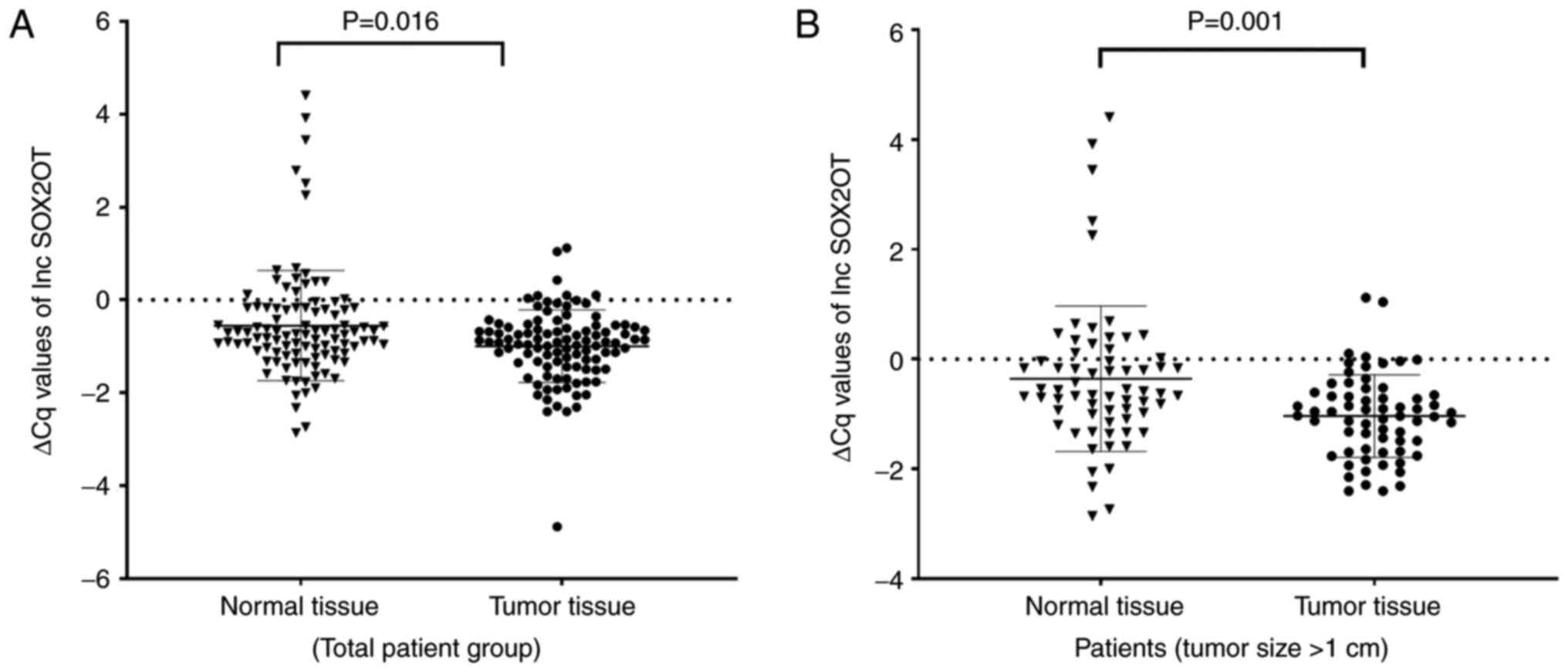
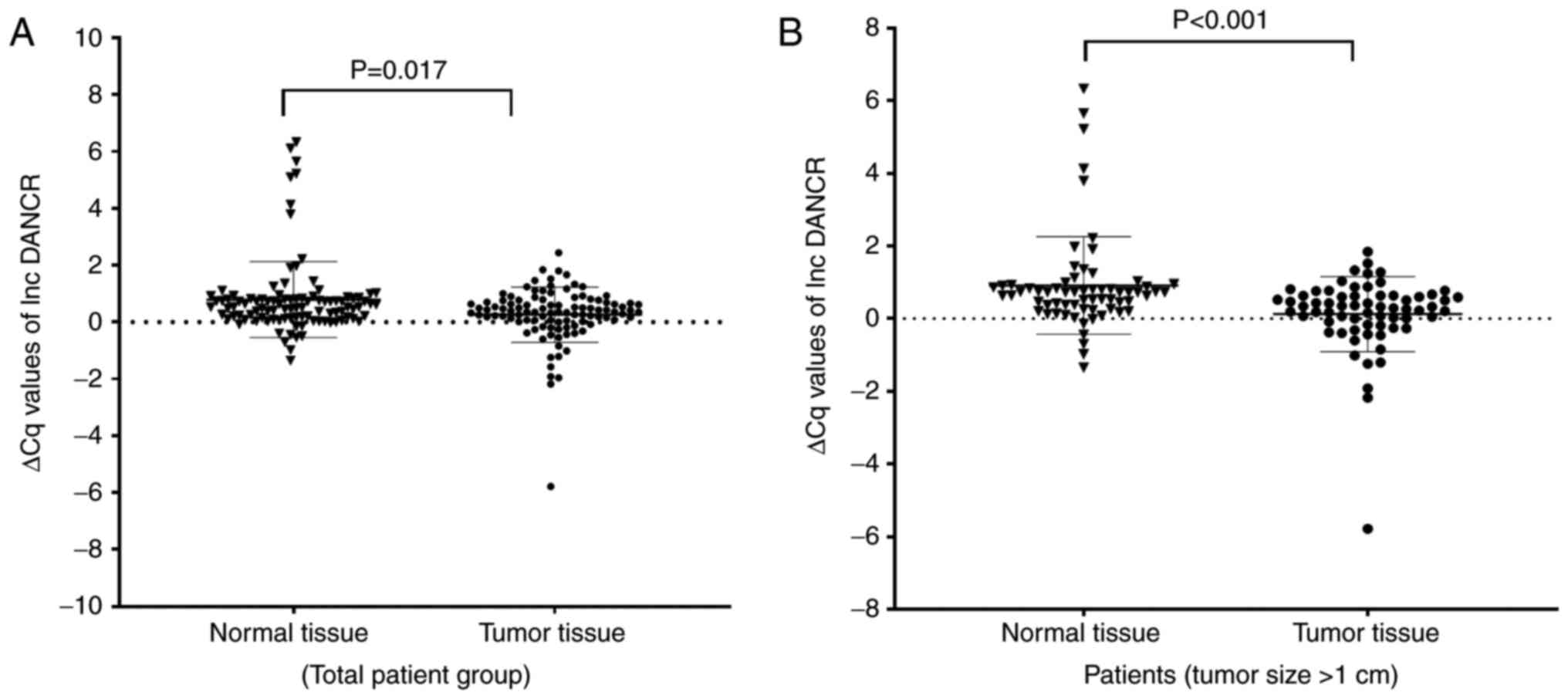
SOX2OT (fold change: 2.03, P=0.016, Z=−2.405) and
DANCR (fold change: 3.23, P=0.017, Z=−2.392) expression
significantly increased in the tumor tissues compared with adjacent
noncancerous thyroid tissues (Figs.
