Introduction
Diabetes mellitus (DM) is one of the major health
issues worldwide and the Kingdom of Saudi Arabia (KSA) has a high
prevalence of DM (1,2). In general, there are two types of
DM: type 1 DM (T1DM) is caused by the destruction of pancreatic β
cells that secrete insulin (3)
and type 2 DM (T2DM) which develops by tissue resistance to insulin
action and pancreatic β cell dysfunction (3). DM is associated with acute
consequences including diabetic ketoacidosis, hyperosmolar
hyperglycemic syndrome and chronic complications such as renal
failure, blindness, cardiovascular disease and diabetic neuropathy
(4). These complications
unfortunately result in high rates of morbidity and mortality. Both
T1DM and T2DM are heterogenous and polygenic in nature with
distinct characteristics (2,5).
Glucokinase (GCK) or hexokinase IV (EC 2.7.1.2)
catalyzes the conversion of glucose to glucose-6-phosphate (step 1
in glycolysis) in the liver and pancreas; and in other cells, this
reaction is catalyzed by hexokinase I (6). In hepatocytes, GCK enhances glucose
uptake for glycogenesis and energy storage, whereas in the
pancreas, GCK senses elevated blood sugar and stimulates the
insulin release by pancreatic β cells (6,7).
GCK activators enhance the pancreatic secretion of insulin and
hence increase hepatic glycogenesis (7,8).
The elevated liver glucose output is the main hepatic dysfunction
associated with T2DM (8). Genetic
variants of the GCK gene have been implicated in gestational
diabetes mellitus (GDM) (9–11),
neonatal diabetes (12) and T2DM
(13–15). GCK SNP rs4607517 T > C
has been reported to cause T2DM in American Indians (16), and rs1799884 G > A has been
associated with T2DM in Dutch (17), French (18) and Moroccan (19) populations.
MicroRNAs (miRNAs/miRs), small non-coding RNA
molecules, regulate gene expression and are involved in important
physiologic processes (20).
miRNA dysfunctions have been implicated in several diseases, such
as cancer, cardiovascular disease and diabetes (21–26). It has been reported that
MIR-196A-2 is involved in the regulation of insulin
signaling pathways (27) and that
gene variation in MIR-196A-2 can induce T2DM through the
regulation of body fat distribution (28). The miR-423 blood levels are
significantly decreased in cases with proliferative diabetic
retinopathy (29). The inhibition
of miR-423-5p decreases gluconeogenesis, reduces insulin resistance
and decreases blood glucose (13). In contrast, overexpression of
liver miR-423-5p increases gluconeogenesis, elevates blood glucose,
and enhances the deposition of fat in mice (30).
In the present study, GCK, MIR-196A-2 and
MIR-423 genotyping was conducted using Tetra
primer-amplification refractory mutation system-based polymerase
chain reaction (T-ARMS-PCR) to evaluate the potential clinical
association of GCK rs1799884 G>A, MIR-196A-2 rs11614913
C>T and MIR-423 rs6505162 C>A with the development and
progression of T2DM in individuals in the Asir and Tabuk regions of
Saudi Arabia. This technique is based on the use of
sequence-specific PCR primers that allow amplification of test DNA
only when the target allele is contained within the sample. It
involves a single PCR followed by gel electrophoresis. Designing
primers for the mutant [with single nucleotide polymorphisms
(SNPs)] and normal (without SNP) alleles allows selective
amplification which can be easily analyzed after electrophoresis.
It utilizes four primers viz forward outer (FO), reverse outer
(RO), forward inner (FI) and reverse inner (RI) primers. The FO/RO
primer combination generates the outer fragment of the SNP locus
and acts as an internal control for the PCR. The FI/RO and FO/RI
primer combinations yield allele-specific amplicons depending on
the genotype of the sample used. The inner primers are positioned
unequally from the corresponding outer primer to generate amplicons
with different sizes and hence easily resolvable in a gel and
distinction is made accordingly. T-ARMS PCR is a flexible, rapid
and economical SNP detection tool compared to contemporary
genotyping tools such as allele-specific PCR (31).
Materials and methods
Study population
This population-based case-control, collaborative
study was conducted on 110 T2DM patients and 110 healthy controls.
Specimens were collected from Asir and Tabuk regions of Saudi
Arabia in the following hospitals: Bisha: Diabetic Center, King
Abdullah Hospital, Bisha; Abha: Asir General Hospital, Abha; Tabuk:
King Fahd Specialty Hospital, Tabuk. The recruitment period of the
patients and controls was from March 2021 to October, 2021.
Informed consent was obtained prior to the collection of samples
from all patients and control subjects.
Ethical approval
Ethical approval was obtained from the local RELOC
Committee of the College of Medicine, University of Bisha (ref. no.
UBCOM/H-06-BH-087(04/10), in accordance with the local guidelines
which conformed in essence, to the principles of the Helsinki
Declaration.
Inclusion criteria
All the study subjects were citizens of Saudi Arabia
and included clinically confirmed cases with T2DM (both males and
females). The selected patients included those with fasting plasma
glucose levels >110 mg/dl and/or those clinically confirmed
patients who were on oral hypoglycemic agents or insulin and had
fasting glucose levels <110 mg/dl on the day of blood sampling.
Patients with random blood glucose >200 mg/dl and/or those
clinically confirmed patients who were on oral hypoglycemic agents
or insulin and had random glucose levels <200 mg/dl on the day
of blood sampling were also included.
Exclusion criteria
The T2DM patients with other significant chronic
diseases, such as renal failure, liver cirrhosis and malignancies
were excluded from the study. Type 1 diabetes patients were also
excluded from the study.
Inclusion criteria for controls
The control subjects were healthy volunteers with no
history of diabetes or any major clinical disorders (including
dyslipidemia) and had normal fasting and random plasma glucose
levels.
Data collection
This study included clinically confirmed cases of
T2DM in Saudi Arabia who visited the hospitals in Abha, Bisha and
Tabuk regions. This case-control study enrolled 110 subjects with
T2DM and 110 normal control subjects for each SNP. T2DM was
diagnosed according to the parameters of WHO criteria (who.int/diabetes/publications/Definition%20and%20diagnosis%20of%20diabetes_new.pdf).
The various variables that were analyzed from the T2DM patients and
controls included the case history, age and sex, duration of T2DM
(only for patients), glycated hemoglobin (HbA1c), fasting and
random blood glucose levels, total cholesterol, triacylglycerol
(TG), high-density lipoprotein-cholesterol (HDL-C) and low-density
lipoprotein cholesterol (LDL-C) concentrations, total
cholesterol/HDL-C ratios and serum creatinine. The biochemical
parameters were assayed using standard protocols.
Sample collection from the T2DM
patients
Approximately 3 ml of peripheral blood sample was
collected in an EDTA or Lavender top tube for all T2DM patients.
One aliquot of the blood specimens was immediately stored at −20 to
−30°C until further molecular studies. Another aliquot of blood (~2
ml) was collected in a red top tube and immediately sent for
biochemical analyses.
Sample collection from healthy
controls
All healthy age-matched control specimens were timed
around routine blood draws that were part of the routine workout
and hence did not require additional phlebotomy. All participants
provided a written informed consent form. Approximately 3 ml
peripheral blood was collected in EDTA tubes. The blood specimens
for molecular studies were immediately stored at −20 to −30°C until
further analyses. Another aliquot of blood (~2 ml) was collected in
a red top tube and immediately sent for biochemical analyses.
Genomic DNA extraction
Genomic DNA was extracted using DNeasy Blood K
(Qiagen GmbH) as per the manufacturer's instructions. The extracted
DNA was dissolved in nuclease-free water and stored at 4°C until
use. The quality and integrity of the DNA were checked by NanoDrop™
(Thermo Fisher Scientific, Inc.). All DNA samples from the patients
and controls were screened for purity by measuring optical density
(OD) at 260 nm (OD260) and 280 nm (OD280). The λ260/λ280 ratios
ranged from 1.83-1.99 indicating good quality DNA.
Genotyping of GCK, MIR-19A-2 and
MIR-423 genes by T-ARMS-PCR
The primers for GCK rs1799884 G>A and
MIR-196A-2 rs11614913 C>T were designed by using primer3
software (version 0.4.0, Whitehead Institute of Biomedical
Research). T-ARMS-PCR primers were optimized by gradient PCR. The
ARMS-PCR primers for MIR-423 rs6505162 C>A were prepared
by following previously used standard procedures (32,33). Reference sequence rs1799884 was
used to design the primers for GCK. For MIR-196A-2
rs11614913 C>T and MIR-423 rs6505162 C>T ARMS primers
were designed according to previously used procedures (32,33). The primers for all three SNPs are
depicted in Table I.
 | Table I.Primer sequences of GCK
rs1799884 G>A, MIR-196A-2 rs11614913 C>T and
MIR-423 rs6505162 C>A genes. |
Table I.
