Introduction
Breast cancer surpassed lung cancer as the most
commonly diagnosed cancer in 2020, with an estimated 2.3 million
new cases, representing 11.7% of all cancer cases; in addition, it
is the fifth leading cause of cancer mortality worldwide, with
685,000 deaths (1). Breast cancer
is a life-threatening malignant tumor, which is the major cause of
premature death in women (2).
Despite advances in the diagnosis, drug development and
personalized treatment based on molecular classification of breast
cancer (3,4), diagnostic markers and therapeutic
targets are still lacking. Therefore basic research on this topic
is a hotspot (5,6). Elucidating the function of
metabolism-associated proteins in regulating breast cancer cell
metabolism is a promising prospect for identifying diagnostic and
therapeutic targets for breast cancer (7).
Cancer cells alter their metabolism to promote
survival, proliferation and long-term maintenance. The common
features of this altered metabolism include increased glucose
uptake and fermentation of glucose to lactate. This phenomenon is
observed even in the presence of completely functioning
mitochondria and is known as ‘the Warburg effect’ (8). Cancer cells use glycolysis as the
primary energy source for proliferation and cell cycle progression,
which is different from normal cells that rely on oxidative
phosphorylation in the mitochondria for energy (9). Although this phenomenon is elusive,
accumulating evidence since Warburg reported that cancer cells
metabolize glucose in the 1920s (10), has supported the hypothesis that
glycolysis is the primary energy metabolism method used to meet the
needs of cancer cells for continuous cell proliferation (9,11).
Furthermore, aerobic glycolysis supports tumor progression,
particularly in breast cancer cells (8). Previous studies have reported that
glycolysis affects the proliferation and progression of breast
cancer (12–14). However, the regulatory mechanism
of aerobic glycolysis in breast cancer cells is unknown.
Dicarbonyl/L-xylulose reductase (DCXR) is a highly
conserved enzyme in mammals that catalyzes conversion of L-xylulose
into xylitol (15). The role of
DCXR in L-xylulose metabolism has been known for decades (16); however, the role of DCXR in normal
human physiology and pathophysiology remains to be elucidated.
Previous studies have shown that DCXR expression disorder is
observed in age- and metabolism-associated diseases, especially in
human male infertility, nephropathy and diabetes (17). DCXR has multifunctional properties
with respect to carbonyl reductase and non-catalytic function
(18). Previous studies have
linked the role of DCXR with cell adhesion, indicating its novel
role in tumor progression and metastasis (18,19). Moreover, certain studies have
reported that DCXR is abnormally expressed in cancer tissue; for
example, DCXR is overexpressed in prostate adenocarcinoma and
melanoma (20–22) and downregulated in hepatocellular
carcinoma (23). Furthermore,
these studies reported a correlation between abnormal DCXR
expression levels and cancer progression or poor prognosis but did
not determine the role of the protein. The correlation between DCXR
and cancer is a promising research subject; therefore, the protein
function needs to be assessed further. To the best of our
knowledge, only a few studies have reported the expression of DCXR
in breast cancer tissue (18,24); however, the effect of this protein
on the pathological mechanisms of cancer progression has not yet
been assessed.
The present study evaluated the DCXR expression
pattern in breast cancer tissue by assessing breast cancer
databases and clinical tissue samples. Further functional analysis
focused on the role of DCXR in glycolysis, cell cycle and
proliferation using DCXR-overexpression and -silencing in breast
cancer cell lines. The assessment of the function and pathological
mechanism of DCXR may demonstrate DCXR to be a candidate for cancer
molecular targeting therapy in breast cancer.
Materials and methods
Bioinformatics analysis
DCXR expression data and survival data from patients
with breast cancer [paracancerous (n=113) and tumor (n=1,104)
tissue data] were obtained from The Cancer Genome Atlas (TCGA)
databases (https://tcga-data.nci.nih.gov/tcga/). Gene set
enrichment analysis (GSEA) was performed by the JAVA program
(http://www.broadinstitute.org/gsea)
using MSigDB portal. Enrichment Score (ES) was calculated via the
Kolmogorov-Smirnov test and the calculation of ES significance
level was analyzed by permutation test; finally, the false
discovery rate method was used for multiple hypothesis testing
correction, and |log2fold-change|>1 and P<0.05
were set as the cut-off for enrichment.