1A and 2A). On the other
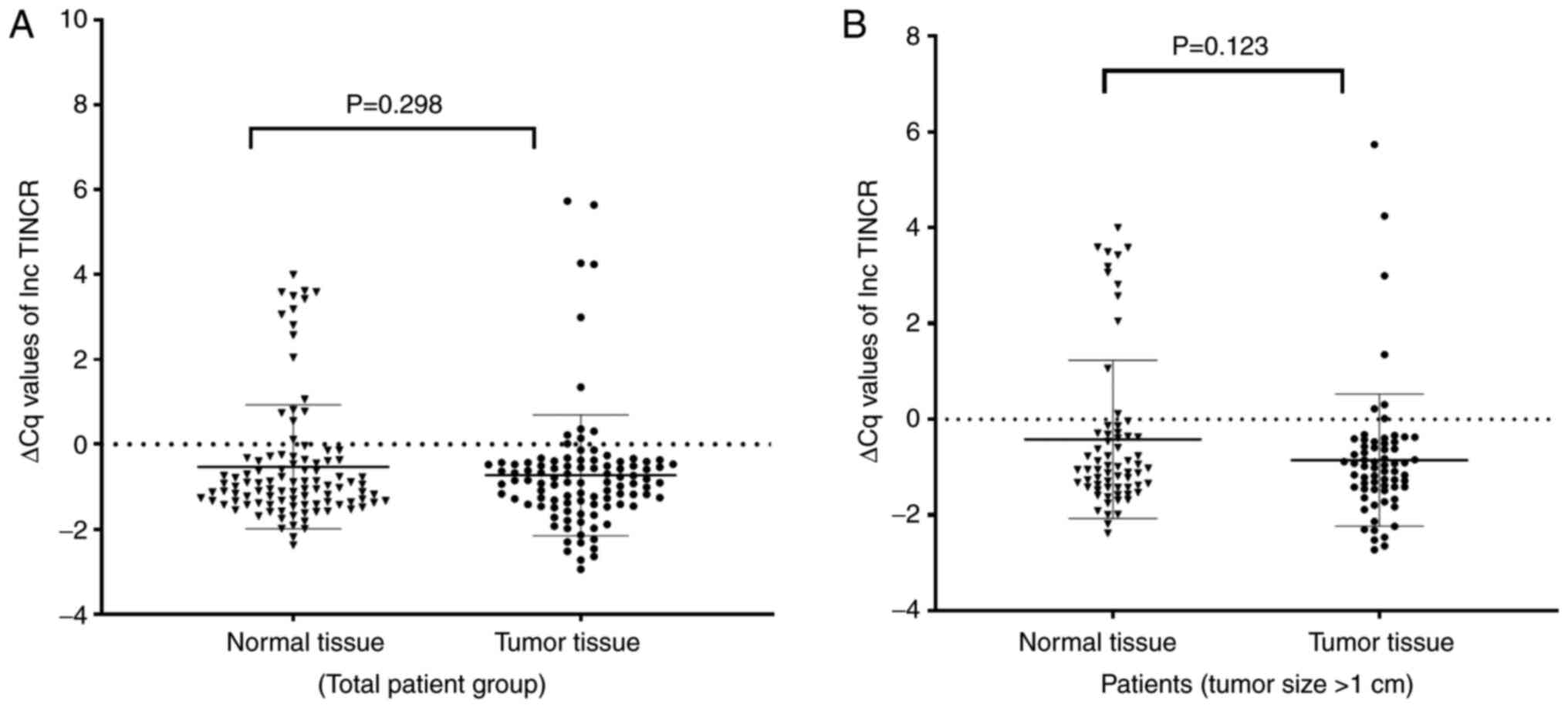
hand, no significant change was observed in the expression of
TINCR (fold change: −1.39, P=0.298, Z=−1.040; Fig. 3A).
The relationship between expression
levels of SOX2OT, DANCR and TINCR and the clinicopathological
features of PTC
The patient group was divided into high and low
expression subgroups according to the median expression values of
lncRNAs in order to assess the relationship between
clinicopathological features and expressions of lncRNAs.
Accordingly, there was a statistically significant correlation
between the SOX2OT expression level and microcarcinoma
(P<0.001), tumor size (P=0.010), primary tumor (P=0.006;
Table I) and between the
DANCR expression level and age (P=0.030), microcarcinoma
(P=0.004) (Table II). A
significant correlation was found between the expression level of
TINCR and primary tumor (P=0.029; Table III). Since the findings of the
present study revealed a significant correlation between the
expression levels of lncRNAs and tumor size, a separate evaluation
was carried out only in patients who had either papillary
microcarcinoma or papillary carcinoma with a tumor size >1 cm.