Primer sequences of GCK
rs1799884 G>A, MIR-196A-2 rs11614913 C>T and
MIR-423 rs6505162 C>A genes.
| A, ARMS primer
sequences of GCK rs1799884 G>A |
|---|
|
|---|
| Gene |
| Amplicon size | Temperature |
|---|
| GCK OF |
5′-GCTTTCTCTCCTGGTTGTGTTGAG-3′ | 390 bp | 59°C |
| GCK OR |
5′-GGTCACTGTAGTGACAAGGCGA-3′ |
|
|
| GCK
IF-C |
5′-CCTGCCAGGGCTTACTGGGC-3′ | 181 bp |
|
| GCK
IR-A |
5′-GACAACCACAGGCCCTCTCAGTAA-3′ | 252 bp |
|
|
| B, ARMS primer
sequences of miR-196a-2 rs11614913 C>T |
|
| Gene |
| Amplicon
size |
Temperature |
|
| MIR-196A-2
OF |
5-ACCCCCTTCCCTTCTCCTCCAGATAGAT-3 | 297 bp | 61°C |
| MIR-196A-2
OR |
5-AAAGCAGGGTTCTCCAGACTTGTTCTGC-3 |
|
|
| MIR-196A-2
IF (T allele) |
5-AGTTTTGAACTCGGCAACAAGAAACGGT-3 | 199 bp |
|
| MIR-196A-2
IR (C allele) |
5-GACGAAAACCGACTGATGTAACTCCGG-3 | 153 bp |
|
|
| C, ARMS primer
sequences of miR-423 rs6505162 C>A genes |
|
| Gene |
| Amplicon
size |
Temperature |
| MIR-423
OF |
5′-TTTTCCCGGATGGAAGCCCGAAGTTTGA-3′ | 336 bp | 62°C |
| MIR-423
OR |
5′-TTTTGCGGCAACGTATACCCCAATTTCC-3′ |
|
|
| MIR-423 IF
(T allele): |
5′-TGAGGCCCCTCAGTCTTGCTTCCCAA-3′ | 228 bp |
|
| MIR-423 IR
(C allele) |
5′-CAAGCGGGGAGAAACTCAAGCGCGAGG-3′ | 160 bp |
|
Preparation of the PCR cocktail
T-ARMS-PCR was performed in a reaction volume of 25
µl containing template DNA (50 ng), 0.25 µl primer stock solution
FO, RO, FI and RI, containing 5 pmol of each primer and 10 µl from
GoTaq® Green Master Mix (cat no M7122; Promega Corp.).
The final volume of 25 µl was adjusted by adding nuclease-free
ddH2O. Finally, 2 µl of DNA was added from each
subject.
Thermocycling conditions
The thermocycling conditions used were at 95°C for
10 min followed by 40 cycles of 95°C for 35 sec, annealing
temperature GCK rs1799884 G→A (59ºC),
MIR-196A-2 rs11614913 C>T (61ºC) and
MIR-423 rs6505162 C>A genes (62ºC), extension
for 72°C for 45 msec and final extension at 72°C for 10 min.
Gel electrophoresis for GCK
amplification
GCK rs1799884 G>A PCR products were
separated on 2% agarose gel stained with 2 µl of SYBR Safe stain
(Thermo Fisher Scientific, Inc.) and visualized on a UV
transilluminator (Bio-Rad Laboratories, Inc.). Primers FO and RO
flank the exon of the GCK rs1799884 G>A gene, resulting
in a band of 390 bp to act as a control for DNA quality and
quantity. Primers FO and RO amplify a wild-type allele (G allele),
generating a band of 181 bp, and primers FO and reverse mutant)
generate a band of 252 bp from the mutant allele (A allele). The
results are depicted in Fig.
1.
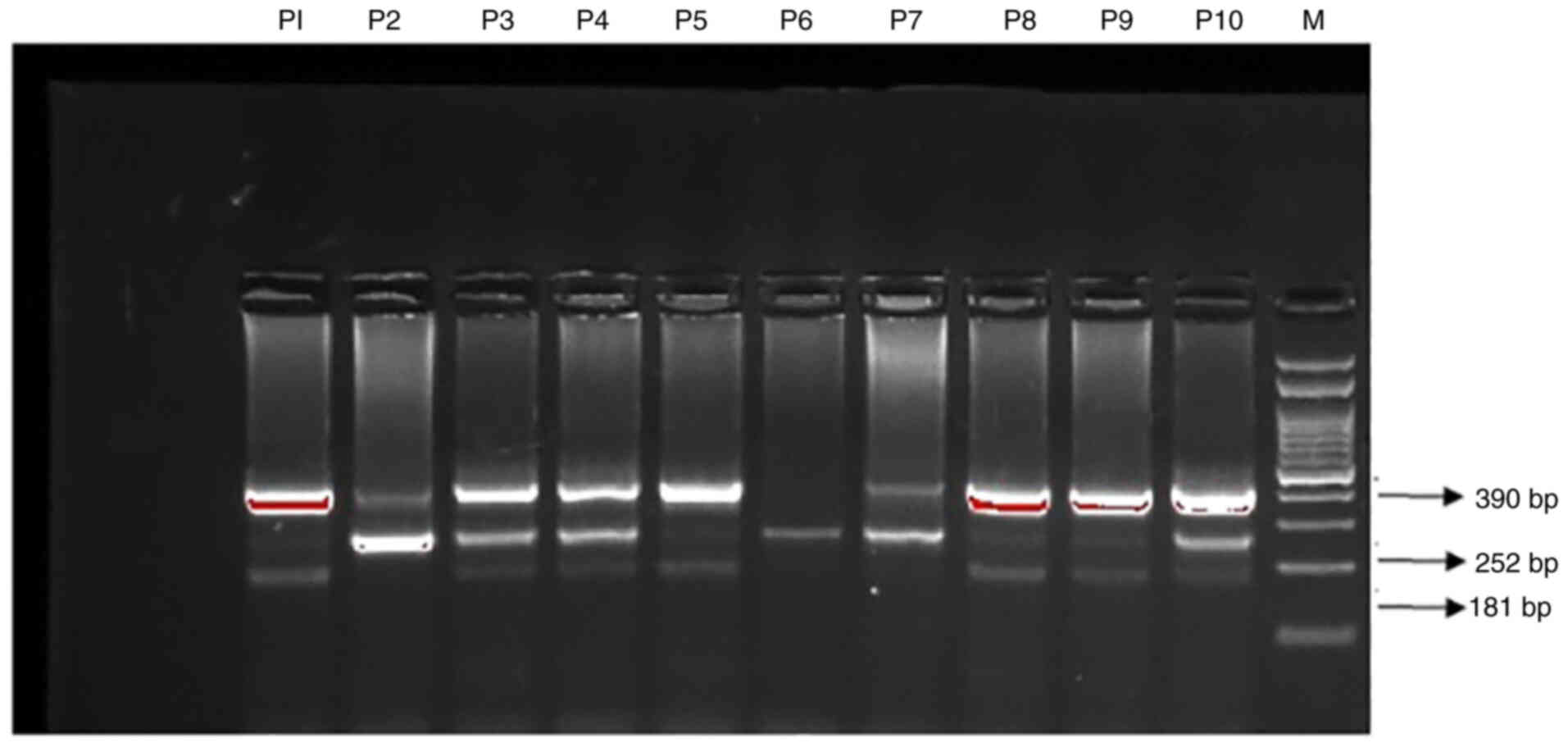 | Figure 1.Detection of GCK rs 1799884
G>A gene polymorphism by T-ARMS-PCR in T2DM patients. Lane M,
marker 100-bp DNA ladder; lanes P3, P4, P8, and P10, heterozygous
patients G/A; lanes, P2, P6, P7, homozygous patients GG allele;
lanes, P1, P5, P9, homozygous patients AA allele. GCK,
glucokinase; T2DM, type 2 diabetes mellitus; T-ARMS-PCR, tetra
primer-amplification refractory mutation system-based polymerase
chain reaction. |
Gel electrophoresis for MIR-196A-2
amplification
The amplification products for MIR-196A-2
rs11614913 C>T amplification were separated by electrophoresis
through 2% agarose gel stained with 0.5 µg/ml ethidium bromide and
visualized on a UV transilluminator. Primers FO and RO flank the
exon of the MIR-196A-2 rs11614913 C>T gene, resulting in
a band of 297 bp to act as a control for DNA quality and quantity.
Primers FO and RO amplify a wild-type allele (C allele), generating
a band of 153 bp, and primers FO and RI generate a band of 199 bp
from the mutant allele (T allele). The electrophoresis gel image is
shown in Fig. 2.
