Introduction
With an increasing global population and the
problems of the aging population, prostate cancer (PCa) has
remained a major public health challenge affecting men worldwide
(1). It is a highly prevalent
malignancy, the second most common cancer, and the leading cause of
cancer-related deaths in men, accounting for an estimated 1.6
million cases and 366,000 deaths annually (2).
The high risk of PCa is mainly due to its aggressive
metastatic nature. Due to the silent nature of this tumor, early
diagnosis and treatment is difficult. In many cases, by the time of
diagnosis, the tumor tissue has already developed extraprostatic or
even bone metastasis (3,4). The global incidence of PCa has
continued to increase in recent years, largely due to increased
diagnosis owing to the widespread use of prostate-specific antigen
testing; which has allowed the detection of more early-stage
cancers. In addition, PCa prevalence increases with age; at
present, more than half of Caucasian and Asian men aged >80
years-old have an indolent PCa (5).
PCa is considered a highly heterogeneous cancer
characterized by multiple genomic alterations. Accordingly, tumors
are graded by clinical hazards ranging from indolent to highly
aggressive. Clinicians dealing with PCa patients need to
distinguish between PCa and benign prostatic hyperplasia and
determine the aggressiveness and metastatic nature of the tumor
(1). Hormone therapy, or more
accurately, androgen-deficiency treatment (or testosterone
therapy), was shown to be effective in the early stages of PCa.
However, advanced PCa usually progresses despite androgen ablation,
develops castration resistance, and progresses to lethal PCa, which
is considered incurable (4,6–8).
Therefore, more effective and lasting treatment for PCa is urgently
needed. Currently, proteomics, gene therapy and exosome research,
among other approaches are the focus of cancer research. With the
recent and growing progress of epigenetics, several researchers
have focused on PCa.
N6-methyladenosine (m6A): New
hope for PCa
With the advancement in technology for detecting
epigenetic modifications, the study of DNA methylation and histone
modifications, which are directly linked to tumors, has progressed
significantly. Meanwhile, non-coding RNAs have also been
increasingly studied (1,9–11).
Consequently, the relationship between RNA modification and PCa was
also revealed recently. In particular, the m6A as
methylation modification garnered much attention.
m6A is a modification at the sixth
position of adenine (A) bases in RNA and occurs in several species
(12–14). Initially reported in 1974, it did
not receive much attention until the detection method was proposed
(3). m6A modifications
are abundant within the long internal exons, 3′ untranslated (UTR)
regions of linear RNAs, and around stop codons. They occur mostly
in the RRACH sequence (R=G or A; H=A, C, or U) (15). Similar to other RNA modifications,
the m6A modification is regulated by three protein
types: methyltransferases, demethylases and binding proteins-more
commonly-writers, erasers and readers (16). m6A is involved in
various aspects of mRNA metabolism, including mRNA structure,
maturation, stabilization, splicing, output, translation and decay.
It also affects the cell cycle and differentiation and influences
the maintenance of circadian rhythms (17). Besides, m6A can
influence tumor occurrence and development via various mechanisms.
Furthermore, m6A regulation can affect the progression
of cancer and other diseases (18–20).
Multiple possibilities: Some mechanisms
currently known in PCa databases
The Cancer Genome Atlas and various genomic
databases are particularly beneficial for researchers to analyze
mRNAs and find targets for characteristic m6A
modifications. In previous studies, it was found that approximately
all the m6A regulatory factors were associated with
androgen receptor (AR), a primary oncogene driver of PCa. Of these
regulatory factors, the expression of methyltransferase-like
(METTL) 14, fat mass and obesity-associated protein (FTO) and human
AlkB homolog H5 (ALKBH5) was reduced, while that of METTL3, YTH
domain-containing protein 2 (YTHDC2), YTHDF1, and YTHDF2 was
elevated in patients with PCa at different Gleason grades. At
advanced pathological stages, the expression levels of Vir-like
m6A methyltransferase-associated (VIRMA) and YTHDF3 mRNA
were significantly increased (3,21).
Recurrence-free survival of PCa was also influenced by IGF2BP3,
hnRNP A2/B1, METTL14 and ALKBH5 (22). In AR-dependent and
castration-resistant target genes, Somasekharan et al
(23) identified that AR mRNA
translation is coordinately regulated by the RNA binding proteins
YTHDF3 and G3BP1. AR-regulated PCa cell lines subjected to AR
pathway inhibition (ARPI) stress showed the recruitment of
m6A-modified AR mRNA from actively translating polysomes
to RNA-protein stress granules, leading to reduced AR mRNA
translation. YTHDF3 or G3BP1 silencing could block ARPI-induced
stress granule formation and decrease PCa cell death resulting from
ARPI stress (23). However,
further research is required to validate these results. A precise
understanding of these mechanisms may provide insights into the
prevention and treatment of recurrent tumors (24–26). In addition, drug development
targeting corresponding targets may improve the clinical outcomes
of CRPC. Studies focusing on both m6A modification and
the tumor immune microenvironment in PCa can lead to more effective
immunotherapy approaches (27).
Although database mining resolves several problems,
it cannot explain the specific mechanism of m6A
methylation in PCa development and progression, especially related
molecular mechanisms. Therefore, further experimental exploration
is required to determine therapeutic targets for PCa.
Three parts of m6A: Functional
proteins and cancer
Collectively, three distinct proteins-readers,
writers and erasers-affect cancer development and tumor cell
growth. During m6A methylation modification, they
cooperate to regulate the position of m6A. They are
important targets or components of important pathways in the
development of cancer, and should be carefully considered in the
field of tumor therapy.
Writers
First, we researched the term ‘writers,’ among
which, METTL3 is actively being investigated. METTL3 was the first
m6A writer identified, followed by other components of
the methylation complex, namely, METTL14, METTL4, METTL16, Wilms
tumor 1-associating protein (WTAP), KIAA1429/VIRMA, and RNA binding
motif protein 15 (RBM15, RBM15B) (3). METTL3/14 is found in the nucleus
localized to nuclear speckles. METTL3 and METTL14 are hypothesized
to form an m6A-generating heterodimeric enzyme complex
on mRNAs, while WTAP functions as the splicing regulator (Fig. 1A) (28). The other writers similarly
influence the regulation of m6A modification.