There was no significant difference in the expression levels of
SOX2OT (P=0.198, Z=−1.288), DANCR (P=0.162,
Z=−1.398), or TINCR (P=0.637, Z=−0.471) between the tumor
and adjacent normal tissues in the patients with papillary
microcarcinoma. On the other hand, when evaluating only papillary
carcinoma samples (tumor size >1 cm), it was observed that
differences in SOX2OT (P=0.001, Z=−3.440) and DANCR
(P<0.001, Z=−3.830) expression levels between tumor and normal
tissues became more significant (Figs. 1B and 2B). Again, no significant change was
observed in the expression level of TINCR (P=0.123,
Z=−1.543; Fig. 3B).
 | Table I.Association between expression level
of SOX2OT and clinicopathological characteristics of
patients with PTC. Low/high decided by the median expression of
SOX2OT. Chi square test was used for statistical
analysis. |
Table I.
Association between expression level
of SOX2OT and clinicopathological characteristics of
patients with PTC. Low/high decided by the median expression of
SOX2OT. Chi square test was used for statistical
analysis.
|
|
| SOX2OT
expression |
|
|
|---|
|
|
|
|
|
|
|---|
|
Characteristics | Number of
patients | Low | High | P-value | Odds ratio (95%
CI) |
|---|
| Age, years |
|
|
| 0.759 | 1.143
(0.490-2.680) |
|
<45 | 30 | 16 | 14 |
|
|
|
≥45 | 72 | 36 | 36 |
|
|
| Sex |
|
|
|
|
|
|
Female | 81 | 42 | 39 | 0.730 | 1.185
(0.450-3.100) |
|
Male | 21 | 10 | 11 |
|
|
| Microcarcinoma |
|
|
|
<0.001a | 0.203
(0.080-0.500) |
| No | 66 | 25 | 41 |
|
|
|
Yes | 36 | 27 | 9 |
|
|
| Histological
type |
|
|
| 0.129 |
|
|
Classical PTC | 17 | 1 | 10 |
|
|
|
Follicular PTC | 62 | 31 | 31 |
|
|
|
Classical-Follicular PTC | 5 | 1 | 4 |
|
|
|
Unknown | 18 | 13 | 5 |
|
|
| Tumor size
(cm) |
|
|
| 0.010a | 2.940
(1.290-6.720) |
|
<2 | 62 | 38 | 24 |
|
|
| ≥2 | 40 | 14 | 26 |
|
|
| Lymphovascular
invasion |
|
|
| 0.391 | 1.551
(0.570-4.250) |
| No | 83 | 44 | 39 |
|
|
|
Yes | 19 | 8 | 11 |
|
|
| Primary tumor |
|
|
| 0.006a |
|
| T1 | 78 | 46 | 32 |
|
|
| T2 | 20 | 6 | 14 |
|
|
| T3 | 4 | 0 | 4 |
|
|
| TNM stage |
|
|
| 0.114 | 0.475
(0.390-0.580) |
| I | 99 | 52 | 47 |
|
|
| II | 3 | 0 | 3 |
|
|
| Lymph node
metastasis |
|
|
| 1.000 | 0.510
(0.050-5.810) |
| No | 50 | 49 |
|
|
|
|
Yes | 2 | 1 |
|
|
|
| Extrathyroidal
extension |
|
|
| 0.488 | 0.489
(0.120-2.070) |
| No | 93 | 46 | 47 |
|
|
|
Yes | 9 | 6 | 3 |
|
|
|
Multicentricity |
|
|
| 0.238 | 1.629
(0.720-3.68) |
| No | 65 | 36 | 29 |
|
|
|
Yes | 37 | 16 | 21 |
|
|
| Multifocality |
|
|
| 0.712 | 1.160
(0.530-2.550) |
| No | 31 | 28 |
|
|
|
|
Yes | 21 | 22 |
|
|
|
 | Table II.Association between expression level
of DANCR and clinicopathological characteristics of patients
with PTC. Low/high decided by the median expression of
DANCR. Chi square test was used for statistical
analysis. |
Table II.
Association between expression level
of DANCR and clinicopathological characteristics of patients
with PTC. Low/high decided by the median expression of
DANCR. Chi square test was used for statistical
analysis.