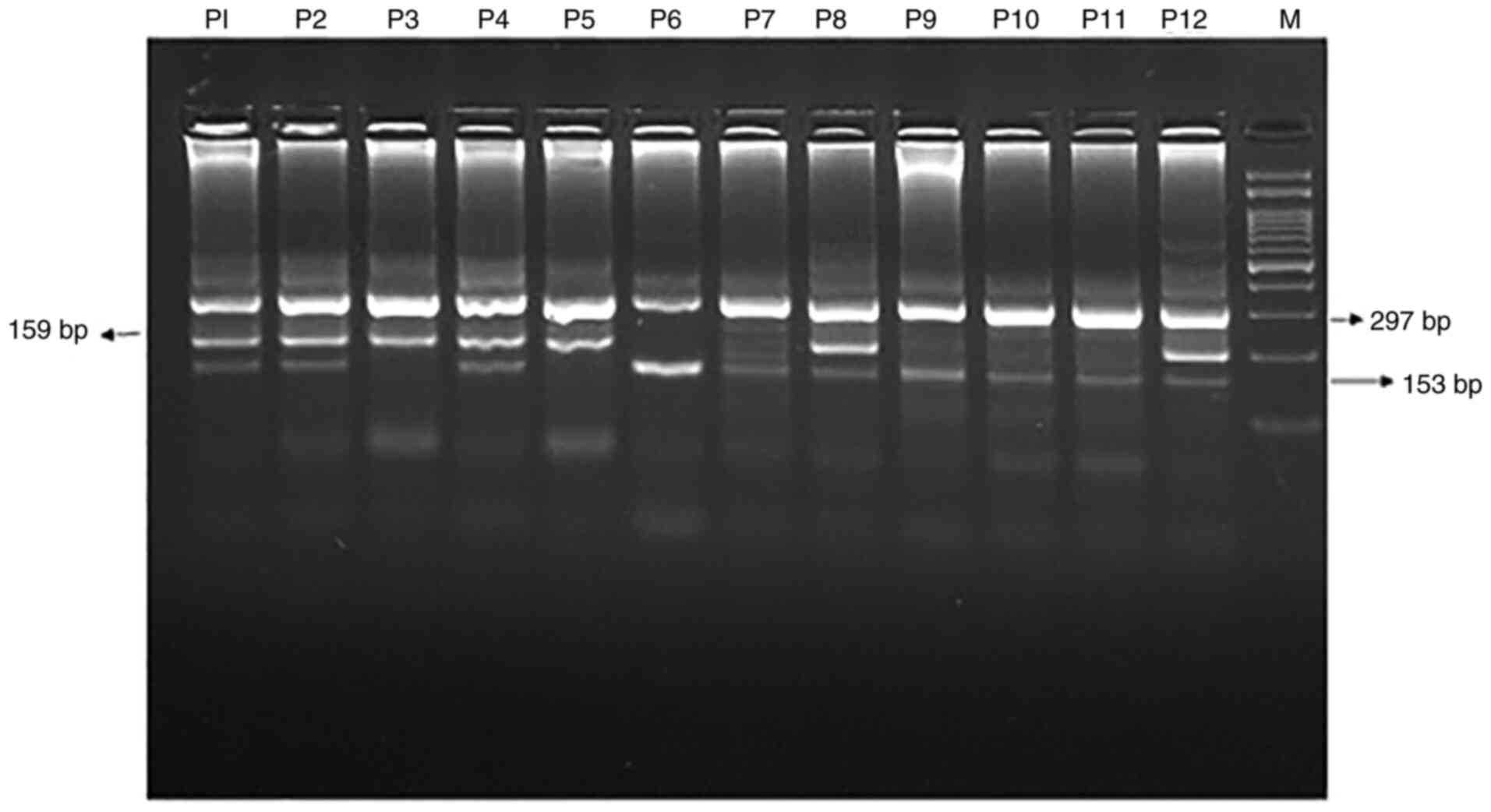 | Figure 2.Detection of MIR-196A-2
rs11614913 C>T gene polymorphism by T-ARMS-PCR in T2DM patients.
Lane M, marker 100-bp DNA ladder; lanes P1, P2, P4, P8, P12,
heterozygous patients; lanes P3, P5, homozygous TT patients; lanes
P6, P7, P9, P10, P11, homozygous CC patients. T2DM, type 2 diabetes
mellitus; T-ARMS-PCR, tetra primer-amplification refractory
mutation system-based polymerase chain reaction. |
Gel electrophoresis for MIR-423
amplification
PCR products were separated on 2% agarose gel
stained with 2 µl of SYBR Safe stain and visualized on a UV
transilluminator. Primers FO and RO flank the exon of the
MIR-423 rs6505162 C>T gene, resulting in a band of 336 bp
to act as a control for DNA quality and quantity. Primers FO and RO
amplify a wild-type allele (C allele), generating a band of 160 bp,
and primers FO and RO generate a band of 228 bp from the mutant
allele (T allele).
Statistical analysis
Group differences were compared using the Student's
two-sample t-test or one-way analysis of variance (ANOVA) for
continuous variables and the Chi-squared test for categorical
variables. Differences in the GCK rs1799884 G>A,
MIR-196A-2 rs11614913 C>T and MIR-423 rs6505162
C>A allele and genotype frequencies between groups were
evaluated using the Chi-square test. The associations between GCK,
MIR-196A-2 and MIR-423 genotypes with the risk of
T2DM were estimated by computing the odds ratios (ORs), risk ratios
(RRs) and risk differences (RDs) with 95% confidence intervals
(CIs). OR was calculated by dividing the odds of the first group by
the odds in the second group. The interpretation of the OR depends
on whether the predictor is categorical or continuous. ORs that are
>1 indicate that the event is more likely to occur as the
predictor increases. Odds ratios that are <1 indicate that the
event is less likely to occur as the predictor increases. OR
>1.0 indicates that the odds of exposure among patients are
greater than the odds of exposure among controls. For example, an
OR of 1.2 is above 1.0, but is not a strong association while as an
OR of 10 suggests a stronger association. Deviation from
Hardy-Weinberg disequilibrium (HWD) was calculated by Chi-square
(χ2) ‘goodness of fit test’. Allele frequencies among
patients and controls were evaluated by using the Chi-square
Hardy-Weinberg equilibrium test. A P-value <0.05 was considered
as indicative of a statistically significant difference. The
univariate and multivariate analyses were calculated by using
MedCalc software, version 20.027
(medcalc.org/calc/odds_ratio.php)/SPSS 16.0 (SPSS, Inc.).
Results
Demographic features and baseline
characteristics
The demographic features and the baseline
characteristics of the 110 consecutive T2DM patients are summarized
in Table II. Of the 110
consecutive patients, 61 were males and 49 were females, 20
patients were ≤40 years of age and 23 were >40 years of
age. The age range of the patients was 24–77 years with a mean of
50.32 years. The age range of the control group was 26–77 years
with a mean age of 51.46 years. Among the 110 T2DM patients, 28 had
fasting glucose ≤110 mg/dl and 82 had glucose >110 mg/dl.
Random blood glucose (RBG) was ≤200 mg/dl in 56 and >200
mg/dl in 54 patients respectively. A total of 60 T2DM patients had
total cholesterol ≤200 mg/dl and 50 had total cholesterol
>200 mg/dl. Among the 110 T2DM patients, 64 had triglycerides
(TG) >150 mg/dl and 46 had triglycerides ≤150 mg/dl. The
HDL-cholesterol was ≤55 mg/dl in 82 while it was >55
mg/dl in 28 patients, respectively. The LDL-cholesterol was
≤100 mg/dl in 30 while it was >100 mg/dl in 75 patients,
respectively. Differences in the mean of the serum lipid profile
for HDL-C, LDL-C, total cholesterol and TG were significant between
the patient and controls (P=0.0001). A total of 80 T2DM patients
had HbA1c >6% and 30 had HbA1c ≤6%. The serum creatinine
values were ≤1.35 mg/dl in 83 and >1.35 in 27 patients,
respectively.
 | Table II.Demographic features and baseline
characteristics of the T2DM patients and controls. |
Table II.
Demographic features and baseline
characteristics of the T2DM patients and controls.
|
| T2DM group | Control group |
|---|
|
|
|
|
|---|
| Subject
characteristics | n | % | n | % |
|---|
| Sex
distribution |
|
|
|
|
|
Males | 61 | 55.45 | 65 | 59.09 |
|
Females | 49 | 44.55 | 45 | 40.91 |
| Age distribution
(years) |
|
|
|
|
| Age
≤40 | 20 | 18.18 | 23 | 20.91 |
| Age
>40 | 23 | 81.82 | 87 | 79.09 |
| Fasting blood
glucose (mg/dl) |
|
|
|
|
| Glucose
≤110 | 28 | 25.45 | 96 | 87.27 |
| Glucose
>110 | 82 | 74.55 | 14a | 12.73 |
| Association with
RBG (mg/dl) |
|
|
|
|
| RBG
≤200 | 56 | 50.91 | 110 | 100 |
| RBG
>200 | 54 | 49.09 | 0 | 0 |
| Total cholesterol
(mg/dl) |
|
|
|
|
|
Cholesterol ≤200 | 60 | 54.55 | 104 | 94.55 |
|
Cholesterol >200 | 50 | 45.46 | 6b | 05.45 |
| HDL-C (mg/dl) |
|
|
|
|
| HDL-C
≤55 | 82 | 74.55 | 110 | 100 |
| HDL-C
>55 | 28 | 25.45 | 0 | 0 |
| LDL-C (mg/dl) |
|
|
|
|
| LDL
≤100 | 30c | 28.57 | 107 | 97.27 |
| LDL
>100 | 75c | 71.43 | 3d | 02.73 |
| TG (mg/dl) |
|
|
|
|
| TG
≤150 | 46 | 41.82 | 110 | 100 |
| TG
>150 | 64 | 58.18 | 0 | 0 |
| HbA1c |
|
|
|
|
| HbA1c
≤6% | 30 | 27.27 | 110 | 0 |
| HbA1c
>6% | 80 | 72.73 | 0 | 0 |
| Creatinine
(mg/dl) |
|
|
|
|
|
Creatinine ≤1.35 | 83 | 75.45 | 110 | 100 |
|
Creatinine >1.35 | 27 | 24.55 | 0 | 0 |
Statistical comparisons of GCK
(rs1799884 G>A) genotypes in the T2DM patients and controls
At the time of analysis, all of the 110 T2DM
patients displayed results in gel electrophoresis whereas only 107
healthy controls displayed sharp bands in the gel. As such only 107
results were included for the analyses. The results indicated that
there were significant differences in genotype distribution of the
GCK rs1799884 G>A genotypes between T2DM patients and
controls (P<0.015) (Table
III). The frequency of the genotypes (GG, GA and AA) between
the patients and controls was 23.7% 39 and 37.3 and 28, 52.3 and
19.7%, respectively. A higher frequency of the A allele (0.57) was
reported in T2DM patients in comparison to the healthy controls
(0.46).
 | Table III.Statistical comparisons of GCK
(rs1799884 G>A) genotypes in the T2DM patients and controls. |
Table III.
