Introduction
The complex and characteristic structures of
dendrites in neurons are critical to their function of receiving
signals (1). Correct development
of dendrites is essential for neuronal function, which depends on
the coordinated organization of the cytoskeleton (2,3).
Rho GTPases are well-established regulators of the actin
cytoskeleton in various biological processes, including cell
migration, cell polarity and cell cycle progression (4). RhoA, Ras-related C4 botulinum toxin
substrate 1 (Rac1) and cell division control protein 42 homologue
(Cdc42) are extensively characterized members of the Rho family
that control dendrite growth in mammalian neurons by regulating the
growth dynamics of the cytoskeleton (5–8).
To date, the most common view is that Rac1 and Cdc42 positively
regulate dendrite growth and dynamics, whereas RhoA acts as a
negative regulator of dendrite arbor growth (5,9,10).
Cdc42 is highly expressed in the nervous system and
has emerged as a key regulator in dendrite branching and spine
development and morphology (11–13). A previous report revealed that
activated mutant Cdc42 expression inhibits dendritic arbor
morphology in the Drosophila peripheral nervous system
(14), while another study found
that loss-of-function mutations of Cdc42 result in normal
complexity but increased dendritic length, as well as defects in
dendrite caliber and stereotyped dendritic branch positions in
Drosophila mushroom body neurons, suggesting that Cdc42 is a
regulator of multiple aspects of dendritic development (15). In Xenopus central neurons,
it has been found that enhanced Cdc42 activity selectively
increased branch additions and retractions (16), and another study found that both
dominant-negative and constitutively active Cdc42 decrease the
total number of dendritic end tips (17). In mammals, dominant-negative Cdc42
expression in cortical neurons causes a marked reduction in the
number of primary dendrites. Conversely, Cdc42 overexpression in
cortical neurons leads to increased dendrite branching (10). However, one study reported that
knockdown of Cdc42 expression with short hairpin RNA (shRNA)
constructs increase dendritic branching in primary hippocampal
neurons (18). Another study also
demonstrated that inactivation Cdc42 by the GTPase-activating
protein NOMA-GAP is necessary for dendritic branching in an
analysis performed with transgenic mice (19). These contradictory reports suggest
that the role of Cdc42 may vary in different types of neurons.
However, the regulatory proteins of Cdc42 activity, such as Cdc42
effector protein, in regulating neuronal morphology are largely
unknown.
Cdc42 effector protein-4 (CEP4), a binder of Rho
GTPase-4, is ubiquitously expressed in all adult tissues (20,21). It has been confirmed that Cdc42
binds to CEP4 at its 16-amino acid Cdc42/Rac interaction binding
domain by site-specific mutagenesis (20). Previous studies have focused on
CEP4′s role in cancer (22–24), fibroblasts and epithelial cells
(25). However, the physiological
function of CEP4 in dendrite growth is unknown. In the present
study, the expression of CEP4 in hippocampal neurons was
investigated via western blotting and immunofluorescence (IF), and
the role of CEP4 in dendrite growth was investigated by observing
morphological changes in primary neurons following overexpression
or knockdown of CEP4.
Materials and methods
Mice
A total of 8 pregnant ICR mice and 6 adult mice
(age, 8–12 weeks; weight, 18–22 g) used in this study were
purchased from the Shanghai SLAC Laboratory Animal Co., Ltd. The
mice were bred and housed under standard conditions (temperature,
22 ± 2°C; humidity, 50±5%) with 12-h light/dark cycles and free
access to standard food and water. All animal procedures were
monitored by the Ethics Committee of the Soochow University and
were performed according to the guidelines of the Animal Care
Committee of Soochow University (approval no. SUDA20200116A01;
Suzhou, China). The E18 embryos used in this study were sacrificed
by decapitation with surgical scissors. The P1 and P3 neonatal pups
were sacrificed by cervical dislocation following anesthetized with
ice. The mice at P7, P14, P28, the pregnant females and adults were
sacrificed by cervical dislocation following anesthesia with 4%
isoflurane in 70% N2O and 30% O2.
Plasmid construction
Total RNA was extracted from hippocampal cDNA of
adult ICR mice using an RNeasy Mini Kit (Qiagen, Inc.). The
first-strand cDNA was obtained using Superscript II reverse
transcriptase (Vazyme Biotech Co., Ltd.) and reverse transcription
was performed according to the manufacturer's protocol. The CEP4
was amplified by PCR using the following primers: Forward
5′-CAGCTCTGTGAACTCGAAGC-3′ and reverse,
5′-GCTAGTGAGGAAAGACGTGTCC-3′. The following thermocycling
conditions were used: 95°C for 3 min; followed by 34 cycles of 95°C
for 30 sec, 55°C for 50 sec and 72°C for 2 min; and finally 72°C
for 15 min. The full-length CEP4 cDNA fragment was inserted into
Plv-IRES-ZsGREEN1 plasmid [Jiman Biotechnology (Shanghai) Co.,
Ltd.]. The CEP4 shRNA sequences were as follows: shRNA1,
5′-CAGCACTGTTTGTCAAGAATG-3′; shRNA2, 5′-GGAAGATGAGATCCGAGTTTG-3′;
shRNA3, 5′-CTCAGCTCAATGAGAAGGAAG-3′; and scramble shRNA (shSCR),
5′-TTCTCCGAACGTGTCACGT-3′. All shRNAs mentioned above were inserted
into the pGMLV-SC5-EGFP vector [Jiman Biotechnology (Shanghai) Co.,
Ltd.].
HT22 cell culture and
transfection
Mouse HT22 hippocampal neuronal cells were acquired
from the Cell Bank of Shanghai Institute of Cells, Chinese Academy
of Science. HT22 cells (2×105/well) were seeded in a
6-well culture plate and incubated in DMEM (Gibco, Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.) for 24 h prior to transfection. Subsequently,
Lipofectamine® 3000 transfection reagent (Invitrogen;
Thermo Fisher Scientific) was used for plasmid transfection (2,500
ng) according to the manufacturer's protocol. At 6–8 h
post-transfection, the culture media was replaced with DMEM
supplemented with 10% FBS and 100 U/ml penicillin/streptomycin
(Gibco; Thermo Fisher Scientific, Inc.). Cells were then grown for
48 h prior to harvesting.