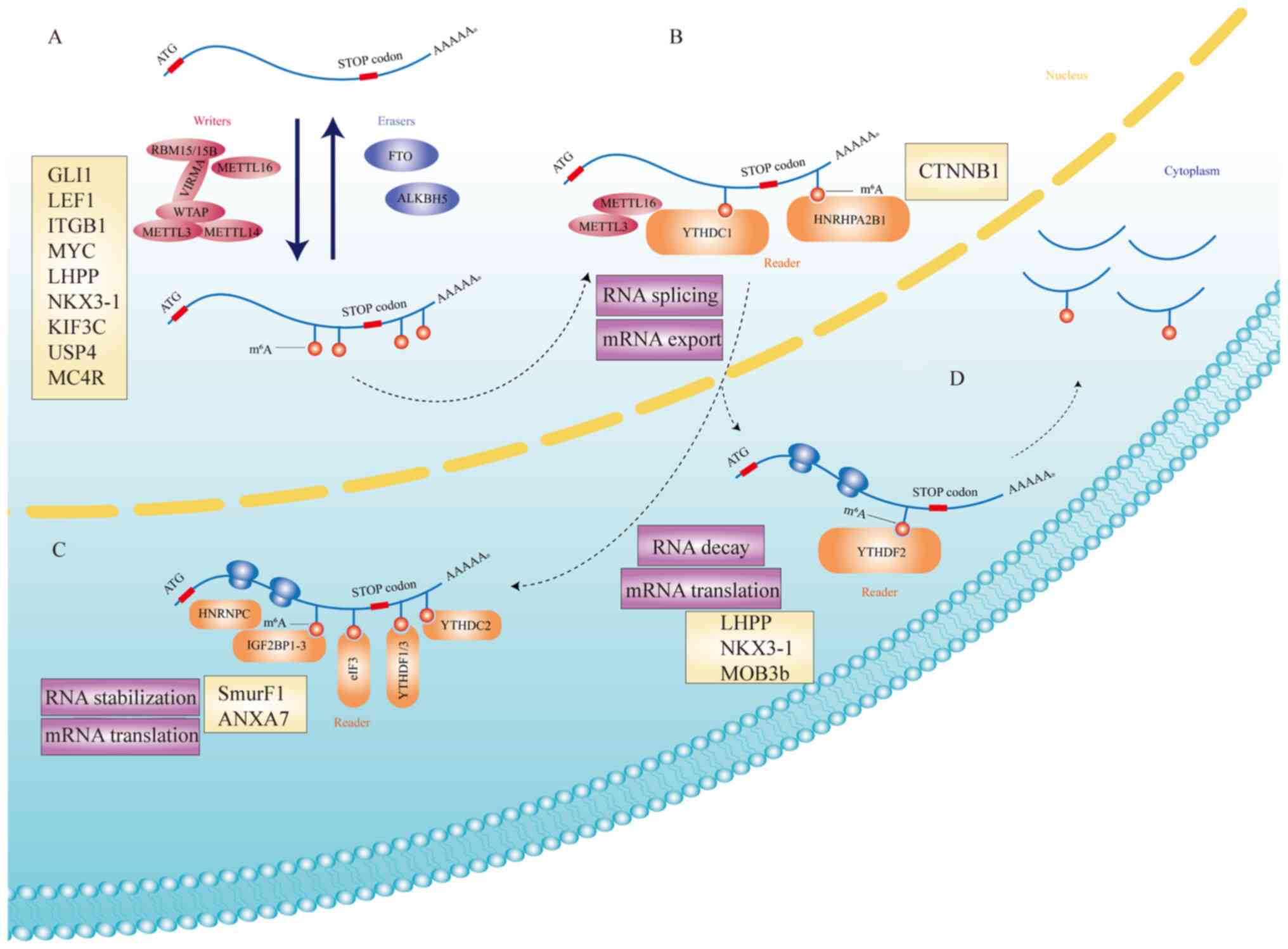 | Figure 1.The mechanism of m6A and
its roles in cells. (A) m6A is deposited by an
m6A multiprotein ‘writer’ complex (METTL3, METTL14,
METTL16, WTAP, VIRMA and RBM 15/15B) and removed by ‘eraser’
demethylases (FTO and ALKBH5). Targets of m6A
multiprotein ‘writer’ complex and ‘eraser’ demethylases (GLI1,
LEF1, ITGB1, MYC, LHPP, NKX3-1, KIF3C, USP4, and MC4R)
can affect the progress of PCa. (B) METTL3, METTL16 and part of
‘reader’ protein (YTHDC1 and hnRHP A2/B1) affect RNA splicing and
mRNA export. CTNNB1 is the target of hnRHP A2/B1 in PCa. (C)
In the cytoplasm, m6A modifications are recognized by
‘reader’ proteins (hnRNPC, IGF2BP1-3, eIF3, YTHDF1/3, and YTHDC2),
resulting in mRNA stabilization and enhanced translation.
SmurF1 and ANXA7 are targets of some of ‘reader’
proteins in PCa. (D) YTHDF2 can regulate mRNA translation and
mediate RNA decay. LHPP, NKX3-1, and MOB3b are main
targets of YTHDF2. m6A, N6-methyladenosine; METTL,
methyltransferase-like; WTAP, Wilms tumor 1-associating protein;
VIRMA, Vir-like m6A methyltransferase-associated; RBM,
RNA binding motif protein; FTO, fat mass and obesity-associated
protein; ALKBH5, human AlkB homolog H5; PCa, prostate cancer;
YTHDC1, YTH domain-containing protein 1; CTNNB1, catenin β1. |
Writers are associated with some altered pathways.
In urologic malignancies, the low expression of METTL3 and METTL14
can negatively regulate cell growth-related pathways (mTOR, EMT,
and P2XR6) and positively regulate cell death-related pathways or
tumor suppressors such as P53, PTEN, and Notch1 (Fig. 1A). Furthermore, METTL3 positively
regulated proliferation-related pathways (NK-kB and SHH-GL1) and
negatively regulated PTEN (29).