Statistical comparisons of GCK
(rs1799884 G>A) genotypes in the T2DM patients and controls.
| Subjects | N | GG (%) | GA (%) | AA (%) | χ2 | Df | G | A | P-value |
|---|
| T2DM patients | 110 | 26 (23.7) | 43 (39) | 41 (37.3) | 8.4 | 2 | 0.43 | 0.57 | 0.0150a |
| Controls | 107 | 30 (28) | 56 (52.33) | 21 (19.62) |
|
| 0.54 | 0.46 |
|
Multivariate analysis to estimate the
association between GCK genotypes and risk to T2DM
The presented study, significantly, yielded the
following results. a) The AA genotype was associated with T2DM with
OR=2.25 (1.071 to 4.737), RR=1.58 (1.034 to 2.418), P<0.0320
(Table IV). b) The A allele of
the rs1799884 G>A was associated with T2DM with an OR=1.55
(1.066 to 2.274), 1.25 (1.032 to 1.515), P<0.0210 (Table IV). (c) There was a significant
difference (P<0.05) in genotype distribution of rs1799884 G>A
between males and females (Table
V). d) There were significant differences in rs1799884 G>A
genotype distribution between patients with normal and elevated
fasting and random glucose and HbA1c (P<0.05) (Table V). e) Finally, there were
significant differences in the rs1799884 G>A genotype
distribution between patients with normal and abnormal lipid
profiles (Table V).
 | Table IV.Statistical comparisons between T2DM
patients and controls for GCK (rs1799884 G>A) genotypes
using multivariate analysis. |
Table IV.
Statistical comparisons between T2DM
patients and controls for GCK (rs1799884 G>A) genotypes
using multivariate analysis.
| Mode of
inheritance | Controls
(N=107) | Patients
(N=110) | OR (95% CI) | RR (95% CI) | P-value |
|---|
| Co-dominant |
|
|
|
|
|
|
GCK-GG | 30 | 26 | (ref.) | (ref.) |
|
|
GCK-GA | 56 | 43 | 0.86 (0.46 to
1.71) | 0.94 (0.71 to
1.27) | 0.7100 |
|
GCK-AA | 21 | 41 | 2.25 (1.07 to
4.74) | 1.58 (1.03 to
2.42) | 0.0320a |
| Dominant |
|
|
|
|
|
|
GCK-GG | 30 | 26 | (ref.) | (ref.) |
|
|
GCK-(GA+AA) | 77 | 84 | 1.25 (0.68 to
2.32) | 1.12 (0.84 to
1.50) | 0.4500 |
| Recessive |
|
|
|
|
|
|
GCK (GG+GA) | 86 | 69 | (ref.) | (ref.) |
|
|
GCK-AA | 21 | 41 | 2.43 (1.32 to
4.49) | 1.63 (1.13 to
2.38) | 0.0045a |
| Allele |
|
|
|
|
|
|
GCK-G | 116 | 95 | (ref.) | (ref.) |
|
|
GCK-A | 98 | 125 | 1.55 (1.07 to
2.27) | 1.25 (1.03 to
1.52) | 0.0210a |
 | Table V.Statistical comparisons of the
clinical features of the T2DM patients with GCK rs1799884
G>A genotypes. |
Table V.
Statistical comparisons of the
clinical features of the T2DM patients with GCK rs1799884
G>A genotypes.
| Subject
charactetistics | N=110 | GG | GA | AA | χ2 | df | P-value |
|---|
| Association with
sex |
|
|
|
|
|
|
|
|
Males | 61 | 10 | 40 | 11 | 11.8 | 2 | 0.0027b |
|
Females | 49 | 20 | 16 | 13 |
|
|
|
| Association with
age (years) |
|
|
|
|
|
|
|
| Age
≤40 | 20 | 10 | 6 | 4 | 2.12 | 2 | 0.3400 |
| Age
>40 | 90 | 28 | 44 | 18 |
|
|
|
| Fasting glucose
(mg/dl) |
|
|
|
|
|
|
|
| Glucose
≤110 | 28 | 13 | 6 | 9 | 14.52 | 2 | 0.0007b |
| Glucose
>110 | 82 | 20 | 50 | 12 |
|
|
|
| Association with
RBG (mg/dl) |
|
|
|
|
|
|
|
| RBG
≤200 | 35 | 20 | 10 | 05 | 22.0 | 2 | 0.0001b |
| RBG
>200 | 72 | 10 | 46 | 16 |
|
|
|
| Association with
total cholesterol (mg/dl) |
|
|
|
|
|
|
|
|
Cholesterol ≤200 | 60 | 22 | 32 | 06 | 9.82 | 2 | 0.0070b |
|
Cholesterol >200 | 50 | 10 | 24 | 16 |
|
|
|
| Association with
HDL-C (mg/dl) |
|
|
|
|
|
|
|
| HDL-C
≤55 | 82 | 10 | 50 | 22 | 28.8 | 2 | 0.0001b |
| HDL-C
>55 | 28 | 09 | 14 | 05 |
|
|
|
| Association with
LDL-C (mg/dl) |
|
|
|
|
|
|
|
| LDL-C
≤100 | 30a | 10 | 8 | 12 | 3.65 | 2 | 0.1600 |
| LDL-C
>100 | 75a | 20 | 45 | 10 |
|
|
|
| Association with TG
(mg/dl) |
|
|
|
|
|
|
|
| TG
≤150 | 46 | 10 | 28 | 08 | 14.2 | 2 | 0.0008b |
| TG
>150 | 64 | 10 | 42 | 12 |
|
|
|
| Association with
HBA1c % |
|
|
|
|
|
|
|
| HBA1c
≤6 | 30 | 16 | 06 | 08 | 13 | 2 | 0.0013b |
| HBA1c
>6 | 80 | 20 | 50 | 10 |
|
|
|
| Association with
creatinine (mg/dl) |
|
|
|
|
|
|
|
|
Creatinine ≤1.35 | 83 | 18 | 49 | 16 | 4.35 | 2 | 0.1100 |
|
Creatinine >1.35 | 27 | 12 | 07 | 08 |
|
|
|
Association of MIR-196A-2 rs11614913
C>T genotypes with T2DM
At the time of analysis, out of 110, only 100 T2DM
patient samples gave sharp bands in gel electrophoresis for
MIR-196A-2 rs11614913 C>T genotyping. Similarly, for
controls only 100 displayed sharp bands. The results in this
analysis indicated there was a significant difference (P<0.0190)
in the MIR-196A-2 rs11614913 C>T genotype between
patients and controls (Table
VI).
 | Table VI.Distribution of MIR-196A-2
rs11614913 C>T SNP genotypes in T2DM patients and controls. |
Table VI.
Distribution of MIR-196A-2
rs11614913 C>T SNP genotypes in T2DM patients and controls.
| Subjects | N | CC (%) | CT (%) | TT (%) | Df | χ2 | C | T | P-value |
|---|
| Patients | 100a | 51 (51) | 43 (43) | 6 (6) | 2 | 7.84 | 0.73 | 0.27 | 0.0190b |
| Controls | 100a | 70 (70) | 25 (25) | 5 (5) |
|
| 0.85 | 0.15 |
|
Multivariate analysis to estimate the
association between MIR-196A-2 rs11614913 C>T genotypes and T2DM
risk
Results showed that MIR-196A-2 rs11614913 CT
genotype was associated with T2DM with OR=2.36 (1.2816 to 4.348),
RR=1.57 (1.1124 to 2.225), P=0.0059 (Table VII). The T allele of the
MIR-196A-2 rs11614913 was associated with T2DM with OR=1.74
(1.0787 to 2.807), RR=1.35 (1.0217 to 1.787), P=0.023 (Table VII).
 | Table VII.Statistical comparisons between T2DM
patients and controls forMIR-196A-2 rs11614913 C>T
genotypes using multivariate analysisa. |
Table VII.