Primary neuronal cell cultures and
transfection
To obtain primary isolated hippocampal neurons from
mice, E18 embryo brains were used; for each culture, one ICR
pregnant female mouse was sacrificed and 5–6 E18 embryos were used.
The E18 embryos were separated from the sacrificed nursing mother's
uterus after the death of the pregnant mouse was confirmed. The
hippocampus tissue was separated from the brain under a microscope
and cut into small pieces. After centrifugation (194 × g, 5 min,
4°C) to remove the supernatant, 1 ml Papin (1 mg/ml; cat. no.
p4752; Sigma-Aldrich; Merck KGaA) and DNase (5 mg/ml; 20 µl; cat.
no. DN25; Sigma-Aldrich; Merck KGaA) were added to the tube and
incubated at 37°C for 30 min. Subsequently, a single cell
suspension was pipetted, centrifuged to remove the upper clear
liquid and then added to complete medium. Isolated primary neurons
(5–10×104/ml) were seeded onto poly-D-lysine and
cultured in Neurobasal-A medium (cat. no. 10888022; Gibco; Thermo
Fisher Scientific, Inc.) supplemented with B27 (cat. no. 17504;
Gibco; Thermo Fisher Scientific, Inc.), 1% FBS (cat. no. 10099;
Gibco; Thermo Fisher Scientific, Inc.) and penicillin/streptomycin
mix (Gibco; Thermo Fisher Scientific, Inc.). For dendritic
complexity, 7-day-old cultured hippocampal neurons in 24-well
culture plates were transfected with different plasmids (shSCR,
shCEP4-1 or 2; Vector or CEP4-HA; 500 ng/well) for 7 days using the
calcium phosphate precipitation method (26). For IF, 7-day-old cultured
hippocampal neurons were transfected with the aforementioned
plasmids for 5 days.
Western blotting
When analyzing the expression of CEP4 in the
hippocampi from mice at different ages, 6 mice at P1, 6 mice at P3,
3 mice at P7, 3 mice at P14 and 3 mice at P28 were used in the
present study. To detect the CEP4 expression in the central nervous
system (CNS), the brain, spine, eye, optic nerve and different
regions of brain were collected from mice following transcardial
perfusion with cold PBS. Cultured HT22 cells, primary neurons
[cultured 0 to 21 days in vitro (DIV)] and hippocampal
regions from mice at different ages were harvested and lysed at 4°C
in RIPA lysis buffer (cat. no. P0013; Beyotime Institute of
Biotechnology) supplemented with 1 mM PMSF and protease inhibitor
cocktail (cat. no. 78425; Thermo Fisher Scientific, Inc.). Nuclear
and cytosol proteins were obtained using nuclear extraction kit
(cat. no. P0028; Beyotime Institute of Biotechnology). The protein
concentrations were analyzed using a Bradford assay. Protein
extracts were denatured for 5–8 min at 95°C in sample buffer.
Protein extract (20 µg) was loaded onto denatured 10% SDS gels
(EpiZyme), then transferred onto PVDF membranes (MilliporeSigma).
Membranes were subsequently blocked with 5% bovine serum albumin
(Beyotime Institute of Biotechnology) for 1 h at room temperature
and incubated with primary antibodies overnight at 4°C. The
following primary antibodies were used: Rabbit anti-CEP4 (1:1,000;
cat. no. LS-C804922; Lifespan Biosciences); mouse anti-GAPDH
(1:5,000; cat. no. G8795; Sigma-Aldrich; Merck KGaA); rabbit
anti-α-tubulin (1:2,000; cat. no. 2148S; Cell Signaling Technology,
Inc.); mouse anti- HA Tag (1:5,000; cat. no. M20003; Abmart, Inc.);
and rabbit anti-Lamin B (1:5,00; cat. no. AF1408; Beyotime
Institute of Biotechnology). On the next day, the membranes were
incubated with HRP-conjugated anti-mouse or anti-rabbit secondary
antibodies (1:5,000; cat. nos. A0216 and A0208, respectively;
Beyotime Institute of Biotechnology) at room temperature for 2 h
following washing with TBST (0.05% Tween). After washing three
times with TBST, the HRP signals were detected with enhanced
chemiluminescence reagents (Beyotime Institute of Biotechnology).
Quantification was performed by analyzing the relative density of
the immunoreactive bands using ImageJ software (version K 1.45;
National Institutes of Health).
IF
Primary cultured neurons were fixed in 0.01 M PBS
(pH 7.4) containing 4% PFA for 30 min at 4°C. For brain slices, 30
µm coronal brain slices were generated following transcardial
perfusion of mice with 4% PFA and fixed at 4°C overnight. The cells
or brain slices were then blocked with blocking buffer (0.3% Triton
X-100 in PBS) containing 10% donkey serum (Jackson ImmunoResearch
Laboratories, Inc.) for 1 h at room temperature. Then, the cells
were incubated with primary antibodies in blocking buffer
containing 2% goat serum overnight at 4°C. The following primary
antibodies were used for dual IF staining (same as used for western
blotting if not stated otherwise): Anti-CEP4 (1:1,000 for cells,
1:200 for brain slices); chicken anti-microtubule-associated
protein 2 (Map2; 1:10,000 for cells, 1:1,000 for slices; cat. no.