The elevated expression of METTL3 in PCa tumor cells promoted the
expression of GLI1 in the hedgehog pathway, the growth of PCa, and
the motility of cancer cells (30). Similarly, decreased METTL3
expression inhibited LEF1 in the Wnt pathway, thereby preventing
tumor cell migration (31). In
tumorigenesis, METTL3 was shown to enhance MYC (c-myc) expression
by increasing m6A levels of MYC mRNA transcript,
triggering PCa (32). Another
study revealed that in promoting the proliferation of PCa cells,
like YTHDF3, METTL3 could inhibit corresponding mRNA degradation by
targeting LHPP and NKX3-1, regulating AKT phosphorylation to induce
cancer progression (33).
METTL3 induces m6A modification on
Kinesin Family Member 3C (KIF3C) mRNA, promoting the stabilization
of KIF3C mRNA by IGF2BP1. Tumor-suppressor factor miR-320d inhibits
KIF3C expression by targeting METTL3 and restrains PCa growth,
migration and invasion (34).
METTL3 mediates m6A modification of ubiquitin-specific
peptidase 4 (USP4) mRNA at A2696, and m6A reader protein
YTHDF2 binds to and induces the degradation of USP4 mRNA by
recruiting RNA-binding protein heterogeneous nuclear
ribonucleoprotein D (HNRNPD) to the mRNA. Decreased USP4 levels do
not remove the ubiquitin group from ELAV like RNA binding protein 1
(ELAVL1), resulting in a reduction in ELAVL1 protein, which
increases Rho GDP dissociation inhibitor alpha (ARHGDIA)
expression, promoting the migration and invasion of PCa cells
(35). Furthermore, reader
proteins and methyltransferase complexes, METTL14 inclusive, can
cause poor prognosis by affecting subcellular protein localization
(22). Li et al (36) found that METTL3 could enhance the
expression of ITGB1 and the adhesion of cancer cells and type I
collagen bone matrix, promoting bone metastasis in PCa.
WTAP was shown to affect the development of urinary
tumors heterogeneously. It interacted with the Wilms tumor
suppressor (WT1) and was also a regulator of the m6A
methylation complex, which was responsible for regulating mRNA
stability. In addition, binding sites for signal transducer and
activator of transcription 1, forkhead box protein O1, interferon
regulatory factor 1, glucocorticoid receptor, and peroxisome
proliferator-activated receptor γ transcription factor exist in the
upstream region of WTAP, which may affect the function of WTAP in
tumor formation (37). However,
to the best of our knowledge, no detailed reports exist on the
mechanism of WTAP action in PCa.
Erasers
The term ‘erasers’ refers to demethylases and mainly
comprises two kinds of proteins-FTO and human ALKBH5. These two
proteins regulate m6A modification and render the RNA
modification dynamic and reversible (Fig. 1A) (3,16).
Increasing evidence suggested that FTO is highly expressed in some
types of cancer and is associated with a poor prognosis. However,
FTO also acts as a tumor suppressor in thyroid cancer. Low protein
expression of FTO was consistent with high tumor grade and
increased lymph node metastasis (20). ALKBH5, another m6A
demethylase, was shown to either inhibit or promote tumorigenesis.
Both FTO and ALKBH5 belong to the AlkB family; the differential
recognition and interactions between them and RNA largely result
from different conformational outcomes in RNAs, which are induced
by m6A. In conclusion, m6A may serve as a
conformational marker in regulating the changes in FTO and ALKBH5
expression (38,39).
FTOs are a class of eraser proteins that are
downregulated in PCa tissues and cell lines (40). They can downregulate the
m6A level and thus inhibit tumor invasion and migration
in PCa by regulating total m6A levels (41). For years, FTO mutations rs9939609
and rs9930506 have been reported in the tumor tissues of patients
with PCa, and rs9939609 has been negatively associated with overall
PCa cases (42–44). Li and Cao revealed that FTO could
restrain the proliferation, migration and invasion of PCa by
downregulating the expression of melanocortin 4 receptor (MC4R)
(45). ALKBH5 is also an
important reader in cancers, but it remains unexplored in PCa.
Studies on m6A erasers are undoubtedly limited to
meta-studies, and research on the specific mechanism of FTO and
ALKBH5 remains a hot topic.
Readers
‘Readers’ include YTH domain-containing protein 1
(YTHDC1), YTHDC2, YTHDF family proteins (YTHDF1, YTHDF2, and
YTHDF3), eukaryotic initiation factor 3 (eIF3), heterogeneous
nuclear ribonucleoprotein C (hnRNP C), hnRNP A2/B1 and IGF2BP
family proteins (IGF2BP1, IGF2BP2 and IGF2BP3) that bind
m6A in RNA to regulate the fate of the corresponding RNA
and adjust downstream functions (Fig.
1B-D).
The YTH family is divided into the following three
major classes: DC1, DC2 and DF (16). They contain an RNA-binding domain,
which is a conserved aromatic ring that can recognize the
m6A modification (46). YTHDF1 and YTHDF3 can both promote
the translation of m6A RNA, while YTHDF2 interferes with
the stability of m6A RNA and causes RNA decay.
Additionally, YTHDF3 can contribute to mRNA degradation (Fig. 1C and D). By contrast, YTHDC1 is
enriched in the nucleus and functions in regulating RNA splicing.
Along with the complex functions of YTHDC2, it regulates RNA
stability and promotes RNA degradation and translation (47). Furthermore, YTHDC1 modulates mRNA
splice site selection in a concentration-dependent manner (Fig. 1B). Another reader, the subunit of
eukaryotic initiation factor 3 (eIF3), is closely related to cancer
occurrence and development. IGF2BP family proteins recognize and
bind the GG(m6A)C sequence via their K homology domains
(16). IGF2BP1 (IMP-1)-a
non-catalytic post-transcriptional enhancer of tumor growth-is
upregulated and associated with adverse prognosis in solid cancers.
It shortened the G1 phase of the tumor cells by relying on 3′UTR-,
miRNA- and m6A-dependent regulations (48). Like the rest of the family, the
overexpression of IGF2BP3 (IMP3) was associated with cancer
progression and survival. Using a new RNA sequencing technique, it
was found that hnRNPC functioned as an RNA nucleosome in RNA
packaging and masking decoy splice (49). While hnRNP A2/B1 was an important
cleavage factor, it was an independent prognostic factor, mainly
affecting the cell cycle by delaying or promoting cancer
progression (50,51). Besides, multiple hnRNP complexes
triggered abnormal transcription and splicing of annexin-A7
(ANXA7), a tumor suppressor, thus affecting hnRNP A2/B1 function
(52).