Statistical comparisons between T2DM
patients and controls forMIR-196A-2 rs11614913 C>T
genotypes using multivariate analysisa.
| Genotypes | Healthy
controls | T2DM cases | OR (95% CI) | RR | P-value |
|---|
| Codominant |
(N=100)b |
(N=100)b |
|
|
|
|
MIR-196A-2-CC | 70 | 51 | 1 (ref.) | 1 (ref.) |
|
|
MIR-196A-2-CT | 25 | 43 | 2.36 (1.28 to
4.35) | 1.57 (1.11 to
2.23) | 0.0059c |
|
MIR-196A-2-TT | 05 | 06 | 1.64 (0.48 to
5.69) | 1.27 (0.65 to
2.47) | 0.4300 |
| Dominant |
|
|
|
|
|
|
MIR-196A-2-CC | 70 | 51 | 1 (ref.) | 1 (ref.) |
|
|
miR-196-CT+TT) | 30 | 49 | 2.24 (1.25 to
4.01) | 1.52 (1.11 to
2.09) | 0.0060c |
| Recessive |
|
|
|
|
|
|
MIR-196A-2-(CC+CT) | 95 | 98 | 1 (ref.) | 1 (ref.) |
|
|
MIR-196A-2-TT | 05 | 06 | 1.16 (0.34 to
3.94) | 1.08 (0.56 to
2.10) | 0.8000 |
| Allele |
|
|
|
|
|
|
MIR-196A-2-C
allele | 165 | 149 | 1 (ref.) | 1 (ref.) |
|
|
MIR-196A-2-T
allele | 35 | 55 | 1.74 (1.08 to
2.81) | 1.35 (1.02 to
1.78) | 0.0230 |
| Over-dominant |
|
|
|
|
|
|
MIR-196A-2-CC+TT | 75 | 57 | 1 (ref.) | 1 (ref.) |
|
|
MIR-196A-2-CT | 25 | 43 | 2.26 (1.24 to
4.13) | 1.54 (1.09 to
2.18) | 0.0070c |
Statistical correlation of MIR-196A-2
rs11614913 C>T genotypes with patient characteristics
The results indicated that there were significant
differences in the MIR-196A-2 rs11614913 genotype
distribution between patients with normal and those with elevated
random blood glucose (RBG) and HbA1c (P=0.0050 and =0.0380,
respectively) (Table
VIII).
 | Table VIII.Association of MIR-196A-2
rs11614913 C>T SNP genotypes with the T2DM patient
characteristics. |
Table VIII.
Association of MIR-196A-2
rs11614913 C>T SNP genotypes with the T2DM patient
characteristics.
| Subject
characteristics | N=100 | CC | CT | TT | χ2 | df | P-value |
|---|
| Association with
sex |
|
|
|
|
|
|
|
|
Males | 61 | 30 | 27 | 04 | 0.24 | 2 | 0.8800 |
|
Females | 39 | 21 | 16 | 02 |
|
|
|
| Association with
age (years) |
|
|
|
|
|
|
|
| Age
≤40 | 18 | 8 | 8 | 2 | 2.4 | 2 | 0.3000 |
| Age
>40 | 82 | 40 | 36 | 6 |
|
|
|
| Fasting glucose
(mg/dl) |
|
|
|
|
|
|
|
| Glucose
≤110 | 21 | 13 | 6 | 02 | 2.6 | 2 | 0.2900 |
| Glucose
>110 | 79 | 38 | 37 | 04 |
|
|
|
| Association with
RBG (mg/dl) |
|
|
|
|
|
|
|
| RBG
≤200 | 52 | 22 | 23 | 07 | 10.55 | 2 | 0.0050a |
| RBG
>200 | 48 | 26 | 15 | 07 |
|
|
|
| Association with
total cholesterol (mg/dl) |
|
|
|
|
|
|
|
|
Cholesterol ≤200 | 57 | 19 | 34 | 4 | 19.89 | 02 | 0.0002a |
|
Cholesterol >200 | 43 | 32 | 09 | 02 |
|
|
|
| Association with
HDL-C (mg/dl) |
|
|
|
|
|
|
|
| HDL-C
≤55 | 71 | 16 | 13 | 02 | 03 | 2 | 0.9800 |
| HDL-C
>55 | 29 | 35 | 30 | 04 |
|
|
|
| Association with
LDL-C (mg/dl) |
|
|
|
|
|
|
|
| LDL
≤100 | 28 | 21 | 05 | 02 | 10.11 | 2 | 0.0060a |
| LDL
>100 | 72 | 30 | 38 | 04 |
|
|
|
| Association with TG
(mg/dl) |
|
|
|
|
|
|
|
| TG
≤150 | 37 | 24 | 10 | 03 | 6.13 | 2 | 0.0470a |
| TG
>150 | 63 | 27 | 33 | 03 |
|
|
|
| Association with
HBA1c % |
|
|
|
|
|
|
|
| HBA1c
≤6 | 24 | 10 | 10 | 4 | 6.54 | 2 | 0.0380 |
| HBA1c
>6 | 76 | 41 | 33 | 2 |
|
|
|
| Association with
creatinine (mg/dl) |
|
|
|
|
|
|
|
|
Creatinine ≤1.35 | 76 | 29 | 33 | 14 | 4.11 | 2 | 0.1200 |
|
Creatinine >1.35 | 24 | 22 | 10 | 02 |
|
|
|
Association of MIR-423 rs6505162
C>A gene variation with T2DM
At the time of analysis, out of 110, only 100 T2DM
patient samples gave sharp bands in gel electrophoresis. Similarly,
for the controls only 100 displayed sharp bands. As such only 107
results were included for the analyses. The results indicated that
there was a significant difference in genotype distribution of the
MIR-423 rs6505162 C>A genotypes between T2DM patients and
controls (P<0.0240) (Table
IX). The frequency of CC, CA and AA genotypes was 23, 67 and
10% for patients, and 35, 48 and 17% for the controls,
respectively. A higher frequency of C allele (0.62) was reported in
T2DM patients than among the healthy controls (0.59 (Table IX).
 | Table IX.Association of MIR-423
rs6505162 C>A gene variation in T2DM patients and controls. |
Table IX.
Association of MIR-423
rs6505162 C>A gene variation in T2DM patients and controls.
| Subjects | N | CC (%) | CA (%) | AA (%) | Df | χ2 | C | A | P-value |
|---|
| Patients | 100 | 23 (23) | 67 (67) | 10 (10) | 2 | 7.44 | 0.62 | 0.38 | 0.0240a |
| Controls | 100 | 35 (35) | 48 (48) | 17 (17) |
|
| 0.59 | 0.41 |
|
Multivariate analysis to estimate the
association between MIR-423 rs6505162 C>A gene genotypes and
risk to T2DM
The results showed that the CA genotype was
associated with T2DM with OR=2.12 (1.1160 to 4.0426), RR=1.44
(1.0708 to 1.952), P<0.0210 in the codominant model (Table X). The results also indicated that
there were no significant differences in the MIR-423
rs6505162 C>A genotypes in dominant, recessive and over-dominant
alleles (Table X).
 | Table X.Multivariate analysis to estimate the
association between MIR-423 rs6505162 C>A gene genotypes
and risk to T2DM. |
Table X.
Multivariate analysis to estimate the
association between MIR-423 rs6505162 C>A gene genotypes
and risk to T2DM.
| Genotypes | Healthy controls
(N=100) | T2DM patients
(N=100) | OR (95% CI) | RR | P-value |
|---|
| Codominant |
|
|
|
|
|
|
MIR-423-CC | 35 | 23 | 1 (ref.) | 1 (ref.) |
|
|
MIR-423-CA | 48 | 67 | 2.12 (1.12 to
4.04) | 1.44 (1.07 to
1.95) | 0.0210a |
|
MIR-423-AA | 17 | 10 | 0.89 (0.35 to
2.29) | 0.95 (0.67 to
1.37) | 0.8100 |
| Dominant |
|
|
|
|
|
|
MIR-423-CC | 35 | 23 | 1 (ref.) | 1 (ref.) |
|
|
MIR-423-(CA+AA) | 65 | 77 | 1.80 (0.97 to
3.35) | 1.31 (1.01 to
1.74) | 0.6300 |
| Recessive |
|
|
|
|
|
|
MIR-423-(CC+CA) | 83 | 90 | 1 (ref.) | 1 (ref.) |
|
|
MIR-423-AA | 17 | 10 | 0.54 (0.24 to
1.25) | 0.76 (0.55 to
1.06) | 0.1500 |
| Allele |
|
|
|
|
|
|
MIR-423-C | 118 | 113 | 1 (ref.) | 1 (ref.) |
|
|
MIR-423-A | 82 | 87 | 1.10 (0.75 to
1.65) | 1.05 (0.86 to
1.29) | 0.6100 |
| Over-dominant |
|
|
|
|
|
|
MIR-423-CC+AA | 52 | 33 | 1 (ref.) | 1 (ref.) |
|
|
MIR-423-A | 17 | 10 | 0.92 (0.38 to
2.27) | 0.97 (0.69 to
1.36) | 0.8600 |
Association of MIR-423 rs6505162
C>A with T2DM patient characteristics
The statistical comparisons (P-values) of
MIR-423 rs6505162 C>A genotypes with comorbid conditions
and T2DM severity was conducted by using multivariate analysis
based on logistic regression such as odds ratio (OD) and risk ratio
(RR) with 95% confidence intervals (CI) (Table XI). A significant correlation was
reported between the MIR-423 rs6505162 C>A genotypes and
the age of the subjects (P<0.0001). A significant correlation
was reported between the MIR-423 rs6505162 C>A genotypes
with biochemical parameters such as fasting glucose, RBG, total
serum cholesterol, LDL-C, TG and HbA1c.
 | Table XI.Association of MIR-423
rs6505162 C>A with the T2DM patient characteristics. |
Table XI.