MAP; Aves Labs); and mouse anti-HA (1:1,000). The cells or slices
were then incubated with 488-conjugated donkey anti-rabbit antibody
(cat. no. A-21206; Invitrogen; Thermo Fisher Scientific, Inc.),
555-conjugated donkey anti-rabbit antibody (cat. no. A-31572;
Invitrogen; Thermo Fisher Scientific, Inc.), 647-conjugated donkey
anti-mouse antibody (cat. no. A-31571; Invitrogen; Thermo Fisher
Scientific, Inc.) or Cy3-conjugated donkey anti-chicken antibody
(cat. no. 703-605-155; Jackson ImmunoResearch Laboratories, Inc.)
at room temperature for 2 h. Prior to visualization, the cells were
incubated with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.)
for 5 min at room temperature, then washed with PBST. A confocal
microscope (Leica SP8; Leica Microsystems GmbH) was used to capture
images of cultured neurons and brain slices.
Image analysis and quantification
For the analysis of dendritic morphology, single
neuronal images were obtained with a confocal microscope
(magnification, ×40; SP8; Leica Microsystems GmbH) and 6–8 fields
of view were observed. Morphometric analysis and quantification
were performed as previously described (27). Briefly, a z-series of 6–12 images
with a 0.5-1 µm depth interval was taken at 1,024×1,024-pixel
resolutions. MetaMorph image analysis software version 7.1.3.0
(Molecular Devices, LLC) and the NeuronJ plugin in ImageJ was used
to analyze and semi-quantify neuronal morphology as previously
described (28–30). For Sholl analysis, a concentric
circle with a diameter of 15 µm was drawn around the cell body, and
the number of dendrites passing through each circle was manually
calculated.
Statistical analysis
Data are represented as the mean ± SEM from at least
three biological replicates for experiments. Statistical
differences were determined by Student's t-test for two-group
comparisons or ANOVA followed by Tukey's test for multiple
comparisons among >2 groups. All data statistical analyses were
performed using GraphPad Prism (version 8; GraphPad Software,
Inc.).
Results
CEP4 is highly expressed in
hippocampal neuron dendrites
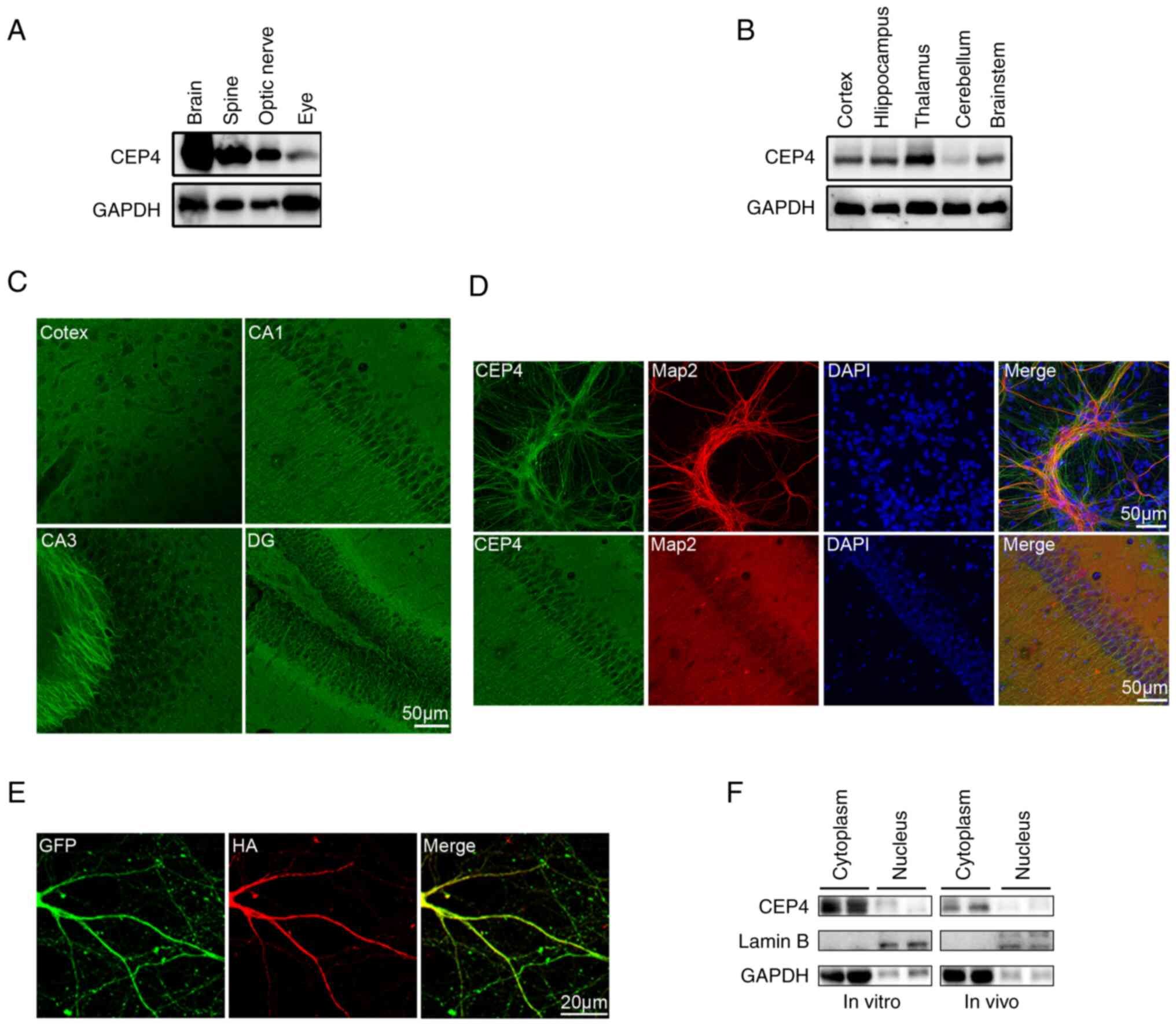
Initially, CEP4 expression was investigated in CNS.
As shown in Fig. 1A, CEP4 was
highly expressed in the brain compared with its expression in the
spine and optic nerve (Fig. 1A).