Human samples
A total of 80 pairs of paracancerous and cancer
tissue samples (1 cm distance) were obtained from patients (mean
age, 49 years; age range, 30–70 years) who underwent breast tumor
resection surgery at Seventh People's Hospital of Shanghai
University of Traditional Chinese Medicine (Shanghai, China)
between September 2020 and August 2021. Patients who received
chemotherapy or radiation prior to resection were excluded from the
present study. Resected tissues were stored at −80°C until
examination. All patients provided written informed consent. Tissue
DCXR mRNA and protein expression levels were assessed using reverse
transcription-quantitative PCR (RT-qPCR) and immunohistochemistry.
Protocols using human samples were approved by the Independent
Ethics Committee of The Shanghai Seventh People's Hospital
(approval no. 2020-7th-HIRB-031) and followed the Declaration of
Helsinki.
Cell culture
MDA-MB-231, BT-474, T47D, MCF-7 and ZR751 human
breast cancer cell lines were purchased from the The Cell Bank of
Type Culture Collection of The Chinese Academy of Sciences. MCF-10A
human normal mammary epithelium cell line (Procell Life Science
& Technology Co., Ltd.) was used as the control. Cells were
grown in DMEM (Thermo Fisher Scientific, Inc.), with 10% fetal calf
serum (Thermo Fisher Scientific, Inc.), L-glutamine (2 mM) and 1%
penicillin/streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.). All cells were cultured with 5% CO2 at
37°C.
RT-qPCR
From the 80 patients, a total of 30 pairs of
paracancerous and cancer tissue samples were randomly selected.
Tissue and cell DCXR mRNA expression levels were assessed by
RT-PCR. Total RNA was extracted from BT-474, ZR751, MCF-7,
MDA-MA-231, T47D and MCF-10A cells, and human breast cancer and
adjacent tissues using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.). After determination of purity and
quality, RT was performed using Maxima SYBR Green/ROX Qpcr
Pre-mixed solution (2X) (cat. no. K0233; Thermo Fisher Scientific,
Inc.) at 42°C for 1 h, followed by 70°C for 15 min. The Cdna
product was used for Qpcr with the following thermocycling
conditions: Initial denaturation of 95°C for 10 min; followed by 40
cycles of 95°C for 15 sec and 60°C for 45 sec (Takara Biotechnology
Co., Ltd.). The primers were as follows: DCXR forward,
5′-GAATGTCTCCAGCCAGTG-3′ and reverse, 5′-GGATTCGGTTCAGCATAG-3′;
GAPDH forward, 5′-CAAATTCCATGGCACCGTCA-3′ and reverse,
5′-GCATCGCCCCACTTGATTTT-3′. GAPDH was used as an internal control.
Relative DCXR Mrna expression levels were calculated using the
2−ΔΔCq method in three replicate experiments (25).
Western blotting
From the 80 patients, a total of 30 pairs of
paracancerous and cancer tissue samples were randomly selected.
Tissue and cell DCXR protein expression levels were assessed by
western blotting. Total proteins were extracted from MDA-MB-231,
BT-474, T47D, MCF-7 and ZR751 human breast cancer cell lines using
RIPA lysis buffer (Thermo Fisher Scientific, Inc.). The protease
inhibitor (Thermo Fisher Scientific, Inc.) was pre-dissolved in the
lysis buffer to prevent proteolysis. The obtained protein samples
underwent quantitative analysis with an enhanced BCA protein assay
kit (Thermo Fisher Scientific, Inc.) and stored at −20°C until
subsequent experimentation. A mixture of loading buffer with an
equal amount of protein samples (25 µg) was loaded onto 10%
SDS-PAGE to separate proteins of different molecular weights.
Protein was then transferred to nitrocellulose membranes
(MilliporeSigma). Non-specific protein on the membrane was blocked
using 5% non-fat dried milk dissolved in 1X PBS containing 0.05%
Tween-20 for 1 h at room temperature. The membranes were then
incubated with the blocking buffer-diluted primary antibodies
overnight at 4°C. The primary antibodies used are as follows: DCXR
(1:1,000; cat. no. ab110283; Abcam) and β-actin (1:2,000; cat. no.