Many studies revealed that some readers, including
YTHDC1, eIF3f, eIF3S3, and IGF2BP3 (IMP-3), play a corresponding
role in influencing PCa progression. Luxton et al found that
the oncogene metadherin collocated with YTHDC1 subnuclear spots and
regulated the ability of YTHDC1 to affect PCa progression (53). Similarly, the downregulation of
eIF3f expression reduces Akt levels, inhibiting PCa growth and
progression (54). In some of the
earliest studies of advanced PCa, researchers found that
upregulated eIF3S3 gene expression was a common phenomenon,
suggesting that eIF3S3 overexpression promoted tumor growth
(55–57). Furthermore, IMP3 was overly
altered in tumor tissue, which increased the ubiquitination of PTEN
mediated by SMAD-specific E3 ubiquitin-protein ligase 1 (SmurF1)
and ultimately activated the PI3K/Akt/mTOR pathway and promoted PCa
progression (58).
YTHDF2, eIF3d, EIF3h and IGF2BP1(IMP-1) are all
associated with the invasion and proliferation of PCa. YTHDF2 was a
direct target of miR-495 and miR-493-3p. In the lysine demethylase
5a (KDM5a)/miRNA-495/YTHDF2/ m6A-MOB3b axis, YTHDF2
recognized the m6A of MOB3b mRNA and induced the
degradation of MOB3b mRNA to inhibit its expression. miR-493-3p
inhibited the expression of YTHDF2 and thus, increased
m6A levels. Thus, high levels of YTHDF2 promoted the
proliferation, migration and invasion of PCa cells (59,60). Moreover, eIF3d knockout inhibited
the proliferation, invasion and colony formation of tumor cells and
arrested the cell cycle in the G2/M phase (61), while EIF3h functions by affecting
mRNA translation. High levels of eIF3h directly stimulated protein
synthesis and played a key role in establishing and maintaining a
malignant state in cells (62).
In PCa, 8S-lipoxygenase (8S-LOX) and 15S-LOX-2 inhibited the c-myc
mRNA coding region on the determinant-binding protein/insulin-like
growth factor-2 mRNA-binding protein 1 (CRD-BP/IMP-1), thereby
inhibiting the proliferation of the PCa cell line PC-3 (63).
PCa prognosis was also associated with readers,
namely, eIF3b, eIF3c, eIF3L, IGF2BP3 and hnRNP A2/B1. Among them,
eIF3b is a strong oncogenic factor and can affect PCa prognosis
(64,65), while eIF3c regulates the
PI3K/Akt/NF-кB signaling pathway (66). eIF3b silencing leads to a
significant increase in tumor suppressor genes PTEN, DIT3
and CDKN1B and a significant decrease in oncogenic genes
IRS1 and CDH1 (65). In cancer cells, eIF3b depletion
inhibits G1-S cell cycle transformation by altering the expression
of cyclin A, cyclin E, retinoblastoma and p27Kip1 proteins, but not
RNA. eIF3b depletion also inhibits the migration of cancer cells
and destroys their actin cytoskeleton and local adhesions (64). Furthermore, studies showed that
androgen-induced eIF3L could facilitate the early diagnosis of PCa
disease. A high level of androgen-induced palmitoylation of eIF3L
is an obvious marker of PCa. Moreover, as eIF3L acts as an
initiation factor, palmitoylated eIF3L may cooperate with the
initiation complex and enhance mRNA translation (67), palmitoylated eIF3L can be used to
treat castration-resistant PCa (CRPC) (68). Case studies showed that IGF2BP3
was associated with invasive recurrence of tumors, which mainly
included extracapsular extension, seminal vesicle invasion,
lymphovascular invasion, and a high pathological Gleason score
(69). Cheng et al
(70) reasoned that hnRNP A2/B1
mainly promoted proliferation, and its high expression in CRPC
cells worsens PCa prognosis. Moreover, hnRNP A2/B1 enables
CTNNB1 3′-UTR mRNA regulation to alter the expression of
β-catenin and other cancer-relevant genes to influence cancer cell
phenotypes (71).
Readers can also affect bone metastasis in PCa. Lin
et al (72) found that
penta-o-galloyl-β-D-glucose, could inhibit the PI3K/Akt/mTOR
pathway and reduce epidermal growth factor (EGF) levels to induce
the expression of eIF3i and reduce the rate of bone metastasis
(72). Moreover, IGF2BP3 was
hypothesized to be associated with recurrence and bone metastasis
in PCa (73). During PCa
metastasis, IGF2BP3 physically binds to circular
RNAhsa_circ_0003258 in the cytoplasm to enhance HDAC4 mRNA
stability, activate ERK signaling pathway, and trigger EMT
programming, ultimately accelerating metastasis (74).
Although the mechanism of some reader proteins
remains unexplored, some evidence suggests that reader proteins and
their subunits could regulate tumor cell proliferation and
development in an m6A-dependent manner, and may be
targeted for tumor diagnosis and treatment. For instance, in the
IgG reactivity screening of two independent patient cohorts, the
response to antigen IgF2BP2 in patients with advanced PCa was
higher than that in patients with early PCa, which suggested the
possibility of new drug development (75). All of the aforementioned molecular
relationships are presented in Table
I.
 | Table I.Review of the literature regarding
m6A modification related proteins, main target and
pathway in PCa. |
Table I.
Review of the literature regarding
m6A modification related proteins, main target and
pathway in PCa.