|
|
| DANCR
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Number of
patients | Low | High | P-value | Odds ratio (95%
CI) |
|---|
| Age |
|
|
| 0.030a | 2.645
(1.090-6.450) |
|
<45 | 30 | 20 | 10 |
|
|
|
≥45 | 72 | 31 | 41 |
|
|
| Sex |
|
|
| 0.221 | 1.839
(0.690-4.910) |
|
Female | 81 | 43 | 38 |
|
|
|
Male | 21 | 8 | 13 |
|
|
| Microcarcinoma |
|
|
| 0.004a | 0.286
(0.120-0.680) |
| No | 66 | 26 | 40 |
|
|
|
Yes | 36 | 25 | 11 |
|
|
| Histological
type |
|
|
| 0.562 |
|
|
Classical PTC | 17 | 10 | 7 |
|
|
|
Follicular PTC | 62 | 31 | 31 |
|
|
|
Classical-Follicular PTC | 5 | 1 | 4 |
|
|
|
Unknown | 18 | 9 | 9 |
|
|
| Tumor size
(cm) |
|
|
| 0.685 | 1.179
(0.530-2.610) |
|
<2 | 62 | 32 | 30 |
|
|
| ≥2 | 40 | 19 | 21 |
|
|
| Lymphovascular
invasion |
|
|
| 0.799 | 1.138
(0.420-3.090) |
| No | 83 | 42 | 41 |
|
|
|
Yes | 19 | 9 | 10 |
|
|
| Primary tumor |
|
|
| 0.693 |
|
| T1 | 78 | 41 | 37 |
|
|
| T2 | 20 | 8 | 12 |
|
|
| T3 | 4 | 2 | 2 |
|
|
| TNM stage |
|
|
| 1.000 | 2.041
(0.180-23.240) |
| I | 99 | 50 | 49 |
|
|
| II | 3 | 1 | 2 |
|
|
| Lymph node
metastasis |
|
|
| 1.000 | 2.041
(0.180-23.240) |
| No | 99 | 50 | 49 |
|
|
|
Yes | 3 | 1 | 2 |
|
|
| Extrathyroidal
extension |
|
|
| 0.487 | 0.469
(0.110-1.990) |
| No | 93 | 45 | 48 |
|
|
|
Yes | 9 | 6 | 3 |
|
|
|
Multicentricity |
|
|
| 0.837 | 0.919
(0.410-2.060) |
| No | 65 | 32 | 33 |
|
|
|
Yes | 37 | 19 | 18 |
|
|
| Multifocality |
|
|
| 0.160 | 0.567
(0.260-1.270) |
| No | 59 | 26 | 33 |
|
|
|
Yes | 43 | 25 | 18 |
|
|
 | Table III.Association between expression level
of TINCR and clinicopathological characteristics of patients
with PTC. Low/high decided by the median expression of TINCR. Chi
square test was used for statistical analysis. |
Table III.
Association between expression level
of TINCR and clinicopathological characteristics of patients
with PTC. Low/high decided by the median expression of TINCR. Chi
square test was used for statistical analysis.
|
|
| TINCR
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | Number of
patients | Low | High | P-value | Odds ratio (95%
CI) |
|---|
| Age, years |
|
|
| 0.385 | 1.462
(0.620-3.450) |
|
<45 | 30 | 17 | 13 |
|
|
|
≥45 | 72 | 34 | 38 |
|
|
| Sex |
|
|
| 0.807 | 0.887
(0.340-2.320) |
|
Female | 81 | 40 | 41 |
|
|
|
Male | 21 | 11 | 10 |
|
|
| Microcarcinoma |
|
|
| 0.214 | 0.595
(0.260-1.350) |
| No | 66 | 30 | 36 |
|
|
|
Yes | 36 | 21 | 15 |
|
|
| Histological
type |
|
|
| 0.148 |
|
|
Classical PTC | 17 | 5 | 12 |
|
|
|
Follicular PTC | 62 | 31 | 31 |
|
|
|
Classical-Follicular PTC | 5 | 4 | 1 |
|
|
|
Unknown | 18 | 11 | 7 |
|
|
| Tumor size
(cm) |
|
|
| 0.417 | 0.719
(0.320-1.600) |
|
<2 | 62 | 29 | 33 |
|
|
| ≥2 | 40 | 22 | 18 |
|
|
| Lymphovascular
invasion |
|
|
| 0.075 | 2.566
(0.890-7.400) |
| No | 83 | 45 | 38 |
|
|
|
Yes | 19 | 6 | 13 |
|
|
| Primary tumor |
|
|
| 0.029a |
|
| T1 | 78 | 35 | 43 |
|
|
| T2 | 20 | 15 | 5 |
|
|
| T3 | 4 | 1 | 3 |
|
|
| TNM stage |
|
|
| 1.000 | 2.041
(0.180-23.240) |
| I | 99 | 49 | 50 |
|
|
| II | 3 | 2 | 1 |
|
|
| Lymph node
metastasis |
|
|
| 1.000 | 2.041
(0.180-23.240) |
| No | 50 | 49 |
|
|
|
|
Yes | 1 | 2 |
|
|
|
| Extrathyroidal
extension |
|
|
| 0.160 | 3.900
(0.770-19.760) |
| No | 93 | 49 | 44 |
|
|
|
Yes | 9 | 2 | 7 |
|
|
|
Multicentricity |
|
|
| 0.837 | 0.919
(0.410-2.060) |
| No | 65 | 32 | 33 |
|
|
|
Yes | 37 | 19 | 18 |
|
|
| Multifocality |
|
|
| 0.547 | 0.785
(0.360-1.730) |
| No | 59 | 28 | 31 |
|
|
|
Yes | 43 | 23 | 20 |
|
|
Discussion
Although prognosis of PTC is good, diagnostic
biomarkers and prognostic predictors are still required (28). Recent studies have focused on
lncRNAs as one of the most important molecular biomarkers for
various types of cancer and revealed that lncRNAs have important
roles in the development and progression of certain cancers
(29,30). However, there are few studies
specifically investigating the relationship between thyroid cancer
and lncRNAs (23,26,28). In the present study, the
relationship between the development and clinicopathological
features of PTC and expression levels of SOX2OT, DANCR and
TINCR was assessed. To the best of the authors' knowledge,
the present study is the first attempt to investigate the
association between expression level of SOX2OT and PTC.
Some studies have indicated that SOX2OT
lncRNA contributes to tumorigenesis via different mechanisms
(11,31). For example, it regulates the
expression of the SOX2 gene, which is located in its
intronic region and has a key role in regulating the pluripotency
properties of embryonic stem cells (32). In addition, SOX2OT acts as
a miRNA sponge. In a number of studies, it has been reported that
it regulates the expression of genes involved in tumorigenesis,
especially by binding to tumor suppressor miRNAs (12,14,33). Li et al (11) reported that SOX2OT
interacts with epigenetic regulators or directly binds to the
transcription factors and destabilizes them. Studies have revealed
that SOX2OT is an oncogene and its expression increases in
esophageal squamous cell, bladder, nasopharyngeal, prostate,
non-small-cell lung, gastric and hepatocellular cancer alongside
glioma (10,11). Similarly, the present study
revealed that SOX2OT expression increased significantly in
the PTC tumor samples compared with the adjacent normal tissues.