4970; Cell Signaling Technologies, Inc.). After rinsing with
Tris-HCl buffer (pH 7.4, 20 mM), membranes were incubated at room
temperature with the corresponding secondary antibodies bound to
horseradish peroxidase (anti-rabbit, 1:2,000, cat. no. A0208;
anti-mouse, 1:2,000, cat. no. A0216; both from Beyotime Institute
of Biotechnology) for 2 h. The probed targeted proteins were
visualized using a Tanon-5200 Multi-Imaging System (Tanon Science
and Technology Co., Ltd.) following treatment with an Tanon™ ECL
chemiluminescence substrate kit (cat. no. 180-501; Tanon Science
and Technology Co., Ltd.). The relative protein levels were
semi-quantified using ImageJ V1.8.0 software (National Institutes
of Health).
DCXR knockdown and overexpression
vector construction and transduction into breast cancer cells
Three short hairpin (sh)RNAs targeting three
different human DCXR gene loci were synthesized (shDCXR-1, shDCXR-2
and shDCXR-3). The shRNA sequences were: shDCXR-1,
5′-CACCGGCCTTTGACAGATCCTTTGACGAATCAAAGGATCTGTCAAAGGCC-3′; shDCXR-2,
5′-CACCGCGGGCAGTAACTAACCATAGCGAACTATGGTTAGTTACTGCCCGC-3′; shDCXR-3,
5′-CACCGAATCCCACTTGGCAAGTTTGCGAACAAACTTGCCAAGTGGGATTC-3′. A
scrambled sequence was used as shRNA negative control (shNC).
shDCXR and shNC were constructed in lentiviral plasmids (pLKO.1)
using a 2nd generation system and 293T cells (The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences) were
transfected with these plasmids to produce lentiviruses using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.); the ratio of plasmids, pMD2.G and psPAX2
(Shanghai GenePharma Co., Ltd.) was 2:1:2. All shRNAs were
purchased from Shanghai Majorbio Bio-Pharm Technology Co., Ltd. For
overexpression of DCXR in the breast cancer cell lines, recombinant
overexpression DCXR (oeDCXR) was generated using the pLVX-puro
lentiviral plasmid constructed with DCXR (NM_016286.4) cDNA (2nd
generation system). An empty plasmid was used as oeNC. The
overexpression lentiviral plasmids were generated in 293T cells
according to the aforementioned protocol for shDCXR and shNC.
oeDCXR and oeNC were purchased from Shanghai GenePharma Co., Ltd.
The quantity of lentiviral plasmid used for transfection was 5 µg
(108 TU/ml). For breast cancer transduction, MDA-MB-231
cells were seeded in a 24-well plate and cultured to 70–80%
confluency at 37°. shDCXRs or oeDCXR were added in the presence of
Lipofectamine 2000 (26). The
concentration of purinomycin used for screening was 7 µg/ml, and
after 48 h transduction at 37°C, the cells were harvested for
analysis.
Cell Counting Kit-8 (CCK-8) assay for
cell proliferation
Cell proliferation was evaluated using CCK-8 assay
(Signalway Antibody LLC). Briefly, followed by treatment with shRNA
at 0, 12, 24 or 48 h transduction, cells were incubated with CCK-8
solution (1:10) for 1 h and the absorbance was measured at 450 nm
using a microplate reader (BioTek Corporation).
Flow cytometry
Changes to the cell cycle were evaluated using
propidium iodide (PI) staining on a flow cytometer (Accuri C6; BD
Biosciences). ZR751 and BT-474 cells (1×106) were
suspended in PBS and fixed with 70% ethanol for 2 h at 20°C. RNase
A (Beijing Solarbio Science & Technology Co., Ltd.) was used to
treat cells for 15 min at 37°C. Cells were stained using PI (7Sea
PharmTech Co., Ltd.) for 30 min in the dark at 4°C. The DNA content
of each sample was analyzed by flow cytometry. The percentage of
cells at G0/G1, S and G2/M phase was calculated using FlowJo v10.8
(FlowJo LLC).
Chemical detection of ATP and lactate
dehydrogenase (LD)
Cell ATP levels were assessed using a
luciferase-based ATP assay kit (Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Briefly,
breast cancer cells were transduced with or without shRNA, the
cells were scraped off with a cell scraper, and cell precipitates
were collected by centrifugation at 272 × g for 10 min at room
temperature. Subsequently, the cell precipitates were added to the
lysis buffer of the ATP assay kit at a ratio of 50–100 µl of lysis
buffer to each well of a 24-well plate. The samples were then
vortexed, after which, they were centrifuged at 12,000 × g for 5
min at 4°C, and the supernatant was obtained for subsequent
determination. In 24-well plates, 100 µl ATP detection reagent was
added to 100 µl supernatant at room temperature for 5 min.