| Gene symbol | Type of enzyme | Role | Regulatory
factors | Main target | Pathway | Expression in
cancer | Impact in PCa | (Refs.) |
|---|
| METTL3 | Writer | Oncogene | - | GLI1 | Hedgehog | Upregulated | Growth and
movement | (23) |
|
|
| Oncogene | - | LEF1 | Wnt | Upregulated | Migration | (24) |
|
|
| Oncogene | - | ITGB1 | - | Upregulated | Bone
metastasis | (29) |
|
|
| Oncogene | - | MYC | - | Upregulated | Formation | (25) |
|
|
| Oncogene | - | LHPP,
NKX3-1 | - | Upregulated | Promote AKT
phosphorylation and progression of cancer | (26) |
|
|
| Oncogene | miR-320d | KIF3C | - | Upregulated | Growth, migration
and invasion | (27) |
|
|
| Oncogene | - | USP4 | - | Upregulated | Migration and
invasion | (28) |
| FTO | Erasers | Anti-oncogene | - | MC4R | - | Downregulated | Proliferation,
migration and invasion | (38) |
| YTHDC1 | Reader | Oncogene | Metadherin | - | - | Upregulated | Progression of
PCa | (46) |
| YTHDF2 | Reader | Oncogene | - | LHPP,
NKX3-1 | - | Upregulated | Promote AKT
phosphorylation and progression of cancer | (26) |
|
|
| Oncogene | miR-495 | MOB3b | - | Upregulated | Proliferation,
migration and invasion | (53) |
|
|
| Oncogene |
miR-493-3p | - | - | Upregulated | Proliferation,
migration and invasion | (52) |
| eIF3b | Reader | Oncogene | - | - | - | Upregulated | Poor prognosis | (57, 58) |
| eIF3c | Reader | Oncogene | - | - | PI3K/Akt/
NF-κb | Upregulated | Poor prognosis | (59) |
| eIF3d | Reader | Oncogene | - | - | - | Upregulated | Proliferation,
invasion, colony formation and down-cell cycle in the G2/M
phase | (54) |
| eIF3f | Reader | Oncogene | - | - | - | Upregulated | High Akt level and
progression of PCa | (47) |
| eIF3h | Reader | Oncogene | - | - | - | Upregulated | Malignant state in
cells | (55) |
| eIF3i | Reader | Oncogene | PGG | - | PI3K/Akt/mTOR | Upregulated | Bone
metastasis | (64) |
| eIF3L | Reader | Oncogene | - | - | - | Upregulated | Palmitylation to
treat CRPC | (60) |
| eIF3S3 | Reader | Oncogene | - | - | - | Upregulated | Growth | (48–50) |
|
CRD-BP/IMP-1 | Reader | Oncogene | 8S-LOX,
15S-LOX-2 | - | - | Upregulated | Proliferation | (56) |
| IGF2BP2 | Reader | Oncogene | - | - | - | Upregulated | Advance PCa | (66) |
| IGF2BP3 | Reader | Oncogene | - | - | - | Upregulated | Progression,
recurrence, metastasis and PCa-specific survival | (61,65,73) |
|
|
| Oncogene | - | SmurF1 | PI3K/Akt/mTOR | Upregulated | PTEN
ubiquitination, apoptosis inhibition and proliferation | (51) |
|
HNRNPA2B1 | Reader | Oncogene | - | - | - | Upregulated | Poor prognosis in
CRPC | (62) |
|
|
| Oncogene | - | CTNNB1 | - | Upregulated | High stage of
tumor | (63) |
| HNRNP
complexes | Reader | Oncogene | - | ANXA7 | - | Upregulated | Affects the
function of tumor suppressor factors | (45) |
Discussion
The risk of PCa-a major long-standing public health
problem that affects men worldwide-is mainly due to its aggressive
metastatic nature. Castration resistance PCa and advancement to
lethal PCa are considered incurable. Understanding the relationship
between RNA modification and PCa may lead to the development of new
strategies for PCa treatment and thus, m6A modification
in the light of PCa is increasingly being studied.
m6A modifications occur in every step of
mRNA transcription, splicing, translation and expression; they can
systematically change the expression of specific genes and the
formation of related proteins. In PCa, many functional groups and
regulatory targets show potential as effective treatment. Three
types of proteins associated with m6A can equivalently
affect the occurrence, development and invasion of cancer.
m6A-related mRNAs can be affected by these proteins to
modify their expression, which is necessary for the transformation
into corresponding oncogenic or tumor-suppressor factors. For PCa
treatment, the association between genes and their expressed
proteins and corresponding oncogenic or tumor-suppressor factors,
including multiple protein pathways and their corresponding
targets, have been suggested. Several studies have shown that
various m6A-related gene expression changes (whether up-
or downregulated) affect PCa prognosis and progression. Therefore,
targeted therapies offer great promise (22,76).
However, compared with that of other tumors, the
study of m6A in PCa is not comprehensive, and many
related protein mechanisms are yet to be explored. Therefore,
understanding the precise mechanisms of m6A in PCa,
especially PCa-related proteins and genes, may promote the
development of more effective cancer treatment. For instance,
lysine-specific demethylase 5 (KDM5) family members act as
oncogenic drivers in PCa via activation of the
KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis (59).
The m6A signatures may also serve as an
early diagnostic marker to supplement prostate-specific antigen
diagnosis, which would improve PCa diagnosis. m6A may
also be used as an indicator to evaluate treatment outcome and
prognosis follow-up. Although the mechanisms of some gene targets
remain unexplored, some writers and readers in m6A have
been revealed to promote or inhibit cancer. The development of
drugs targeting these targets has great potential for improving PCa
treatment.
In summary, the literature on m6A and its
mechanism of action in tumors, especially PCa, suggests the rapidly
advancing epigenetics approach for cancer treatment, which will
benefit patients with PCa.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation (grant nos. 81802576, 81902565 and 81372316), the
Jiangsu Provincial Central Administration Bureau (grant no.
YB201827), the Wuxi Commission of Health and Family Planning (grant
nos. T202024, J202012, Z202011, ZM001, J201802 and J201810), the
Science and Technology Development Fund of Wuxi (grant no.
WX18IIAN024 and N20202021), the Jiangnan University Wuxi School of
Medicine (grant no. 1286010242190070), the Wuxi Taihu Lake Talent
Plan, the Supports for Leading Talents in Medical and Health
Profession and the Top Talent Support Program for Young and
Middle-aged People of Wuxi Health Committee (grant no.
BJ2020061).
Availability of data and materials
The datasets used and/or analyzed during this study
are available from the corresponding author on reasonable
request.
Authors' contributions
HYW and YYF were major contributors in writing the
manuscript. JJW determined the specific research direction of the
manuscript and sorted out the data collected. HYW and JJW created
the figure. YYF and JJW performed the literature search. LJZ and
YYM made substantial contributions to the design of the manuscript
and revised it critically for important intellectual content. All
authors have read and approved the final version of the manuscript.