To further explore the distribution of CEP4 in the brain, its
expression pattern was analyzed via immunoblotting and IF. CEP4 was
highly expressed in the cortex, hippocampus, thalamus and
brainstem, while its expression in the cerebellum was substantially
lower (Fig. 1B). Prominent
labeling of CEP4 was also observed in the branching of neurons in
the cortex, CA1, CA3 and the granule cell layer of the dentate
gyrus (Fig. 1C). Expression of
CEP4 was observed in cells positive for the neuronal marker Map2
both in dissociated cultures of hippocampal neurons and in
hippocampal neurons in vivo, indicating that CEP4 is mainly
expressed in the dendrites of neurons (Fig. 1D). To avoid the uncertainty of the
results caused by polyclonal antibodies, an HA-tagged CEP4 plasmid
was used to overexpress CEP4 in cultured neurons, which confirmed
that CEP4 was mainly expressed in the cytoplasm and branching of
neurons, as determined by IF staining with monoclonal anti-HA
antibodies (Fig. 1E). Based on
these findings, cytoplasmic and nuclear proteins were separately
analyzed to determine the subcellular distribution of CEP4 both
in vitro and in vivo. As shown in Fig. 1F, CEP4 was mainly expressed in the
cytoplasm both in vitro and in vivo.
CEP4 is expressed in late
developmental stages in dissociated cultures of hippocampal neurons
and in the hippocampus
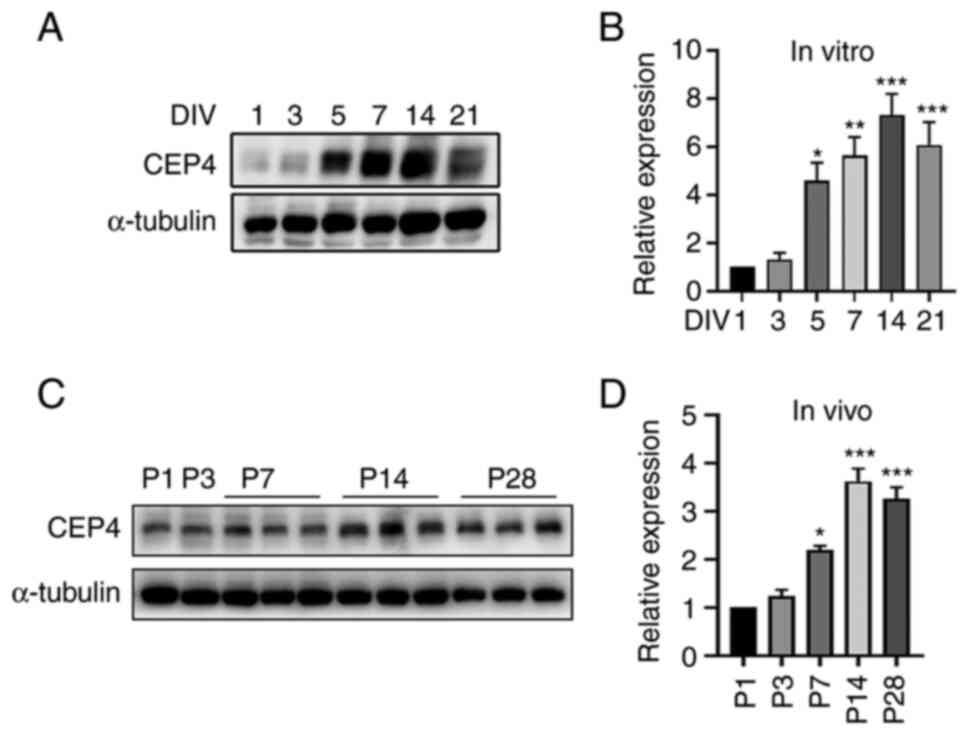
The expression of CEP4 was subsequently evaluated in
dissociated cultures of hippocampal neurons and in mouse brains at
various stages of development via western blotting; α-tubulin was
used as an internal standard. As shown in Fig. 2, CEP4 was present in dissociated
hippocampal neurons from 0 to 21 DIV, displaying upregulation from
day 5 onwards compared with day 1. In terms of CEP4 expression
in vivo, CEP4 was detected in extracts from mouse
hippocampus throughout development (P1 to P28), with increased
expression at P7-28 compared with P1.
Knockdown of CEP4 using shRNA
suppresses dendrite growth in primary cultured mouse hippocampal
neurons
To investigate the neuronal function of CEP4, three
shRNAs against CEP4 were designed: shCEP4-1, shCEP4-2 and shCEP4-3.
CEP4 knockdown was first performed in HT22 cells. All the shRNAs
induced efficient knockdown of CEP4, as determined via western
blotting, with >50% knockdown at the protein level, whereas
shSCR had no effect on the expression of endogenous CEP4 (Fig. 3A). To further determine the
effectiveness of the shCEP4 vectors, IF was performed using a
specific antibody against CEP4. shCEP4-1 markedly decreased CEP4
protein level compared with shSCR-transfected cells (Fig. 3B). The effects of CEP4 knockdown
on the dendritic arbor morphology of mouse hippocampal neurons were
subsequently explored. Hippocampal neurons cultured for 7 DIV were
used in this study, as intensive dendritogenesis occurs at this
time point; shRNA plasmids were transfected into hippocampal
neurons at 7 DIV, which were fixed at 14 DIV. Sholl analysis, which
measures the number of dendrites crossing the circle at different
radial distances from the cell soma, was used to quantify the
branching pattern. In principle, a leftward and/or downward shift
in Sholl analysis represents shrinkage of dendritic branching,
whereas a rightward or upward shift represents increased complexity
of the dendritic arbor. In the present study, a notable leftward
and downward shift was observed in shCEP4-transfected hippocampal
neurons compared with the shSCR group, indicating that CEP4 has a
specific and important role in the dendrites of neurons (Fig. 3C and D). The introduction of shRNA
against CEP4 markedly reduced the complexity of the dendritic
arbors of transfected cells. Transfection with shCEP4-1 or shCEP4-2
significantly decreased the total number of dendritic tips (TNDT)
compared with neurons transfected with shSCR (Fig. 3C and E). Changes in the total
dendrite length (TDL) evoked by CEP4 knockdown were also evaluated.