Luminescence was measured using a Safire II monochromator
microplate reader (Tecan Group Ltd.). Standard curves were obtained
by diluting standards using ATP diluents. The concentration of ATP
in each sample was calculated from the standard curve. In order to
eliminate the error caused by the difference in protein amount in
sample preparation, the protein concentration of each treatment
group was assessed using Bradford Protein Assay.
LD levels were assessed using breast cancer cells at
the logarithmic growth stage transduced with shRNA or
overexpression lentiviral particles; the blank control group was
cultured the same way. The supernatant was collected by
centrifugation at 12,000 × g for 5 min and diluted with 400 µl
normal saline at room temperature. The assay was performed
according to the manufacturer's protocol (Lactic Acid assay kit;
Nanjing Jiancheng Bioengineering Institute), heated in a 37°C water
bath for 10 min and stop solution was then added. After mixing, the
absorbance was measured at 530 nm (ELx800™; BioTek
Corporation).
Glycolysis and mitochondrial
respiration assay
MDA-MB-231 cells (5×105/well) were
cultured in 24-well plates. The extracellular acidification rate
(ECAR) assay was used to evaluate glycolysis. Briefly, cells were
grown in XF base medium (cat. no. 103575; Seahorse Bioscience;
Agilent Technologies, Inc.) supplemented with 2 mM L-glutamine and
25 mM glucose. Glycolysis inhibitor 2-deoxy-D-glucose (2-DG; 10
mmol/l; Beijing Solarbio Science & Technology Co., Ltd.) and
oligomycin (1 µM; Beijing Solarbio Science & Technology Co.,
Ltd.) were used to treat cells at 37°C for 1 h to suppress
glycolytic metabolism and the effects on breast cancer cells were
assessed. For oxygen consumption rate (OCR) assay, cells were
cultured in XF base medium with 1.0 oligomycin, 0.5 FCCP (a potent
uncoupler of mitochondrial oxidative phosphorylation) and 0.45 µM
rotenone/antimycin A to assess mitochondrial oxidative
phosphorylation. ECAR and OCR examination were performed using an
XFe24 Extracellular Flux Analyzer (Seahorse Bioscience; Agilent
Technologies, Inc.).
Immunohistochemistry (IHC) and
fluorescence staining
Human tissues were embedded in paraffin after fixing
with 10% formalin at 4°C for 24 h. The sections (2 µm) were dewaxed
and rehydrated before IHC staining. Subsequently, the slides were
soaked in 3% hydrogen peroxide solution for 15 min at room
temperature to quench the endogenous peroxidase activity. To avoid
producing non-specific binding, 2.5% goat serum (cat. no. ab7481;
Abcam) was used to block the sections at room temperature for 1 h.
A primary antibody against DCXR (1:500, cat. no. ab110283; Abcam)
was used to incubate the sections at room temperature for 1 h,
followed by incubation with the secondary antibodies (1:2,000;
anti-mouse; cat. no. ab205719; Abcam) at room temperature for 30
min, and washing with TBS. Immunoreactivity was visualized using
DAB and hematoxylin was used for counterstaining at room
temperature for 1 min. Protein expression levels were assessed
using a light microscope (ECLIPSE E100; Nikon Corporation) and
imaged. For IHC, the scoring system was composed of staining
intensity (A) and positive areas (B). A was scored as no staining
(0), weak staining (1+), moderate staining (2+) or strong staining
(3+). B was classified as 0% (0+), 1–10% (1+), 11–25% (2+), 26–50%
(3+) or 51–100% (4+). The total score of A and B was 0–9 for each
specimen; 0–3 was defined as low protein expression and 4–9 defined
as high protein expression of DCXR.