Data authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sugiura M, Sato H, Kanesaka M, Imamura Y,
Sakamoto S, Ichikawa T and Kaneda A: Epigenetic modifications in
prostate cancer. Int J Urol. 28:140–149. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang G, Zhao D, Spring DJ and DePinho RA:
Genetics and biology of prostate cancer. Genes Dev. 32:1105–1140.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lobo J, Barros-Silva D, Henrique R and
Jerónimo C: The emerging role of epitranscriptomics in cancer:
Focus on urological tumors. Genes (Basel). 9:5522018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rebello RJ, Oing C, Knudsen KE, Loeb S,
Johnson DC, Reiter RE, Gillessen S, Van der Kwast T and Bristow RG:
Prostate cancer. Nat Rev Dis Primers. 7:92021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kimura T, Sato S, Takahashi H and Egawa S:
Global trends of latent prostate cancer in autopsy studies. Cancers
(Basel). 13:3592021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maitland NJ: Resistance to antiandrogens
in prostate cancer: Is it inevitable, intrinsic or induced? Cancers
(Basel). 13:3272021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Chen J, Wu Z, Ding W, Gao S, Gao Y
and Xu C: Mechanisms of enzalutamide resistance in
castration-resistant prostate cancer and therapeutic strategies to
overcome it. Br J Pharmacol. 178:239–261. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lowrance WT, Breau RH, Chou R, Chapin BF,
Crispino T, Dreicer R, Jarrard DF, Kibel AS, Morgan TM, Morgans AK,
et al: Advanced prostate cancer: AUA/ASTRO/SUO guideline PART I. J
Urol. 205:14–21. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Borque-Fernando A, Espilez R, Miramar D,
Corbatón D, Rodríguez A, Castro E, Mateo J, Rello L, Méndez A and
Gil Sanz MJ: Genetic counseling in prostate cancer: How to
implement it in daily clinical practice? Actas Urol Esp (Engl Ed).
45:8–20. 2021.(In English, Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nowacka-Zawisza M and Wiśnik E: DNA
methylation and histone modifications as epigenetic regulation in
prostate cancer (review). Oncol Rep. 38:2587–2596. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cimadamore A, Gasparrini S, Scarpelli M,
Doria A, Mazzucchelli R, Massari F, Cheng L, Lopez-Beltran A and
Montironi R: Epigenetic Modifications and modulators in prostate
cancer. Crit Rev Oncog. 22:439–450. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang YN, Yu CY and Jin HZ: RNA
N(6)-methyladenosine modifications and the immune response. J
Immunol Res. 2020:63276142020.PubMed/NCBI
|
|
13
|
Desrosiers R, Friderici K and Rottman F:
Identification of methylated nucleosides in messenger RNA from
Novikoff hepatoma cells. Proc Natl Acad Sci USA. 71:3971–3975.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Perry RP, Kelley DE, Friderici K and
Rottman F: The methylated constituents of L cell messenger RNA:
Evidence for an unusual cluster at the 5′ terminus. Cell.
4:387–394. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Meyer KD, Saletore Y, Zumbo P, Elemento O,
Mason CE and Jaffrey SR: Comprehensive analysis of mRNA methylation
reveals enrichment in 3′ UTRs and near stop codons. Cell.
149:1635–1646. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liang Z, Kidwell RL, Deng H and Xie Q:
Epigenetic N6-methyladenosine modification of RNA and DNA regulates
cancer. Cancer Biol Med. 17:9–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Z, Wang T, Wu D, Min Z, Tan J and Yu
B: RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and
angiogenesis in colon cancer. J Exp Clin Cancer Res. 39:2032020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui H, Wang Y, Li F, He G, Jiang Z, Gang X
and Wang G: Quantifying observational evidence for risk of dementia
following androgen deprivation therapy for prostate cancer: An
updated systematic review and meta-analysis. Prostate Cancer
Prostatic Dis. 24:15–23. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B,
Li C, Sun L, Qin J, Xu T, et al: METTL14-mediated
N6-methyladenosine modification of SOX4 mRNA inhibits tumor
metastasis in colorectal cancer. Mol Cancer. 19:1062020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Niu Y, Lin Z, Wan A, Chen H, Liang H, Sun
L, Wang Y, Li X, Xiong XF, Wei B, et al: RNA N6-methyladenosine
demethylase FTO promotes breast tumor progression through
inhibiting BNIP3. Mol Cancer. 18:462019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Q, Xie X, Huang Y, Meng S, Li Y, Wang H
and Hu Y: N6-methyladenosine RNA methylation regulators contribute
to the progression of prostate cancer. J Cancer. 12:682–692. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ji G, Huang C, He S, Gong Y, Song G, Li X
and Zhou L: Comprehensive analysis of m6A regulators prognostic
value in prostate cancer. Aging (Albany NY). 12:14863–14884. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Somasekharan SP, Saxena N, Zhang F,
Beraldi E, Huang JN, Gentle C, Fazli L, Thi M, Sorensen PH and
Gleave M: Regulation of AR mRNA translation in response to acute AR
pathway inhibition. Nucleic Acids Res. 50:1069–1091. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wood S, Willbanks A and Cheng JX: The role
of RNA modifications and RNA-modifying proteins in cancer therapy
and drug resistance. Curr Cancer Drug Targets. 21:326–352. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nombela P, Miguel-López B and Blanco S:
The role of m6A, m5C and Ψ RNA modifications
in cancer: Novel therapeutic opportunities. Mol Cancer. 20:182021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Barbieri I and Kouzarides T: Role of RNA
modifications in cancer. Nat Rev Cancer. 20:303–322. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Z, Zhong J, Zeng J, Duan X, Lu J, Sun
X, Liu Q, Liang Y, Lin Z, Zhong W, et al: Characterization of the
m6A-Associated tumor immune microenvironment in prostate cancer to
aid immunotherapy. Front Immunol. 12:7351702021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schöller E, Weichmann F, Treiber T, Ringle
S, Treiber N, Flatley A, Feederle R, Bruckmann A and Meister G:
Interactions, localization, and phosphorylation of the
m6A generating METTL3-METTL14-WTAP complex. RNA.
24:499–512. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tao Z, Zhao Y and Chen X: Role of
methyltransferase-like enzyme 3 and methyltransferase-like enzyme
14 in urological cancers. PeerJ. 8:e95892020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai J, Yang F, Zhan H, Situ J, Li W, Mao Y
and Luo Y: RNA m6A methyltransferase METTL3 promotes the
growth of prostate cancer by regulating hedgehog pathway. Onco
Targets Ther. 12:9143–9152. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma XX, Cao ZG and Zhao SL: m6A
methyltransferase METTL3 promotes the progression of prostate
cancer via m6A-modified LEF1. Eur Rev Med Pharmacol Sci.