This suggested that the increase in the expression of SOX2OT
may be associated with the tumorigenesis process of PTC.
Expression of miR-204 is downregulated in PTC,
breast and cervical cancer and it acts as a tumor suppressor
(34). On the other hand,
SOX2OT contains a binding site for miR-204 and acts as a
sponge for this miRNA (11).
Similarly, some studies have reported that miR-132 and miR-122 have
tumor suppressor roles in PTC by suppressing FOXA1 and
DUSP4 expression, respectively (35,36). SOX2OT binds to these two
miRNAs and suppresses their expression (11). When the results of the present
study were compared with those of the aforementioned studies, it
was hypothesized that SOX2OT might be involved in the
papillary thyroid carcinoma process as it acts a miRNA sponge for
miRNA-204, miRNA-122 and miRNA-132.
Xue et al (37) reported that the enhancer of
zestehomolog 2 (EZH2) expression was higher in tumor samples
of PTC patients compared with the normal thyroid tissue samples.
SOX2OT is known to recruit EZH2 which induces H3K27me3 and
downregulates PTEN expression (10). Therefore, SOX2OT may
downregulate the expression of PTEN by upregulating
EZH2 and thus might contribute to tumorigenesis of PTC.
SOX2OT expression in various types of cancer has also been
found to be associated with the clinicopathological characteristics
of the patients. A study by Teng et al (38) correlated exosomal SOX2OT
expression with tumor size, lymph node metastasis and TNM stage in
patients with lung squamous cell carcinoma. Shi and Teng (39) determined that SOX2OT
expression was associated with histological grade, tumor number and
vein invasion status in hepatocellular carcinoma. In the present
study, it was found that elevated SOX2OT expression was
associated with tumor type, tumor size and primary tumor. When the
expression of SOX2OT was assessed only in micropapillary
carcinoma samples (tumor size ≤1 cm), no difference was found
between the tumor and normal tissues. On the other hand, when the
papillary carcinoma samples (tumor size >1 cm) were examined, it
was found that the expression difference between the tumor and
normal tissues was greater than in the total sample. Therefore, it
might be concluded that the expression of SOX2OT increased
significantly in PTC; however, the increase in tumors of 1 cm or
below was insignificant.
In recent years, an increasing number of studies
have reported abnormal DANCR expression in a number of types
of cancer (18–20). It has been suggested that this
abnormal expression is also associated with clinicopathological
features (19,21). Almost all of the studies conducted
on other types of cancer (e.g. osteosarcoma, nasopharyngeal,
glioma, non-small cell lung, cervical, ovarian, bladder, prostate,
breast, colorectal, gastric) have reported that DANCR
expression increased in tumor tissues compared with adjacent normal
tissues and this elevated expression level was associated with poor
prognosis (21,40–43). For example, in patients with
breast and colorectal cancer, DANCR expression is higher in tumor
compared with normal tissue and high DANCR expression is associated
with poor prognosis (19,20). In the current study, DANCR
expression increased significantly in the tumor samples of the 102
patients with PTC compared with their adjacent normal tissue. In
addition, DANCR expression was associated with age and
presence of microcarcinoma. Findings of the present study are
compatible with those of the literature.
In some studies, it was found that expressions of
miRNA-138, miRNA-199a, miRNA-335-5p and miRNA214 were
decreased in PTC tumor samples compared with adjacent normal tissue
samples and these miRNAs were identified as tumor suppressor miRNAs
that served a role in suppressing proliferation, inducing apoptosis
and mediating suppression of migration and invasion (34,44). On the other hand, it is known that
DANCR acts as a sponge for these miRNAs and reduces their
expression (21). Therefore, it
is hypothesized that DANCR may be involved in PTC
tumorigenesis process by acting as a competing endogenous RNA
(ceRNA) for these miRNAs.
Abnormal activation of the Wnt pathway and increased
expression of β-catenin may cause PTC to develop (45). Some studies have suggested that
DANCR may even have an oncogenic role by activating the
β-catenin signaling pathway in various cancers (16,18,43,46). AXL, a receptor tyrosine kinase,
shows an oncogenic activity by helping regulate a number of
different processes related to cancer development and progression
(47). AXL is highly
expressed in various types of cancer including thyroid cancer
(48). In some studies it has
been reported that DANCR activates the PI3K/AKT/NF-κB
signaling pathway by upregulating AXL (16). Therefore, DANCR may be
involved in the development and progression of PTC by interacting
with genes in the Wnt and PI3K/AKT pathways.
A few studies have suggested that low DANCR
expression may be associated with the development of cancer
(16,21). Zhang et al (23) reported that DANCR
expression is downregulated in tumor samples compared with the
adjacent normal tissues in 76 patients with PTC and they found that
DANCR expression was correlated with T grade and TNM stage.
The findings of the present study are not compatible with that
data. It is known that the roles of lncRNAs can differ from one
form of cancer to another (16,49). In addition, there are studies in
the literature reporting different results in the same cancer type
(16,24). Furthermore, expressions of lncRNAs
may differ in different populations (50). However, the relatively small
number of patients included in such studies may also be the reason
for the inconsistency between the results. In the present study,
similar to SOX2OT, when the expression of DANCR was
analyzed only in micropapillary carcinoma samples (tumor size ≤1
cm), no difference was observed between the tumor and normal
tissues. However, when only papillary carcinoma samples (tumor size
>1 cm) were examined, it was observed that the difference
between the tumor and normal tissues was greater compared with the
total sample evaluation. Based on findings of the present study, it
may be concluded that the expression of DANCR had increased
significantly in PTC, but it did not significantly increase in
tumors of ≤1.