Association of MIR-423
rs6505162 C>A with the T2DM patient characteristics.
| Subject
characteristics | N=100 | CC | CA | AA | χ2 | df | P-value |
|---|
| Association with
sex |
|
|
|
|
|
|
|
|
Males | 61 | 8 | 47 | 06 | 1.6 | 2 | 0.4400 |
|
Females | 39 | 9 | 24 | 06 |
|
|
|
| Association with
age (years) |
|
|
|
|
|
|
|
| Age
≤40 | 18 | 7 | 7 | 4 | 18.66 | 2 | 0.0001a |
| Age
>40 | 82 | 14 | 52 | 14 |
|
|
|
| Fasting glucose
(mg/dl) |
|
|
|
|
|
|
|
| Glucose
≤110 | 21 | 9 | 7 | 5 | 14.12 | 2 | 0.0009a |
| Glucose
>110 | 79 | 14 | 60 | 5 |
|
|
|
| Association with
RBG (mg/dl) |
|
|
|
|
|
|
|
| RBG
≤200 | 52 | 10 | 39 | 3 | 7.23 | 2 | 0.0260a |
| RBG
>200 | 48 | 12 | 30 | 6 |
|
|
|
| Association with
total cholesterol (mg/dl) |
|
|
|
|
|
|
|
|
Cholesterol ≤200 | 57 | 7 | 40 | 5 | 9.9 | 2 | 0.0060a |
|
Cholesterol >200 | 43 | 11 | 27 | 5 |
|
|
|
| Association with
HDL-C (mg/dl) |
|
|
|
|
|
|
|
| HDL-C
≤55 | 71 | 21 | 44 | 6 | 0.66 | 2 | 0.7100 |
| HDL-C
>55 | 29 | 07 | 17 | 5 |
|
|
|
| Association with
LDL-C (mg/dl) |
|
|
|
|
|
|
|
| LDL
≤100 | 28 | 9 | 12 | 7 | 13.5 | 2 | 0.0001a |
| LDL
>100 | 72 | 14 | 55 | 3 |
|
|
|
| Association with TG
(mg/dl) |
|
|
|
|
|
|
|
| TG
≤150 | 37 | 15 | 17 | 5 | 12.47 | 2 | 0.0020a |
| TG
>150 | 63 | 8 | 50 | 5 |
|
|
|
| Association with
HBA1c % |
|
|
|
|
|
|
|
| HBA1c
≤6 | 24 | 8 | 11 | 5 | 7.28 | 2 | 0.0260a |
| HBA1c
>6 | 76 | 15 | 56 | 5 |
|
|
|
| Association with
creatinine (mg/dl) |
|
|
|
|
|
|
|
|
Creatinine ≤1.35 | 76 | 18 | 44 | 14 | 4.56 | 2 | 0.1020 |
|
Creatinine >1.35 | 24 | 5 | 16 | 3 |
|
|
|
Discussion
Type 2 diabetes mellitus (T2DM) is a metabolic
disorder characterized by hyperglycemia resulting from impaired
insulin action caused by insulin resistance in the liver, muscles
and adipose tissues (3,34). Insulin resistance leads to
hyperinsulinemia and pancreatic β cell dysfunction (34). Glucokinase is very important for
glucose homeostasis, since it is essential for insulin secretion,
energy storage as glycogen, and gluconeogenesis (35). The rs1799884 SNP is found in the
specific promoter region of the glucokinase (GCK) gene
(14). The results revealed that
there was a significant difference in rs1799884 G>A genotype
distribution between the T2DM patients and the controls. The A
allele of the rs1799884 G>A was also associated with T2DM
(Table V). This result is
consistent with previous studies that indicated the association of
rs1799884 with an increased fasting blood glucose concentration and
susceptibility to T2DM (13–16). Simultaneously, the result is also
in agreement with previous studies as well which reported that i)
rs1799884 influences glucokinase activity and that the reduced
glucokinase activity is associated with T2DM (14,36–39), and ii) rs1799884 SNP is associated
with dyslipidemia and coronary artery disease (CAD) in Han Chinese
and Austrian populations (40,41).
The present results also revealed that rs1799884 GA
and AA genotypes were associated with hyperlipidemia.
Hyperlipidemia and cardiovascular diseases are among the
traditional complications of diabetes mellitus (38–40). This result is substantiated by a
recent study by Ormazabal et al, who reported that the
systemic metabolism of lipids is altered in the insulin resistance
that leads to the so-called lipid triad; hypertriglyceridemia,
reduced HDL and the development of small dense LDL (41). In the stratified analysis by
ethnicity, significant associations have been found in Caucasians
for the polymorphism in all genetic models; while no associations
were detected among Asians (13,19). There are several possible reasons
for such differences. First, the distribution of the A allele
varies extensively between different races, ethnicities, with a
prevalence of ~23% among Asians and ~17% among Caucasians. The
frequency of three genotypes GG, GA, AA between the T2DM patients
and controls was found to be 23, 39 and 37.3% and 28, 52.3 and
19.7% respectively. A higher frequency of A allele (0.57) was
reported in our T2DM cases when compared with the healthy controls
(0.46). Therefore, additional studies are warranted to further
validate the ethnic difference in the effect of this polymorphism
on T2DM risk.
The results on MIRNA SNPs showed that the CT
genotype and the T allele of MIR-196A-2 rs11614913 were
associated with T2DM. This result is in agreement with a recent
study that reported MIR-196A-2 rs11614913 to be associated
with T2DM in a Pakistani population (42). The MIR-196A-2 rs11614913
C>T SNP has been reported to influence the expression of mature
mRNAs by binding with target mRNAs (25,43), and the T allele is associated with
reduced mature miR-196a-2 levels (44). It has been reported that
miR-196a-2 directly targets and inhibits the expression of Scm like
with four Mbt domains 1 (SFMBT1) and homeobox C8
(HOXC8) genes (43,44).
The HOXC8 gene was reported to increase white fat cells and
the susceptibility to obesity (44), while SFMBT1 was demonstrated to be
among the adiponectin level-regulating loci (45). The blood levels of adiponectin are
genetically determined and correlate negatively with the
susceptibility to T2DM and cardiovascular diseases (45). It has also been suggested that a
reduction in the mature miR-196a-2 by rs11614913 T allele increases
the expression of HOXC8 and SFMBT1 leading to obesity
probably by the promotion of white fat cells (44). Obesity is a well-established risk
factor for insulin resistance and the development of T2DM (46). The results indicated that there
were significant differences (P<0.05) in MIR-196A-2
rs11614913 C>T SNP genotype distribution between the subjects
with normal and abnormal lipid profiles.
Human miR-196 (miR-196a-1, miR-196a-2, and miR-196b)
is transcribed from three different genes located on chromosomes
17q21, 12q13, and 7p15, respectively. The nucleotide sequences of
miR-196a-1 and miR-196a-2 are identical, while the sequence of
miR-196b differs from that of miR-196a by only one nucleotide in
the non-seed region. Previous research has shown that the
expression level of mature miR-196a-3p is higher in CC carriers
with lung cancer compared to CT and TT individuals (47). Hoffman et al reported
elevated expression of mature miR-196a-2 forms in MCF-7 cells
transfected with a pre-miR-196a-C vector when compared with cells
transfected with a pre-miR-196a-T vector (43). The potential of rs11614913 in
targeting the function of miR-196a-2 has also been documented by
whole-genome expression microarrays which found different numbers
of dysregulated mRNAs after transfecting cells with a
pre-miR-196a-C or pre-miR-196a-T vector (43). It is plausible to believe that
rs11614913 C®T SNP may affect the binding efficiency of
miR-196a-2 to its target mRNA or it might affect the processing of
the pre-miRNA into its mature form, thereby predisposing the
individuals to T2DM (48).
It was observed that the cases with a CT genotype
had low serum cholesterol and TG values. Since this is a cross
sectional study, it is possible that these cases have received
cholesterol-lowering medications and their normal lipid profiles
were already maintained prior to the sample collection.
This result is rather expected as the
MIR-196A-2 rs11614913 C>T SNP has been associated with
cardiovascular disease (CVD) in previous studies in different
populations (49–52). miR-196a-2 is involved in the
regulation of annexin A1 known for reducing the levels of tumor
necrosis factor-α (TNF-α) (53).
TNF-α has an important role in the induction of CVD (53). Moreover, it has been reported that
miR-196a-2 regulates HOXB8-Shh signaling in fetal cardiac tissues
that is required for cardiac septation, morphogenesis and valve
development. Therefore, dysregulation of miR-196a-2 may lead to CVD
(54,55). The results of this study are
substantiated by a similar study that reports that the CC genotype
of MIR-196A-2 rs11614913 affects the maturation of
miR-196a-2 and its interaction with its target mRNAs (55,56).