As presented in Fig. 3F,
transfection of hippocampal neurons with CEP4 shRNA plasmids
significantly decreased their TDL compared with shSCR. Taken
together, these results indicated that knockdown of CEP4 in
hippocampal neurons suppressed the growth of neuronal
dendrites.
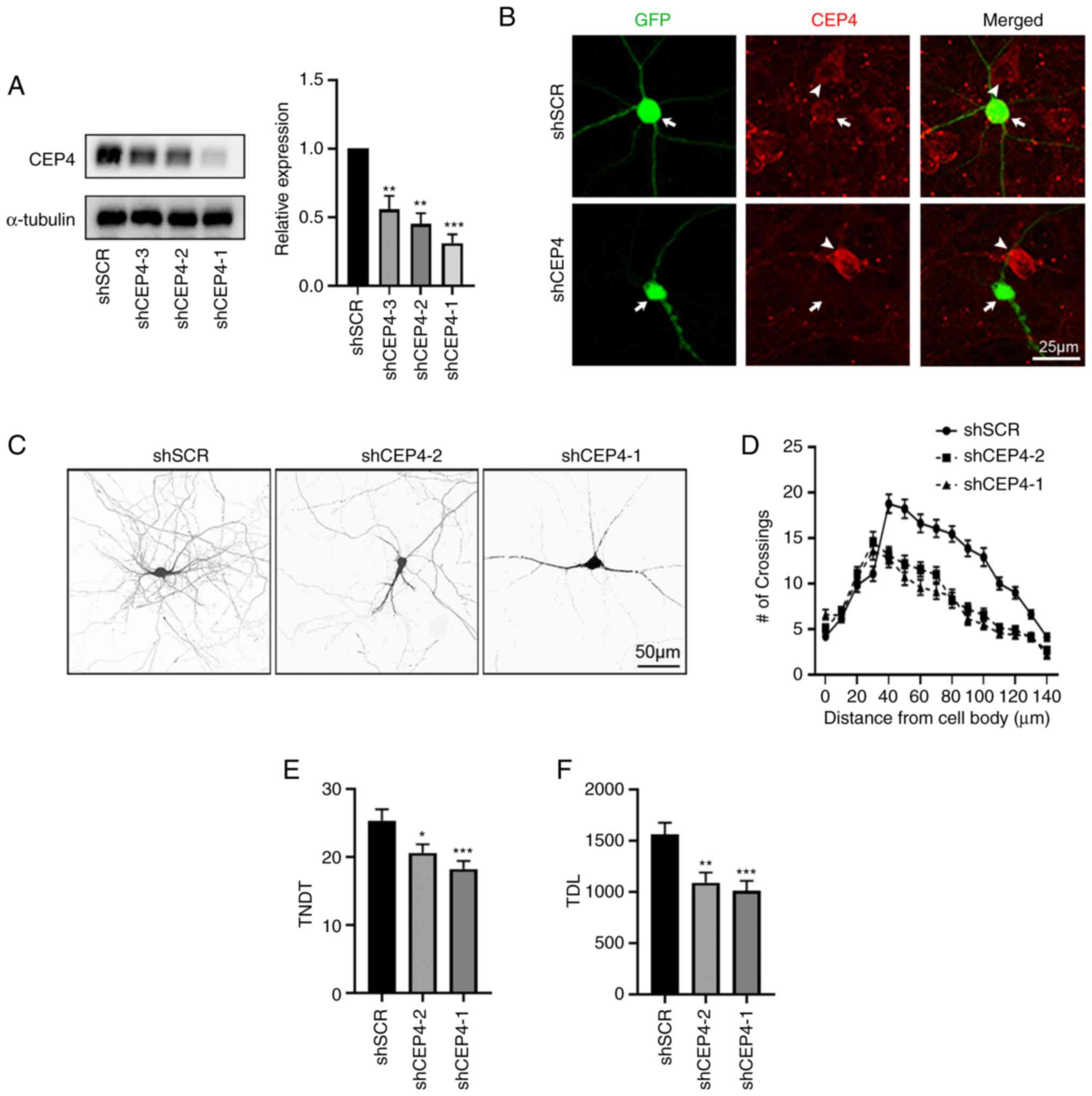 | Figure 3.CEP4 knockdown in hippocampal neurons
promotes the branching of dendrites. (A) HT22 cells were
transfected with shSCR or shCEP4, and the silencing efficiency was
identified via western blotting. The right panel shows
quantification of immunoblotting. (B) Hippocampal neurons cultured
in vitro were transfected on 7 DIV for 5 days with either
shSCR or shCEP4-1. The cells were then stained with an antibody
against CEP4; arrow indicates transfected neurons, whereas
arrowheads indicate non-transfected cells. (C) Representative
images of neurons transfected on 7 DIV for 7 days with shSCR,
shCEP4-1 or shCEP4-2. (D) Sholl analysis of neurons transfected
with shSCR, shCEP4-1 or shCEP4-2 (shSCR, n=48; shCEP4-1, n=48;
shCEP4-2, n=48). (E) TNDT and (F) TDL of hippocampal neurons after
CEP4 knockdown. Cell images were obtained from three culture
batches. Data are presented as the mean ± SEM. *P<0.05,
**P<0.01, ***P<0.001 vs. shSCR. CEP4, cell division control
protein 42 homolog effector protein-4; TNDT, total number of
dendritic tips; TDL, total dendrite length; sh, short hairpin; SCR,
scramble; DIV, days in vitro. |
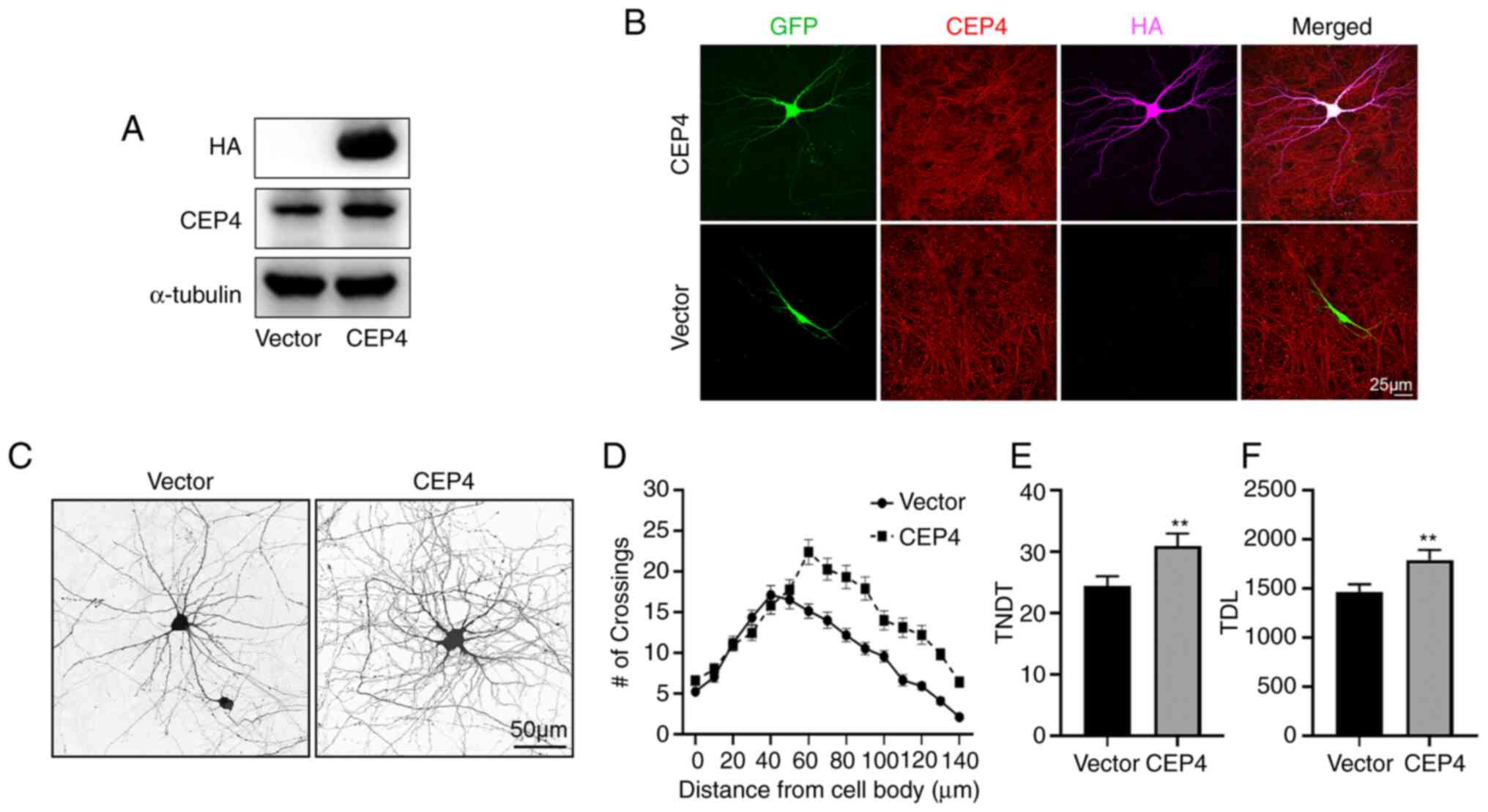
Overexpression of CEP4 in hippocampal
neurons promotes dendrite branching
The finding that CEP4 knockdown in hippocampal
neurons suppressed the growth of dendrites prompted speculation
that CEP4 plays a positive regulatory role in dendrite growth.
Thus, a plasmid construct containing HA-tagged CEP4 cDNA was used
to overexpress CEP4 in cultured hippocampal neurons. The
overexpression of CEP4 in HT22 cells was confirmed via western
blotting (Fig. 4A). In cultured
hippocampal neurons, staining for CEP4 was not notably increased