Numerous studies have reported abnormal TINCR
expression in hepatocellular, colon, breast, bladder, lung,
prostate, gastric, esophageal and oral squamous cell cancer
(25,51–54). The expression of TINCR has
been defined at the last stage of epidermal differentiation and
regulates the expression of ALOXE3, FLG, LOR and
ALOX12B, all of which have important roles in
differentiation at the post-transcriptional level (55). Signaling pathways such as
Wnt/β-catenin, ERK1/2-SP3 and MAPK have been determined to be the
targets of TINCR in different types of cancer (24). In addition, TINCR
expression increases in esophageal, breast, bladder and gastric
cancer, but decreases in retinoblastoma, glioma and prostate cancer
(24,52,54,56–59). On the other hand, it has been
reported that TINCR expression is upregulated in certain
studies while it is downregulated in others conducted in the same
cancer type (24,54,57–59). In their RNA sequencing study, You
et al (26) found that
TINCR expression increased in PTC tumor tissues compared
with adjacent normal tissues. The findings of the present study
however, showed that TINCR expression did not show a
significant difference between the tumor and adjacent normal tissue
samples. You et al (26)
used TCGA data in their study and determined expression levels by
RNA sequencing analysis. In addition, the patient group in the
aforementioned study consisted of stage I, II, III and IV patients.
In the present study, the patient group mostly consisted of stage I
patients. Therefore, the fact that the patients included in the
studies were at different stages, the use of different expression
analysis methods and the limited number of patients may have led to
the difference in the results.
In conclusion, results of the current study
demonstrated that the expression levels of SOX2OT and
DANCR increased in PTC tissues compared with their adjacent
normal tissue. No significant change was observed in the expression
level of TINCR. However, when the micropapillary thyroid
carcinoma was examined, no significant increase was observed in the
expression levels of SOX2OT or DANCR. In addition,
the expression level of SOX2OT was associated with tumor
size and primary tumor. In the light of the results of the present
study, it is concluded that DANCR may contribute to the
development of PTC and SOX2OT may contribute to both the
development and progression of PTC. One of the limitations of the
present study is that the results of the expression analysis were
not confirmed by functional studies. Another limitation is that the
number of patients was relatively low. Future studies with a larger
number of patients in PTC and other subgroups of thyroid cancer
alongside in vitro functional studies will facilitate
supporting the data of the present study.
Acknowledgements
Not applicable.
Funding
This study was supported by the Scientific Research Projects
Committee of Giresun University (project numbers:
SAĞ-BAP-A-150219-39).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FMI, EAk and DS contributed to conception and design
of the study. FMI, EAk, DS and EAl performed experiments and data
collection. FMI, AO and EAl performed data analysis and
interpretation. FMI and AO drafted the paper. All authors
contributed to the manuscript revision and read and approved the
final version. FMI and EAl confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
This study was approved by Giresun University's
Faculty of Medicine Clinical Trials Ethics Committee (approval no:
2018-06-10). All patients provided written informed consent prior
to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Deng Y, Li H, Wang M, Li N, Tian T, Wu Y,
Xu P, Yang S, Zhai Z, Zhou L, et al: Global burden of thyroid
cancer from 1990 to 2017. JAMA Netw Open. 3:e2087592020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cao J, Zhang M, Zhang L, Lou J, Zhou F and
Fang M: Non-coding RNA in thyroid cancer-Functions and mechanisms.
Cancer Lett. 496:117–126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Javed Z, Ahmed Shah F, Rajabi S, Raza Q,
Iqbal Z, Ullah M, Ahmad T, Salehi B, Sharifi-Rad M, Pezzani R, et
al: LncRNAs as potential therapeutic targets in thyroid cancer.
Asian Pac J Cancer Prev. 21:281–287. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peng X, Zhang K, Ma L, Xu J and Chang W:
The Role of long non-coding RNAs in thyroid cancer. Front Oncol.
10:9412020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liyanarachchi S, Gudmundsson J,
Ferkingstad E, He H, Jonasson JG, Tragante V, Asselbergs FW, Xu L,
Kiemeney LA, Netea-Maier RT, et al: Assessing thyroid cancer risk
using polygenic risk scores. Proc Natl Acad Sci USA. 117:5997–6002.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Jin T, Shen H, Yan J, Guan M and
Jin X: Identification of long non-coding RNA expression profiles
and Co-expression genes in thyroid carcinoma based on the cancer
genome atlas (TCGA) database. Med Sci Monit. 25:9752–9769. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ghafouri-Fard S, Mohammad-Rahimi H and
Taheri M: The role of long non-coding RNAs in the pathogenesis of
thyroid cancer. Exp Mol Pathol. 112:1043322020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mahmoudian-Sani MR, Jalali A, Jamshidi M,
Moridi H, Alghasi A, Shojaeian A and Mobini GR: Long non-coding
RNAs in thyroid cancer: Implications for pathogenesis, diagnosis,
and therapy. Oncol Res Treat. 42:136–142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang X, Zhang H, Yang Q and Pang L:
LncRNA SOX2OT affects cervical cancer cell growth, migration and
invasion by regulating SOX2. Cell Cycle. 19:1391–1403. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Wu N, Luo X, Zhang X, Liao Q and
Wang J: SOX2OT, a novel tumor-related long non-coding RNA. Biomed
Pharmacother. 123:1097252020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li PY, Wang P, Gao SG and Dong DY: Long
Noncoding RNA SOX2-OT: Regulations, functions, and roles on mental
Illnesses, cancers, and diabetic complications. Biomed Res Int.