The rs6505162 C>A is located in the pre-miRNA
sequence of MIR-423 that expresses two microRNAs,
MIR-423-3P and MIR-423-5P (52). The current results showed that
there was a significant difference in MIR-423 rs6505162
C>A genotype distribution between T2DM patients and controls,
and that the CA genotype of the MIR-423 rs6505162 C>A was
associated with T2DM. The A allele of rs6505162 has been reported
to increase the expression of the mature miR-423 (53,54). The result of this study is quite
consistent with the study by Yang et al who reported that in
obese diabetic mice suppression of liver miR-423-5p inhibits
gluconeogenesis and ameliorates insulin resistance, and promotes
blood sugar and fatty liver (30). They further reported that the
overexpression of miR-423-5p enhanced gluconeogenesis, increased
blood glucose levels and obesity in healthy mice through the
suppression of the hepatic FAM3A/ATP/Akt pathway (30). However, the result is in
disagreement with a study that reported no association of rs6505162
with the induction of T2DM in the Pakistani population (42). This dissimilarity of findings is
probably due to different subject ethnicity and sample size and
requires further validation. The present results showed that there
were significant differences in rs6505162 genotype distribution in
cases with normal and abnormal lipid profiles. This result is
rather expected as the rs6505162 SNP has been associated with
cardiovascular disease (57,58). The result of this study is also
consistent with studies that demonstrated that the overexpression
of miR-423-5p enhanced fat deposition and that miR-423-5p is
specifically increased in the blood of heart failure subjects
(30,58).
In the present study Tetra primer-amplification
refractory mutation system-based polymerase chain reaction
(T-ARMS-PCR) was successfully used, although the genotyping methods
including high-resolution melting (HRM), pyrosequencing, TaqMan
assay, Mass ARRAY are highly accurate and acknowledged as gold
standard for detecting SNPs but require expensive equipment and
kits. The other alternative methods that could have been used
include quantitative PCR, PCR-RFLP and direct sequencing but
T-ARMS-PCR has been reported to be cost-effective, reliable and
simple (31). The results of
T-ARMS-PCR have been reported to be consistent with DNA sequencing
results by Jin et al (58)
that reiterates our belief that T-ARMS-PCR can offer a viable,
simple and reliable alternative for the detection of SNPs.
To conclude, the SNPs of GCK rs1799884
G>A, MIR-196A-2 rs11614913 C>T, MIR-423
rs6505162 were examined for their association with T2DM in a
section of the Saudi population by using T-ARMS-PCR. The results
indicated that the AA genotype and the A allele of the GCK
rs1799884 G>A were strongly associated with T2DM susceptibility
in the patient population. The results also indicated that the
MIR-196A-2 rs11614913 CT genotype and T allele and
MIR-423 rs6505162 CA genotype were also associated with
T2DM. Since this is the first study of its kind in Saudi Arabia,
the results will help in uncovering more loci that are associated
with T2DM in different ethnic populations and the stratification of
individual susceptible to T2DM. The limitations of this study
include the small sample size and no strict age matching between
patients and healthy controls. More longitudinal studies with
larger sample sizes and in different ethnic populations are
recommended to further validate these observations.
Acknowledgements
The authors extend their appreciation to Dr. Suhail
Ahmed of the English Department, University of Bisha, for language
review and editing.
Funding
The authors extend their appreciation to the ‘Deputyship for
Research and Innovation, Ministry of Education in Saudi Arabia for
funding this research work through the project number 47 of
1442.
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author upon
reasonable request.
Authors' contributions
All the authors were involved in the conception and
planning of the study. MMM, RM, MAAA, MJ, VM and MHA designed the
study. MAAA, JIW, ZUS, MA and AMA were involved in the recruitment
of patients. MMM, RM, MJ and IE performed the experiments. RM and
MMM confirm the authenticity of all the raw data. MMM, RM and IE
wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the local RELOC
Committee of the College of Medicine, University of Bisha (ref. no.
UBCOM/H-06-BH-087(04/10), in accordance with the local guidelines
which conformed in essence, to the principles of the Helsinki
Declaration. Informed consent was obtained prior to the collection
of samples from all patients and control subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
References
|
1
|
Al Mansour MA: The prevalence and risk
factors of type 2 diabetes mellitus (DMT2) in a semi-urban Saudi
population. Int J Environ Res Public Health. 17:72019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gaál Z and Balogh I: Monogenic forms of
diabetes mellitus. Exp Suppl. 111:385–416. 2019.PubMed/NCBI
|
|
3
|
Moin ASM and Butler AE: Alterations in
beta cell identity in type 1 and type 2 diabetes. Curr Diab Rep.
19:832019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Forbes JM and Cooper ME: Mechanisms of
diabetic complications. Physiol Rev. 93:137–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sacks DB and McDonald JM: The pathogenesis
of type II diabetes mellitus. A polygenic disease. Am J Clin
Pathol. 105:149–156. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
De Backer I, Hussain SS, Bloom SR and
Gardiner JV: Insights into the role of neuronal glucokinase. Am J
Physiol Endocrinol Metab. 311:E42–E55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iynedjian PB: Molecular physiology of
mammalian glucokinase. Cell Mol Life Sci. 66:27–42. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Toulis KA, Nirantharakumar K, Pourzitaki
C, Barnett AH and Tahrani AA: Glucokinase activators for type 2
diabetes: Challenges and future developments. Drugs. 80:467–475.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osbak KK, Colclough K, Saint-Martin C,
Beer NL, Bellanné-Chantelot C, Ellard S and Gloyn AL: Update on
mutations in glucokinase (GCK), which cause maturity-onset diabetes
of the young, permanent neonatal diabetes, and hyperinsulinemic
hypoglycemia. Hum Mutat. 30:1512–1526. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
FendLer W, Rizzo M, Borowiec M,
Malachowska B, Antosik K, Szadkowska A, Banach M, Urbanska-Kosinska
M, Szopa M, Malecki M and Mlynarski W: Less but better:
Cardioprotective lipid profile of patients with GCK-MODY despite
lower HDL cholesterol level. Acta Diabetol. 51:625–632. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spyer G, Macleod KM, Shepherd M, Ellard S
and Hattersley AT: Pregnancy outcome in patients with raised blood
glucose due to a heterozygous glucokinase gene mutation. Diabet
Med. 26:14–18. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Njølstad PR, Søvik O, Cuesta-Muñoz A,
Bjørkhaug L, Massa O, Barbetti F, Undlien DE, Shiota C, Magnuson
MA, Molven A, et al: Neonatal diabetes mellitus due to complete
glucokinase deficiency. N Engl J Med. 344:1588–1592. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fu D, Cong X, Ma Y, Cai H, Cai M, Li D, Lv
M, Yuan X, Huang Y and Lv Z: Genetic polymorphism of glucokinase on
the risk of type 2 diabetes and impaired glucose regulation:
Evidence based on 298,468 subjects. PLoS One. 8:e557272013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murad AS, Smith GD, Lewis SJ, Cox A,
Donovan JL, Neal DE, Hamdy FC and Martin RM: A polymorphism in the
glucokinase gene that raises plasma fasting glucose, rs1799884, is
associated with diabetes mellitus and prostate cancer: Findings
from a population-based, case-control study (the ProtecT study).
Int J Mol Epidemiol Genet. 1:175–183. 2010.PubMed/NCBI
|
|
15
|
Li C, Yang Y, Liu X, Li Z, Liu H and Tan
Q: Glucose metabolism-related gene polymorphisms as the risk
predictors of type 2 diabetes. Diabetol Metab Syndr. 12:972020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muller YL, Piaggi P, Hoffman D, Huang K,
Gene B, Kobes S, Thearle MS, Knowler WC, Hanson RL, Baier LJ and
Bogardus C: Common genetic variation in the glucokinase gene (GCK)
is associated with type 2 diabetes and rates of carbohydrate
oxidation and energy expenditure. Diabetologia. 57:1382–1390. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reiling E, van 't Riet E, Groenewoud MJ,
Welschen LM, van Hove EC, Nijpels G, Maassen JA, Dekker JM and 't
Hart LM: Combined effects of single-nucleotide polymorphisms in
GCK, GCKR, G6PC2 and MTNR1B on fasting plasma glucose and type 2
diabetes risk. Diabetologia. 52:1866–1870. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cauchi S, Nead KT, Choquet H, Horber F,
Potoczna N, Balkau B, Marre M, Charpentier G, Froguel P and Meyre
D: The genetic susceptibility to type 2 diabetes may be modulated
by obesity status: Implications for association studies. BMC Med
Genet. 9:452008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cauchi S, Ezzidi I, El Achhab Y, Mtiraoui
N, Chaieb L, Salah D, Nejjari C, Labrune Y, Yengo L, Beury D, et
al: European genetic variants associated with type 2 diabetes in
North African Arabs. Diabetes Metab. 38:316–323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cirillo F, Catellani C, Lazzeroni P,
Sartori C and Street ME: The role of MicroRNAs in influencing body
growth and development. Horm Res Paediatr. 93:7–15. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tan W, Liu B, Qu S, Liang G, Luo W and
Gong C: MicroRNAs and cancer: Key paradigms in molecular therapy.
Oncol Lett. 15:2735–2742. 2018.PubMed/NCBI
|
|
22
|
Raue R, Frank AC, Syed SN and Brüne B:
Therapeutic targeting of MicroRNAs in the tumor microenvironment.
Int J Mol Sci. 22:22102021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fridrichova I and Zmetakova I: MicroRNAs
contribute to breast cancer invasiveness. Cells. 8:13612019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ashrafizadeh M, Ang HL, Moghadam ER,
Mohammadi S, Zarrin V, Hushmandi K, Samarghandian S, Zarrabi A,
Najafi M, Mohammadinejad R and Kumar AP: MicroRNAs and their
influence on the ZEB family: Mechanistic aspects and therapeutic
applications in cancer therapy. Biomolecules. 10:10402020.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mir R, Elfaki I, Khullar N, Waza AA, Jha
C, Mir MM, Nisa S, Mohammad B, Mir TA, Maqbool M, et al: Role of
selected miRNAs as diagnostic and prognostic biomarkers in
cardiovascular diseases, including coronary artery disease,
myocardial infarction and atherosclerosis. J Cardiovasc Dev Dis.