compared with the untransfected cells; however, staining for HA was
only observed in CEP4-overexpressing cells (Fig. 4B). Contrary to the effect of CEP4
knockdown in neurons, CEP4 overexpression in cultures increased
dendrite branching, TNDT and TDL (Fig. 4C). Sholl analysis showed that the
peak of branching in neurons transfected with CEP4 was shifted
rightward (farther from the soma) and upward (Fig. 4C and D). It was also observed that
overexpression of CEP4 significantly increased the TNDT and TDL of
neurons compared with neurons transfected with the empty vector
(Fig. 4C, E and F). Taken
together, these results indicated that CEP4 promoted the growth of
dendrites in neurons.
Discussion
Numerous studies have demonstrated that altering the
activity levels of Rho family proteins affects various aspects of
dendritic development (7,31,32). Cdc42, one of the most investigated
members of the Rho family, has been found to act as a key regulator
of neuronal morphology by controlling cytoskeletal reorganization
(8). CEP4 is a Cdc42 effector
protein that is distributed in various tissues, including the brain
(21). Recently, one study found
that knockdown of CEP4 resulted in an increase in the number of
progenitors in the intermediate zone of the brain in a paired box
protein 6-dependent manner (33);
however, little is known concerning its role in dendrites in the
hippocampus.
Binder of Rho GTPases (Borg)4, a member of the Borg
family of proteins, is expressed ubiquitously in the brain, heart,
kidneys, stomach and other tissues, as demonstrated via reverse
transcription-quantitative PCR (21). Although some dissimilarity exists
in the nucleotide sequences between CEP4 and Borg4, chromosomal
localization supports the idea that CEP4 is an ortholog of mouse
Borg4 (21). Thus, it is
hypothesized that CEP4 is also widely expressed throughout these
tissues. The presence of CEP4 in the brain supports the hypothesis
that CEP4 is highly expressed in the CNS, particularly in the
brain, as determined via western blotting in the present study.