2020:29015892020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhan Y, Chen Z, He S, Gong Y, He A, Li Y,
Zhang L, Zhang X, Fang D, Li X and Zhou L: Long non-coding RNA
SOX2OT promotes the stemness phenotype of bladder cancer cells by
modulating SOX2. Mol Cancer. 19:252020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei CX, Wong H, Xu F, Liu Z, Ran L and
Jiang RD: IRF4-induced upregulation of lncRNA SOX2-OT promotes cell
proliferation and metastasis in cholangiocarcinoma by regulating
SOX2 and PI3K/AKT signaling. Eur Rev Med Pharmacol Sci.
22:8169–8178. 2018.PubMed/NCBI
|
|
14
|
Li Z, Jiang P, Li J, Peng M, Zhao X, Zhang
X, Chen K, Zhang Y, Liu H, Gan L, et al: Tumor-derived exosomal
lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in
pancreatic ductal adenocarcinoma. Oncogene. 37:3822–3838. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kretz M, Webster DE, Flockhart RJ, Lee CS,
Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, et al:
Suppression of progenitor differentiation requires the long
noncoding RNA ANCR. Genes Dev. 26:338–343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan L, Xiao X, Zhao Y, Yin L, Fu M, Zhang
X and Jiang P: The functional roles of long noncoding RNA DANCR in
Human Cancers. J Cancer. 11:6970–6981. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao YP, Qiu JH, Zhang DB and Yu CG: Long
non-coding RNA DANCR, a prognostic indicator, promotes cell growth
and tumorigenicity in gastric cancer. Tumour Biol. Jun
15–2017.(Epub ahead of print). doi: 10.1177/1010428317699798.
View Article : Google Scholar
|
|
18
|
Ma X, Wang X, Yang C, Wang Z, Han B, Wu L
and Zhuang L: DANCR Acts as a diagnostic biomarker and promotes
tumor growth and metastasis in hepatocellular carcinoma. Anticancer
Res. 36:6389–6398. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Y, Zhang M, Liang L, Li J and Chen YX:
Over-expression of lncRNA DANCR is associated with advanced tumor
progression and poor prognosis in patients with colorectal cancer.
Int J Clin Exp Pathol. 8:114802015.PubMed/NCBI
|
|
20
|
Tao W, Wang C, Zhu B, Zhang G and Pang D:
LncRNA DANCR contributes to tumor progression via targetting
miR-216a-5p in breast cancer: LncRNA DANCR contributes to tumor
progression. Biosci Rep. 39:BSR201816182019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan Y, Shi Q, Yuan X, Xue C, Shen S and He
Y: DANCR: An emerging therapeutic target for cancer. Am J Transl
Res. 12:4031–4042. 2020.PubMed/NCBI
|
|
22
|
Li Z, Hou P, Fan D, Dong M, Ma M, Li H,
Yao R, Li Y, Wang G, Geng P, et al: The degradation of EZH2
mediated by lncRNA ANCR attenuated the invasion and metastasis of
breast cancer. Cell Death Differ. 24:59–71. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang K, Lv J, Peng X, Liu J, Li C, Li J,
Yin N, Li H and Li Z: Down-regulation of DANCR acts as a potential
biomarker for papillary thyroid cancer diagnosis. Biosci Rep. Apr
23–2019.(Epub ahead of print). doi: 10.1042/BSR20181618.
|
|
24
|
Ghafouri-Fard S, Dashti S, Taheri M and
Omrani MD: TINCR: An lncRNA with dual functions in the
carcinogenesis process. Non-coding RNA Res. 5:109–115. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sharma U, Barwal TS, Malhotra A, Pant N,
Vive K, Dey D, Gautam A, Tuli HS, Vasquez KM and Jain A: Long
non-coding RNA TINCR as potential biomarker and therapeutic target
for cancer. Life Sci. 257:1180352020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
You X, Zhao Y, Sui J, Shi X, Sun Y, Xu J,
Liang G, Xu Q and Yao Y: Integrated analysis of long noncoding RNA
interactions reveals the potential role in progression of human
papillary thyroid cancer. Cancer Med. 7:5394–5410. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fu XM, Guo W, Li N, Liu HZ, Liu J, Qiu SQ,
Zhang Q, Wang LC, Li F and Li CL: The expression and function of
long noncoding RNA lncRNA-ATB in papillary thyroid cancer. Eur Rev
Med Pharmacol Sci. 21:3239–3246. 2017.PubMed/NCBI
|
|
29
|
Han CG, Huang Y and Qin L: Long Non-Coding
RNA ZFAS1 as a novel potential biomarker for predicting the
prognosis of thyroid cancer. Med Sci Monit. 25:2984–2992. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bolha L, Ravnik-Glavač M and Glavač D:
Long noncoding RNAs as biomarkers in cancer. Dis Markers.
2017:72439682017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Amaral PP, Neyt C, Wilkins SJ,
Askarian-Amiri ME, Sunkin SM, Perkins AC and Mattick JS: Complex
architecture and regulated expression of the Sox2ot locus during
vertebrate development. RNA. 15:2013–2027. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang E and Li X: LncRNA SOX2-OT regulates
proliferation and metastasis of nasopharyngeal carcinoma cells
through miR-146b-5p/HNRNPA2B1 pathway. J Cell Biochem.
120:16575–16588. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Santiago K, Chen Wongworawat Y and Khan S:
Differential MicroRNA-signatures in thyroid cancer subtypes. J
Oncol. 2020:20523962020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu N, Tian Y, Song Y and Zang L:
miR-122-5p suppresses the oncogenesis of PTC by inhibiting DUSP4
expression. Mol Med Rep. 23:3682021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen X, Li M, Zhou H and Zhang L: miR-132
Targets FOXA1 and exerts tumor-suppressing functions in thyroid
cancer. Oncol Res. 27:4312019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xue L, Yan H, Chen Y, Zhang Q, Xie X, Ding
X, Wang X, Qian Z, Xiao F, Song Z, et al: EZH2 upregulation by ERα
induces proliferation and migration of papillary thyroid carcinoma.