For fluorescence staining, tissue samples were fixed
with 4% paraformaldehyde at room temperature for 24 h, embedded in
paraffin and cut into 2-µm sections. Subsequently, the sections
were dewaxed with xylene for 15 min and rehydrated conventionally
using an ethanol gradient (from 99 to 70%, followed by
demineralized water). The endogenous peroxidase activity was
blocked at room temperature using 3% hydrogen peroxide solution for
15 min. After blocking the non-specific protein binding with 5%
skimmed milk at room temperature for 30 min, sections were
incubated with anti-Ki67 antibody (1:500; cat. no. ab15580; Abcam)
at 4°C overnight. Following incubation with Alexa Fluor 488-labeled
goat anti-mouse IgG antibody (1:500; cat. no. A0428; Beyotime
Institute of Biotechnology) for 1 h at room temperature. Images of
the stained sections were captured using a ZEISS fluorescence
microscope (Zeiss AG).
Xenograft model in nude mice
The study protocol for animal experimentation was
approved by the Ethics Committee of Shanghai Seventh People's
Hospital (approval no. 2021-AR-011) and conformed to the ethical
guidelines of the National Institutes of Health Guide for the Care
and Use of Laboratory Animals (NIH Publications No. 8023, revised
1978) (26). Female BALB/c nude
mice (age, 4 weeks; weight, 20±5 g) and ZR751 cells were used for
tumor xenograft experiments. All mice were maintained under
controlled temperature (22±1°C) and humidity (50±5%) in a 12/12-h
light/dark cycle with food and water available ad libitum.
ZR751 cells transduced with different constructs (shDCXR-1 and
shNC) were resuspended in PBS and injected subcutaneously into the
armpits of nude mice (5×106 cells/100 µl; 200 µl). A
total of 12 mice were randomly divided into two groups as follows:
shNC (mice injected subcutaneously with ZR751 cells transduced with
shNC) and shDCXR (mice injected subcutaneously with ZR751 cells
transduced with shDCXR-1). Tumors were measured weekly and the
tumor volume (V) was calculated as follows: V=(length × width
×2)/2. After 33 days, mice were anesthetized using 1% pentobarbital
sodium (40 mg/kg) injected intraperitoneally and sacrificed by
acute exsanguination after they became unconscious. The tumors were
excised for IHC staining.
Statistical analysis
Experimental data are presented as the mean ±
standard error of the mean of a minimum of three independent
experiments. GraphPad Prism 8 (GraphPad Software, Inc.) was used
for statistical analysis. Unpaired Student's t test was used for
comparisons between two groups, and paired Student's t-test was
used to determine the statistical significance of differences in
the expression levels in clinical tissue samples. In addition,
χ2 analysis was performed to assess the association of
DCXR expression with tumor size, tumor stage, AJCC stage, distant
metastasis and ER/PR/HER2-status. Univariate and multivariate Cox
regression analysis were used to evaluate prognostic significance.