24:3565–3571. 2020.PubMed/NCBI
|
|
32
|
Yuan Y, Du Y, Wang L and Liu X: The M6A
methyltransferase METTL3 promotes the development and progression
of prostate carcinoma via mediating MYC methylation. J Cancer.
11:3588–3595. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Xie H, Ying Y, Chen H, Yan H, He L,
Xu M, Xu X, Liang Z, Liu B, et al: YTHDF2 mediates the mRNA
degradation of the tumor suppressors to induce AKT phosphorylation
in N6-methyladenosine-dependent way in prostate cancer. Mol Cancer.
19:1522020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma H, Zhang F, Zhong Q and Hou J:
METTL3-mediated m6A modification of KIF3C-mRNA promotes prostate
cancer progression and is negatively regulated by miR-320d. Aging
(Albany NY). 13:22332–22344. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Y, Pan C, Wang X, Xu D, Ma Y, Hu J,
Chen P, Xiang Z, Rao Q and Han X: Silencing of METTL3 effectively
hinders invasion and metastasis of prostate cancer cells.
Theranostics. 11:7640–7657. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li E, Wei B, Wang X and Kang R: METTL3
enhances cell adhesion through stabilizing integrin β1 mRNA via an
m6A-HuR-dependent mechanism in prostatic carcinoma. Am J Cancer
Res. 10:1012–1025. 2020.PubMed/NCBI
|
|
37
|
Wu LS, Qian JY, Wang M and Yang H:
Identifying the role of Wilms tumor 1 associated protein in cancer
prediction using integrative genomic analyses. Mol Med Rep.
14:2823–2831. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Piette ER and Moore JH: Identification of
epistatic interactions between the human RNA demethylases FTO and
ALKBH5 with gene set enrichment analysis informed by differential
methylation. BMC Proc. 12 (Suppl 9):S592018. View Article : Google Scholar
|
|
39
|
Zou S, Toh JD, Wong KH, Gao YG, Hong W and
Woon EC: N(6)-Methyladenosine: a conformational marker that
regulates the substrate specificity of human demethylases FTO and
ALKBH5. Sci Rep. 6:256772016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu A, Cremaschi P, Wetterskog D, Conteduca
V, Franceschini GM, Kleftogiannis D, Jayaram A, Sandhu S, Wong SQ,
Benelli M, et al: Genome-wide plasma DNA methylation features of
metastatic prostate cancer. J Clin Invest. 130:1991–2000. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhu K, Li Y and Xu Y: The FTO
m6A demethylase inhibits the invasion and migration of
prostate cancer cells by regulating total m6A levels.
Life Sci. 271:1191802021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lewis SJ, Murad A, Chen L, Davey Smith G,
Donovan J, Palmer T, Hamdy F, Neal D, Lane JA, Davis M, et al:
Associations between an obesity related genetic variant (FTO
rs9939609) and prostate cancer risk. PLoS One. 5:e134852010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Khella MS, Salem AM, Abdel-Rahman O and
Saad AS: The association between the FTO rs9939609 variant and
malignant pleural mesothelioma risk: A case-control study. Genet
Test Mol Biomarkers. 22:79–84. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Salgado-Montilla JL, Rodríguez-Cabán JL,
Sánchez-García J, Sánchez-Ortiz R and Irizarry-Ramírez M: Impact of
FTO SNPs rs9930506 and rs9939609 in prostate cancer severity in a
cohort of puerto rican men. Arch Cancer Res. 5:1482017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li S and Cao L: Demethyltransferase FTO
alpha-ketoglutarate dependent dioxygenase (FTO) regulates the
proliferation, migration, invasion and tumor growth of prostate
cancer by modulating the expression of melanocortin 4 receptor
(MC4R). Bioengineered. 13:5598–5612. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu Y, Zhang W, Shen F, Yang X, Liu H, Dai
S, Sun X, Huang J and Guo Q: YTH domain proteins: A family of
m6A readers in cancer progression. Front Oncol.
11:6295602021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu S, Li G, Li Q, Zhang Q, Zhuo L, Chen
X, Zhai B, Sui X, Chen K and Xie T: The roles and mechanisms of YTH
domain-containing proteins in cancer development and progression.
Am J Cancer Res. 10:1068–1084. 2020.PubMed/NCBI
|
|
48
|
Müller S, Bley N, Busch B, Glaß M, Lederer
M, Misiak C, Fuchs T, Wedler A, Haase J, Bertoldo JB, et al: The
oncofetal RNA-binding protein IGF2BP1 is a druggable,
post-transcriptional super-enhancer of E2F-driven gene expression
in cancer. Nucleic Acids Res. 48:8576–8590. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gruber AJ, Schmidt R, Ghosh S, Martin G,
Gruber AR, van Nimwegen E and Zavolan M: Discovery of physiological
and cancer-related regulators of 3′ UTR processing with KAPAC.
Genome Biol. 19:442018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jiang M, Lu Y, Duan D, Wang H, Man G, Kang
C, Abulimiti K and Li Y: Systematic investigation of mRNA N
6-methyladenosine machinery in primary prostate cancer.
Dis Markers. 2020:88334382020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Singh AN and Sharma N: Quantitative
SWATH-based proteomic profiling for identification of
mechanism-driven diagnostic biomarkers conferring in the
progression of metastatic prostate cancer. Front Oncol. 10:4932020.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Torosyan Y, Dobi A, Glasman M, Mezhevaya
K, Naga S, Huang W, Paweletz C, Leighton X, Pollard HB and
Srivastava M: Role of multi-hnRNP nuclear complex in regulation of
tumor suppressor ANXA7 in prostate cancer cells. Oncogene.