8:222021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Elfaki I, Mir R, Duhier FMA, Alotaibi MA,
Alalawy AI, Barnawi J, Babakr AT, Mir MM, Altayeb F, Mirghani H and
Frah EAM: Clinical implications of MiR128, angiotensin I converting
enzyme and vascular endothelial growth factor gene abnormalities
and their association with T2D. Curr Issues Mol Biol. 43:1859–1875.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ibrahim AA, Ramadan A, Wahby AA, Hassan M,
Soliman HM and Abdel Hamid TA: Micro-RNA 196a2 expression and
miR-196a2 (rs11614913) polymorphism in T1DM: A pilot study. J
Pediatr Endocrinol Metab. 32:1171–1179. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhuang GQ and Wang YX: A tiny RNA molecule
with a big impact on type 2 diabetes mellitus susceptibility.
Biomed Environ Sci. 30:855–861. 2017.PubMed/NCBI
|
|
29
|
Blum A, Meerson A, Rohana H, Jabaly H,
Nahul N, Celesh D, Romanenko O and Tamir S: MicroRNA-423 may
regulate diabetic vasculopathy. Clin Exp Med. 19:469–477. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang W, Wang J, Chen Z, Chen J, Meng Y,
Chen L, Chang Y, Geng B, Sun L, Dou L, et al: NFE2 induces
miR-423-5p to promote gluconeogenesis and hyperglycemia by
repressing the hepatic FAM3A-ATP-Akt pathway. Diabetes.
66:1819–1832. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Delvaux N, da Costa VD, da Costa MM and
Lampe E: Comparison of four methods of genotyping IL28B
polymorphisms in chronic hepatitis C patients. J Virol Methods.
220:1–4. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jha CK, Mir R, Elfaki I, Khullar N, Rehman
S, Javid J, Banu S and Chahal SMS: Potential impact of MicroRNA-423
gene variability in coronary artery disease. Endocr Metab Immune
Disord Drug Targets. 19:67–74. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rahim A, Afzal M and Naveed AK: Genetic
polymorphism of miRNA-196a and its target gene annexin-A1
expression based on ethnicity in Pakistani female breast cancer
patients. Pak J Med Sci. 35:1598–1604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hudish LI, Reusch JE and Sussel L: β Cell
dysfunction during progression of metabolic syndrome to type 2
diabetes. J Clin Invest. 129:4001–4008. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matschinsky FM and Wilson DF: The central
role of glucokinase in glucose homeostasis: A perspective 50 years
after demonstrating the presence of the enzyme in islets of
langerhans. Front Physiol. 10:1482019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tam CH, Ho JS, Wang Y, Lee HM, Lam VK,
Germer S, Martin M, So WY, Ma RC, Chan JC and Ng MC: Common
polymorphisms in MTNR1B, G6PC2 and GCK are associated with
increased fasting plasma glucose and impaired beta-cell function in
Chinese subjects. PLoS One. 5:e114282010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weedon MN, Clark VJ, Qian Y, Ben-Shlomo Y,
Timpson N, Ebrahim S, Lawlor DA, Pembrey ME, Ring S, Wilkin TJ, et
al: A common haplotype of the glucokinase gene alters fasting
glucose and birth weight: Association in six studies and
population-genetics analyses. Am J Hum Genet. 79:991–1001. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tremblay J and Hamet P: Biomarkers of
vascular complications in type 2 diabetes. Metabolism. 64 (3 Suppl
1):S28–S32. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qi Q, Wu Y, Li H, Loos RJ, Hu FB, Sun L,
Lu L, Pan A, Liu C, Wu H, et al: Association of GCKR rs780094,
alone or in combination with GCK rs1799884, with type 2 diabetes
and related traits in a Han Chinese population. Diabetologia.
52:834–843. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
März W, Nauck M, Hoffmann MM, Nagel D,
Boehm BO, Koenig W, Rothenbacher D and Winkelmann BR: G(−30)A
polymorphism in the pancreatic promoter of the glucokinase gene
associated with angiographic coronary artery disease and type 2
diabetes mellitus. Circulation. 109:2844–2849. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ormazabal V, Nair S, Elfeky O, Aguayo C,
Salomon C and Zuñiga FA: Association between insulin resistance and
the development of cardiovascular disease. Cardiovasc Diabetol.
17:1222018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Khan MS, Rahman B, Haq TU, Jalil F, Khan
BM, Maodaa SN, Al-Farraj SA, El-Serehy HA and Shah AA: Deciphering
the variants located in the MIR196A2, MIR146A, and MIR423 with
type-2 diabetes mellitus in Pakistani population. Genes (Basel).
12:6642021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hoffman AE, Zheng T, Yi C, Leaderer D,
Weidhaas J, Slack F, Zhang Y, Paranjape T and Zhu Y: microRNA
miR-196a-2 and breast cancer: A genetic and epigenetic association
study and functional analysis. Cancer Res. 69:5970–5977. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ghanbari M, Sedaghat S, de Looper HW,
Hofman A, Erkeland SJ, Franco OH and Dehghan A: The association of
common polymorphisms in miR-196a2 with waist to hip ratio and
miR-1908 with serum lipid and glucose. Obesity (Silver Spring).
23:495–503. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dastani Z, Hivert MF, Timpson N, Perry
JRB, Yuan X, Scott RA, Henneman P, Heid IM, Kizer JR, Lyytikäinen
LP, et al: Novel loci for adiponectin levels and their influence on
type 2 diabetes and metabolic traits: A multi-ethnic meta-analysis
of 45,891 individuals. PLoS Genet. 8:e10026072012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wondmkun YT: Obesity, insulin resistance,
and type 2 diabetes: Associations and therapeutic implications.
Diabetes Metab Syndr Obes. 13:3611–3616. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hu Z, Chen J, Tian T, Zhou X, Gu H, Xu L,
Zeng Y, Miao R, Jin G, Ma H, et al: Genetic variants of miRNA
sequences and non-small cell lung cancer survival. J Clin Invest.
118:2600–2608. 2008.PubMed/NCBI
|
|
48
|
Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY,
Wu KJ, Chiou SH, Lin SC and Chang KW: miR-31 ablates expression of
the HIF regulatory factor FIH to activate the HIF pathway in head
and neck carcinoma. Cancer Res. 70:1635–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fragoso JM, Ramírez-Bello J, Martínez-Ríos
MA, Peña-Duque MA, Posadas-Sánchez R, Delgadillo-Rodríguez H,
Jiménez-Morales M, Posadas-Romero C and Vargas-Alarcón G: miR-196a2
(rs11614913) polymorphism is associated with coronary artery
disease, but not with in-stent coronary restenosis. Inflamm Res.
68:215–221. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sung JH, Kim SH, Yang WI, Kim WJ, Moon JY,
Kim IJ, Cha DH, Cho SY, Kim JO, Kim KA, et al: miRNA polymorphisms
(miR-146a, miR-149, miR-196a2 and miR-499) are associated with the
risk of coronary artery disease. Mol Med Rep. 14:2328–2342. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Agiannitopoulos K, Samara P, Papadopoulou
M, Efthymiadou A, Papadopoulou E, Tsaousis GN, Mertzanos G, Babalis
D and Lamnissou K: miRNA polymorphisms and risk of premature
coronary artery disease. Hellenic J Cardiol. 62:278–284. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Buraczynska M, Zukowski P, Wacinski P,
Ksiazek K and Zaluska W: Polymorphism in microRNA-196a2 contributes
to the risk of cardiovascular disease in type 2 diabetes patients.
J Diabetes Complications. 28:617–620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yuan S, Carter P, Bruzelius M, Vithayathil
M, Kar S, Mason AM, Lin A, Burgess S and Larsson SC: Effects of
tumour necrosis factor on cardiovascular disease and cancer: A
two-sample Mendelian randomization study. EBioMedicine.
59:1029562020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Tian J, An X and Niu L: Role of microRNAs
in cardiac development and disease. Exp Ther Med. 13:3–8. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li XY, Chen K and Lv ZT: APRISMA-compliant
systematic review and meta-analysis determining the association of
miRNA polymorphisms and risk of congenital heart disease. Medicine
(Baltimore). 98:e176532019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ding Y, Sun X and Shan PF: MicroRNAs and
cardiovascular disease in diabetes mellitus. Biomed Res Int.
2017:40803642017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Tijsen AJ, Creemers EE, Moerland PD, de
Windt LJ, van der Wal AC, Kok WE and Pinto YM: MiR423-5p as a
circulating biomarker for heart failure. Circ Res. 106:1035–1039.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jin C, Li Z, Zheng X, Shen K, Chao J, Dong
Y, Huang Q, Yin Q, Deng Y and Zhu W: Development and validation of
T-ARMS-PCR to detect CYP2C19*17 allele. J Clin Lab Anal.
34:e230052020. View Article : Google Scholar : PubMed/NCBI
|
