Further investigation explored the expression of CEP4 in specific
brain regions. CEP4 was co-expressed with the neuronal dendritic
marker Map2 in brain slices and isolated neurons, indicating that
CEP4 is expressed in neurons. It was also found that CEP4 was
highly expressed in the dendrites of neurons and the dendritic
spines of hippocampal neurons by overexpressing the CEP4 sequence
with a fusion HA tag in neurons. Furthermore, it was revealed that
the expression of CEP4 remained high in cultured hippocampal
neurons from 7 DIV and in vivo from P7, a key period during
which dendrites grow and form a wide range of neuron-specific
branching patterns to cover the target area (3). Thus, these results suggested that
CEP4 may have important role in regulating dendrite growth.
shRNA-mediated gene silencing is an important tool
for biological research; these RNAs, through their hairpin-like
structure, can recognize specific mRNAs and interfere with their
degradation or hinder/inhibit their translation into proteins,
ultimately achieving intervention at the mRNA and protein levels
(34,35). As knockdown efficiency varies for
different shRNA target sites, it is necessary to design multiple
shRNAs in the research process to screen the most effective
knockdown vector (36). In the
present study, three shRNA plasmids were designed for CEP4, and it
was found that shCEP4-1 and shCEP4-2 induced effective knockdown of
CEP4 based on protein levels. It was observed that knockdown of
CEP4 significantly decreased TNDT and TDL, which suggested that
CEP4 was necessary for dendrite growth. Conversely, overexpression
of CEP4 increased TNDT and TDL, further supporting this hypothesis.
Therefore, the present study revealed a potentially novel role for
CEP4 in hippocampal neurons whereby CEP4 is necessary for dendritic
morphogenesis.
Several limitations of the current study should be
noted. First, rapid extension and arborization of dendrites occurs
during the development of dendrites (37); therefore, the ability of CEP4
knockdown to inhibit the branching of dendrites may result from
either the decreased formation of new branches or the increased
retraction of existing branches. Further study is necessary to
observe the development of living neurons with altered CEP4
expression on consecutive days using time-lapse microscopy. Second,
changes in electrical potential following knockdown of CEP4 in
hippocampal neurons have not been evaluated in the present study;
electrical potential across the functional membrane is an important
index of neuronal function (38).
Thus, electrophysiological analysis should be conducted in future
studies to evaluate the function of hippocampal neurons after CEP4
knockdown. Third, additional morphological or electrophysiological
evaluations may be required to evaluate the effect of CEP4
knockdown or overexpression on dendritic spines in hippocampal
neurons. Finally, the regulatory mechanism responsible for the
effect of CEP4 on dendrite morphology remains unclear. Therefore,
further elucidation of the related signaling pathways or
interacting proteins of CEP4 is required.
In summary, to the best of our knowledge, this is
the first study to elucidate the role of CEP4 in dendritic
morphogenesis in the hippocampus in vitro. It was
demonstrated that CEP4 was highly expressed in the brain, and that
the expression of CEP4 was gradually upregulated during neuronal
development. Combined with loss-of-function and gain-of-function
analyses in primary cultured hippocampal neurons, the present study
suggested that CEP4 is essential for dendritic growth.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science
Foundation of China for Young Scholars (grant no. 82102307) and the
Biomedical and Engineering Cross Youth Fund of Shanghai Jiao Tong
University (grant no. YG2021QN43).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
GC and YW designed the study. LH, LW and ZZ
performed the experiments and collected the data. LH, LW and WX
interpreted the data and drafted the manuscript. GC, YW and WX
reviewed and edited the manuscript. LH, LW, YW and GC confirmed the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal procedures were monitored by the Ethics
Committee of the Soochow University and conducted according to
protocols approved by the Animal Care Committee of Soochow
University (approval no. SUDA20200116A01).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Takano T, Xu C, Funahashi Y, Namba T and
Kaibuchi K: Neuronal polarization. Development. 142:2088–2093.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Muñoz-Lasso DC, Romá-Mateo C, Pallardó FV
and Gonzalez-Cabo P: Much more than a scaffold: Cytoskeletal
proteins in neurological disorders. Cells. 9:3582020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ledda F and Paratcha G: Mechanisms
regulating dendritic arbor patterning. Cell Mol Life Sci.