29:2457–2466. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Luxton HJ, Simpson BS, Mills IG, Brindle
NR, Ahmed Z, Stavrinides V, Heavey S, Stamm S and Whitaker HC: The
oncogene metadherin interacts with the known splicing proteins
YTHDC1, Sam68 and T-STAR and plays a novel role in alternative mRNA
splicing. Cancers (Basel). 11:12332019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li J, Yu W, Ge J, Zhang J, Wang Y, Wang P
and Shi G: Targeting eIF3f suppresses the growth of prostate cancer
cells by inhibiting Akt signaling. Onco Targets Ther. 13:3739–3750.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Saramäki O, Willi N, Bratt O, Gasser TC,
Koivisto P, Nupponen NN, Bubendorf L and Visakorpi T: Amplification
of EIF3S3 gene is associated with advanced stage in prostate
cancer. Am J Pathol. 159:2089–2094. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Savinainen KJ, Helenius MA, Lehtonen HJ
and Visakorpi T: Overexpression of EIF3S3 promotes cancer cell
growth. Prostate. 66:1144–1150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Savinainen KJ, Linja MJ, Saramäki OR,
Tammela TL, Chang GT, Brinkmann AO and Visakorpi T: Expression and
copy number analysis of TRPS1, EIF3S3 and MYC genes in breast and
prostate cancer. Br J Cancer. 90:1041–1046. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang X, Wang D, Liu B, Jin X, Wang X, Pan
J, Tu W and Shao Y: IMP3 accelerates the progression of prostate
cancer through inhibiting PTEN expression in a SMURF1-dependent
way. J Exp Clin Cancer Res. 39:1902020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Du C, Lv C, Feng Y and Yu S: Activation of
the KDM5A/miRNA-495/YTHDF2/m6A-MOB3B axis facilitates prostate
cancer progression. J Exp Clin Cancer Res. 39:2232020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li J, Meng S, Xu M, Wang S, He L, Xu X,
Wang X and Xie L: Downregulation of N6-methyladenosine
binding YTHDF2 protein mediated by miR-493-3p suppresses prostate
cancer by elevating N6-methyladenosine levels.
Oncotarget. 9:3752–3764. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gao Y, Teng J, Hong Y, Qu F, Ren J, Li L,
Pan X, Chen L, Yin L, Xu D and Cui X: The oncogenic role of EIF3D
is associated with increased cell cycle progression and motility in
prostate cancer. Med Oncol. 32:5182015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang L, Smit-McBride Z, Pan X, Rheinhardt
J and Hershey JW: An oncogenic role for the phosphorylated
h-subunit of human translation initiation factor eIF3. J Biol Chem.
283:24047–24060. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kawakami Y, Kubota N, Ekuni N,
Suzuki-Yamamoto T, Kimoto M, Yamashita H, Tsuji H, Yoshimoto T,
Jisaka M, Tanaka J, et al: Tumor-suppressive lipoxygenases inhibit
the expression of c-myc mRNA coding region determinant-binding
protein/insulin-like growth factor II mRNA-binding protein 1 in
human prostate carcinoma PC-3 cells. Biosci Biotechnol Biochem.
73:1811–1817. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang H, Ru Y, Sanchez-Carbayo M, Wang X,
Kieft JS and Theodorescu D: Translation initiation factor eIF3b
expression in human cancer and its role in tumor growth and lung
colonization. Clin Cancer Res. 19:2850–2860. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Xiang P, Sun Y, Fang Z, Yan K and Fan Y:
Eukaryotic translation initiation factor 3 subunit b is a novel
oncogenic factor in prostate cancer. Mamm Genome. 31:197–204. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hu J, Luo H, Xu Y, Luo G, Xu S, Zhu J,
Song D, Sun Z and Kuang Y: The prognostic significance of EIF3C
gene during the tumorigenesis of prostate cancer. Cancer Invest.
37:199–208. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hershey JW: The role of eIF3 and its
individual subunits in cancer. Biochim Biophys Acta. 1849:792–800.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cui L, Liu M, Lai S, Hou H, Diao T, Zhang
D, Wang M, Zhang Y and Wang J: Androgen upregulates the
palmitoylation of eIF3L in human prostate LNCaP cells. Onco Targets
Ther. 12:4451–4459. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chromecki TF, Cha EK, Pummer K, Scherr DS,
Tewari AK, Sun M, Fajkovic H, Roehrborn CG, Ashfaq R, Karakiewicz
PI and Shariat SF: Prognostic value of insulin-like growth factor
II mRNA binding protein 3 in patients treated with radical
prostatectomy. BJU Int. 110:63–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cheng Y, Li L, Qin Z, Li X and Qi F:
Identification of castration-resistant prostate cancer-related hub
genes using weighted gene co-expression network analysis. J Cell
Mol Med. 24:8006–8017. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Stockley J, Villasevil ME, Nixon C, Ahmad
I, Leung HY and Rajan P: The RNA-binding protein hnRNPA2 regulates
β-catenin protein expression and is overexpressed in prostate
cancer. RNA Biol. 11:755–765. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Lin VC, Kuo PT, Lin YC, Chen Y, Hseu YC,
Yang HL, Kao JY, Ho CT and Way TD: Penta-O-galloyl-β-D-glucose
suppresses EGF-induced eIF3i expression through inhibition of the
PI3K/AKT/mTOR pathway in prostate cancer cells. J Agric Food Chem.
62:8990–8996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xie C, Li Y, Li Q, Chen Y, Yao J, Yin G,
Bi Q, O'Keefe RJ, Schwarz EM and Tyler W: Increased insulin mRNA
binding protein-3 expression correlates with vascular enhancement
of renal cell carcinoma by intravenous contrast-CT and is
associated with bone metastasis. J Bone Oncol. 4:69–76. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yu YZ, Lv DJ, Wang C, Song XL, Xie T, Wang
T, Li ZM, Guo JD, Fu DJ, Li KJ, et al: Hsa_circ_0003258 promotes
prostate cancer metastasis by complexing with IGF2BP3 and sponging
miR-653-5p. Mol Cancer. 21:122022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Pin E, Henjes F, Hong MG, Wiklund F,
Magnusson P, Bjartell A, Uhlén M, Nilsson P and Schwenk JM:
Identification of a novel autoimmune peptide epitope of prostein in
prostate cancer. J Proteome Res. 16:204–216. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang J, Lin H, Zhou M, Xiang Q, Deng Y,
Luo L, Liu Y, Zhu Z and Zhao Z: The m6A methylation regulator-based
signature for predicting the prognosis of prostate cancer. Future
Oncol. 16:2421–2432. 2020. View Article : Google Scholar : PubMed/NCBI
|