BMC Cancer. 19:10942019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Teng Y, Kang H and Chu Y: Identification
of an exosomal long noncoding RNA SOX2-OT in plasma as a promising
biomarker for lung squamous cell carcinoma. Genet Test Mol
Biomarkers. 23:235–240. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shi XM and Teng F: Up-regulation of long
non-coding RNA Sox2ot promotes hepatocellular carcinoma cell
metastasis and correlates with poor prognosis. Int J Clin Exp
Pathol. 8:4008–4014. 2015.PubMed/NCBI
|
|
40
|
Wang S and Jiang M: The long non-coding
RNA-DANCR exerts oncogenic functions in non-small cell lung cancer
via miR-758-3p. Biomed Pharmacother. 103:94–100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pan Z, Wu C, Li Y, Li H, An Y, Wang G, Dai
J and Wang Q: LncRNA DANCR silence inhibits SOX5-medicated
progression and autophagy in osteosarcoma via regulating
miR-216a-5p. Biomed Pharmacother. 122:1097072020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li Q, Jiang Y, Zhong G, Lu Y, Song T,
Zhang Y, Wu J, Zhang M, Liang X, Zhou L, et al: Long noncoding RNA
DANCR regulates cell proliferation by stabilizing SOX2 mRNA in
nasopharyngeal carcinoma. Am J Pathol. 190:2343–2354. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li J and Zhou L: Overexpression of lncRNA
DANCR positively affects progression of glioma via activating
Wnt/β-catenin signaling. Biomed Pharmacother. 102:602–607. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hitu L, Gabora K, Bonci EA, Piciu A, Hitu
AC, Ștefan PA and Piciu D: MicroRNA in papillary thyroid carcinoma:
A systematic review from 2018 to June 2020. Cancers (Basel).
12:31182020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lu HW and Liu XD: UCA1 promotes papillary
thyroid carcinoma development by stimulating cell proliferation via
Wnt pathway. Eur Rev Med Pharmacol Sci. 22:5576–5582.
2018.PubMed/NCBI
|
|
46
|
Pan L, Liang W, Gu J, Zang X, Huang Z, Shi
H, Chen J, Fu M, Zhang P, Xiao X, et al: Long noncoding RNA DANCR
is activated by SALL4 and promotes the proliferation and invasion
of gastric cancer cells. Oncotarget. 9:1915–1930. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Vouri M and Hafizi S: TAM Receptor
tyrosine kinases in cancer drug resistance. Cancer Res.
77:2775–2778. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Collina F, La Sala L, Liotti F, Prevete N,
La Mantia E, Chiofalo MG, Aquino G, Arenare L, Cantile M, Liguori
G, et al: AXL Is a novel predictive factor and therapeutic target
for radioactive iodine refractory thyroid cancer. Cancers (Basel).
11:7852019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu XF, Hao JL, Xie T, Pant OP, Lu CB, Lu
CW and Zhou DD: The BRAF activated non-coding RNA: A pivotal long
non-coding RNA in human malignancies. Cell Prolif. 51:e124492018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pahlevan Kakhki M, Rakhshi N, Emami
Aleagha MS, Abdari M, Alikhah A, Safarian G, Behmanesh M and
Nikravesh A: Differential expression of STAT3 gene and its
regulatory long non-coding RNAs, namely lnc-DC and THRIL, in two
eastern Iranian ethnicities with multiple sclerosis. Neurol Sci.
41:561–568. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen F, Qi S, Zhang X, Wu J, Yang X and
Wang R: lncRNA PLAC2 activated by H3K27 acetylation promotes cell
proliferation and invasion via the activation of Wnt/β-catenin
pathway in oral squamous cell carcinoma. Int J Oncol. 54:1183–1194.
2019.PubMed/NCBI
|
|
52
|
Chen Z, Liu H, Yang H, Gao Y, Zhang G and
Hu J: The long noncoding RNA, TINCR, functions as a competing
endogenous RNA to regulate PDK1 expression by sponging miR-375 in
gastric cancer. Onco Targets Ther. 10:3353–3362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tian F, Xu J, Xue F, Guan E and Xu X:
TINCR expression is associated with unfavorable prognosis in
patients with hepatocellular carcinoma. Biosci Rep.
37:BSR201703012017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu ZJ and He JK: TINCR facilitates
non-small cell lung cancer progression through BRAF-activated MAPK
pathway. Biochem Biophys Res Commun. 497:971–977. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kretz M, Siprashvili Z, Chu C, Webster DE,
Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al:
Control of somatic tissue differentiation by the long non-coding
RNA TINCR. Nature. 493:231–235. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Song L, Qi Y and Lin M: Long noncoding RNA
PLAC2 regulates PTEN in retinoblastoma and participates in the
regulation of cancer cell apoptosis. Oncol Lett. 19:2489–2494.
2020.PubMed/NCBI
|
|
57
|
Xia H, Xiu M, Gao J and Jing H: LncRNA
PLAC 2 downregulated miR-21 in non-small cell lung cancer and
predicted survival. BMC Pulm Med. 19:1722019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu Y, Qiu M, Chen Y, Wang J, Xia W, Mao Q,
Yang L, Li M, Jiang F, Xu L and Yin R: Long noncoding RNA, tissue
differentiation-inducing nonprotein coding RNA is upregulated and
promotes development of esophageal squamous cell carcinoma. Dis
Esophagus. 29:950–958. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang ZY, Lu YX, Zhang ZY, Chang YY, Zheng
L, Yuan L, Zhang F, Hu YH, Zhang WJ and Li XN: Loss of TINCR
expression promotes proliferation, metastasis through activating
EpCAM cleavage in colorectal cancer. Oncotarget. 7:22639–22649.
2016. View Article : Google Scholar : PubMed/NCBI
|