74:4511–4537. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hodge RG and Ridley AJ: Regulating Rho
GTPases and their regulators. Nat Rev Mol Cell Biol. 17:496–510.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen H and Firestein BL: RhoA regulates
dendrite branching in hippocampal neurons by decreasing cypin
protein levels. J Neurosci. 27:8378–8386. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Leemhuis J, Boutillier S, Barth H,
Feuerstein TJ, Brock C, Nürnberg B, Aktories K and Meyer DK: Rho
GTPases and phosphoinositide 3-kinase organize formation of
branched dendrites. J Biol Chem. 279:585–596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stankiewicz TR and Linseman DA: Rho family
GTPases: Key players in neuronal development, neuronal survival,
and neurodegeneration. Front Cell Neurosci. 8:3142014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen C, Wirth A and Ponimaskin E: Cdc42:
An important regulator of neuronal morphology. Int J Biochem Cell
Biol. 44:447–451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Scott GA and Cassidy L: Rac1 mediates
dendrite formation in response to melanocyte stimulating hormone
and ultraviolet light in a murine melanoma model. J Invest
Dermatol. 111:243–250. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Threadgill R, Bobb K and Ghosh A:
Regulation of dendritic growth and remodeling by Rho, Rac, and
Cdc42. Neuron. 19:625–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kalpachidou T, Spiecker L, Kress M and
Quarta S: Rho GTPases in the physiology and pathophysiology of
peripheral sensory neurons. Cells. 8:5912019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kamiyama D and Chiba A: Endogenous
activation patterns of Cdc42 GTPase within Drosophila
embryos. Science. 324:1338–1340. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kreis P, Thévenot E, Rousseau V, Boda B,
Muller D and Barnier JV: The p21-activated kinase 3 implicated in
mental retardation regulates spine morphogenesis through a
Cdc42-dependent pathway. J Biol Chem. 282:21497–21506. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo L, Liao YJ, Jan LY and Jan YN:
Distinct morphogenetic functions of similar small GTPases:
Drosophila Drac1 is involved in axonal outgrowth and
myoblast fusion. Genes Dev. 8:1787–1802. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Scott EK, Reuter JE and Luo L: Small
GTPase Cdc42 is required for multiple aspects of dendritic
morphogenesis. J Neurosci. 23:3118–3123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Z, Van Aelst L and Cline HT: Rho
GTPases regulate distinct aspects of dendritic arbor growth in
Xenopus central neurons in vivo. Nat Neurosci. 3:217–225.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ruchhoeft ML, Ohnuma S, McNeill L, Holt CE
and Harris WA: The neuronal architecture of Xenopus retinal
ganglion cells is sculpted by rho-family GTPases in vivo. J
Neurosci. 19:8454–8463. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Franke K, Otto W, Johannes S, Baumgart J,
Nitsch R and Schumacher S: miR-124-regulated RhoG reduces neuronal
process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO
J. 31:2908–2921. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rosário M, Schuster S, Jüttner R,
Parthasarathy S, Tarabykin V and Birchmeier W: Neocortical
dendritic complexity is controlled during development by
NOMA-GAP-dependent inhibition of Cdc42 and activation of cofilin.
Genes Dev. 26:1743–1757. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Joberty G, Perlungher RR and Macara IG:
The Borgs, a new family of Cdc42 and TC10 GTPase-interacting
proteins. Mol Cell Biol. 19:6585–6597. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Osada N, Kusuda J, Suzuki Y, Sugano S and
Hashimoto K: Sequence analysis, gene expression, and chromosomal
assignment of mouse Borg4 gene and its human orthologue. J Hum
Genet. 45:374–377. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao X and Rotenberg SA: Phosphorylation
of Cdc42 effector protein-4 (CEP4) by protein kinase C promotes
motility of human breast cells. J Biol Chem. 289:25844–25854. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Zhao X, Abeyweera TP and Rotenberg
SA: Analysis of substrates of protein kinase C isoforms in human
breast cells by the traceable kinase method. Biochemistry.
51:7087–7097. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wangsa D, Chowdhury SA, Ryott M, Gertz EM,
Elmberger G, Auer G, Lundqvist EÅ, Küffer S, Ströbel P, Schäffer
AA, et al: Phylogenetic analysis of multiple FISH markers in oral
tongue squamous cell carcinoma suggests that a diverse distribution
of copy number changes is associated with poor prognosis. Int J
Cancer. 138:98–109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hirsch DS, Pirone DM and Burbelo PD: A new
family of Cdc42 effector proteins, CEPs, function in fibroblast and
epithelial cell shape changes. J Biol Chem. 276:875–883. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Passafaro M, Piëch V and Sheng M:
Subunit-specific temporal and spatial patterns of AMPA receptor
exocytosis in hippocampal neurons. Nat Neurosci. 4:917–926. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen K, Zhu L, Guo L, Pan YB and Feng DF:
Maf1 regulates dendritic morphogenesis and influences learning and
memory. Cell Death Dis. 11:6062020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Peng Y, Lee J, Rowland K, Wen Y, Hua H,
Carlson N, Lavania S, Parrish JZ and Kim MD: Regulation of dendrite
growth and maintenance by exocytosis. J Cell Sci. 128:4279–4292.
2015.PubMed/NCBI
|
|
29
|
Jaworski J, Spangler S, Seeburg DP,
Hoogenraad CC and Sheng M: Control of dendritic arborization by the
phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin
pathway. J Neurosci. 25:11300–11312. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Swiech L, Blazejczyk M, Urbanska M,
Pietruszka P, Dortland BR, Malik AR, Wulf PS, Hoogenraad CC and
Jaworski J: CLIP-170 and IQGAP1 cooperatively regulate dendrite
morphology. J Neurosci. 31:4555–4568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Van Aelst L and Cline HT: Rho GTPases and
activity-dependent dendrite development. Curr Opin Neurobiol.
14:297–304. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chan WWR, Li W, Chang RCC and Lau KF:
ARF6-Rac1 signaling-mediated neurite outgrowth is potentiated by
the neuronal adaptor FE65 through orchestrating ARF6 and ELMO1.
FASEB J. 34:16397–16413. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Narayanan R, Pham L, Kerimoglu C, Watanabe
T, Hernandez RC, Sokpor G, Ulmke PA, Kiszka KA, Tonchev AB,
Rosenbusch J, et al: Chromatin remodeling BAF155 subunit regulates
the genesis of basal progenitors in developing cortex. iScience.
4:109–126. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rao DD, Vorhies JS, Senzer N and
Nemunaitis J: siRNA vs. shRNA: Similarities and differences. Adv
Drug Deliv Rev. 61:746–759. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Moazed D: Small RNAs in transcriptional
gene silencing and genome defence. Nature. 457:413–420. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Boettcher M and McManus MT: Choosing the
right tool for the job: RNAi, TALEN, or CRISPR. Mol Cell.
58:575–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Williams DW and Truman JW: Mechanisms of
dendritic elaboration of sensory neurons in Drosophila:
Insights from in vivo time lapse. J Neurosci. 24:1541–1550. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Y, Li X, Wang H, Gao Q, Zhang J, Zhang
W, Zhang Z, Li L, Yu Y and Shuai L: CRISPR/Cas9-edited Pax6-GFP
reporter system facilitates the generation of mouse neural
progenitor cells during differentiation. J Genet Genomics.
45:277–280. 2018. View Article : Google Scholar : PubMed/NCBI
|