Survival rate was analyzed by Kaplan-Meier with log-rank test. To
test statistical significance between multiple groups, one-way
ANOVA followed with Tukey's post hoc test was used. All statistical
tests were two-sided. P<0.05 was considered to indicate a
statistically significant difference.
Results
DCXR is upregulated and associated
with tumor progression in human breast cancer tissue
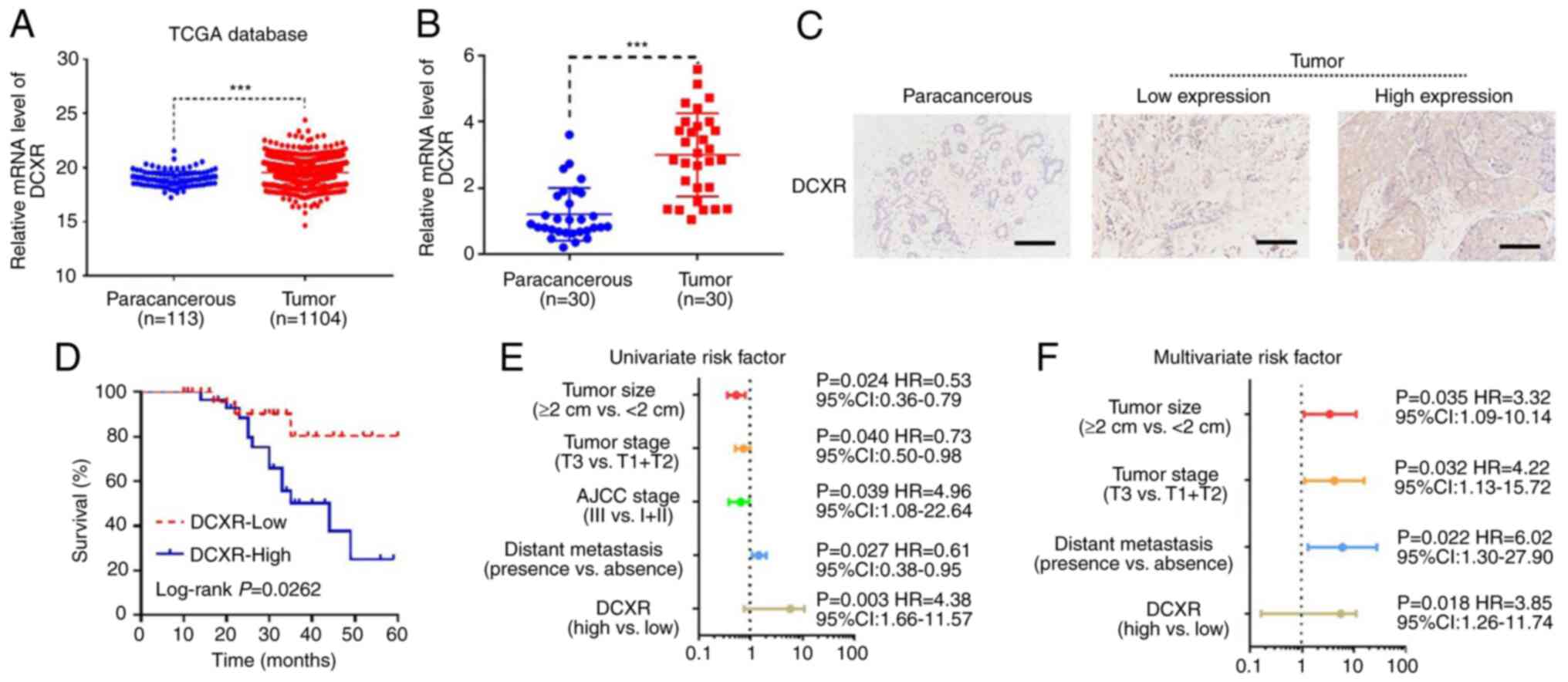
DCXR expression was assessed in 1,104 breast cancer
and 113 paracancerous tissue samples from TCGA database. The
results showed that DCXR was significantly upregulated in breast
cancer tissue (Fig. 1A). A total
of 80 patients' samples and data were collected for univariate and
multivariate risk factor analysis and 30 patients were selected for
RNA and protein expression verification; the expression levels of
DXCR in the 50 patients were determined by IHC. RT-qPCR results
showed that DCXR was significantly upregulated in breast cancer
tissue compared with the paracancerous tissue (Fig. 1B). Furthermore, positive DCXR
protein expression in breast cancer tissue was detected in 30
tissue samples by IHC staining. Subsequently, 30 patients were
grouped into low- and high-expression populations according to the
IHC score: 0–3 was defined as low protein expression and 4–9 was
defined as high protein expression of DCXR (Fig. 1C). To determine the association
between DCXR expression and breast cancer progression, the protein
expression and clinical characteristics of 30 patients were picked
at random to analyze. Survival rate, which was analyzed by
Kaplan-Meier with log-rank test, showed that high expression of
DCXR was associated with poor prognosis in patients with breast
cancer (Fig. 1D). The
χ2 analysis of DCXR expression and tumor indicators
suggested that high protein expression was associated with tumor
size, tumor stage, American Joint Committee on Cancer (AJCC) stage,
ER/PR/HER2-status and distant metastasis (Table I). Univariate regression analysis
showed that DCXR expression, tumor size, tumor stage, AJCC stage
and distant metastasis were significant predictors for breast
cancer (Fig. 1E), whereas
multivariate regression analysis further demonstrated that, with
the exception of AJCC stage, the other four risk factors were
independent predictors of breast cancer aggressiveness, with
significant HRs for predicting clinical outcome (Fig. 1F). These data indicated that
upregulation of DCXR was associated with breast cancer
progression.
 | Table I.Association between DCXR expression
and clinicopathological characteristic in patients with breast
cancer. |
Table I.
Association between DCXR expression
and clinicopathological characteristic in patients with breast
cancer.
|
|
| DCXR
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | Number of
cases | High (%) | Low (%) |
P-valuea |
|---|
| Age, years |
|
|
| 0.2007 |
|
≥50 | 32 | 14 (43.8) | 18 (56.2) |
|
|
<50 | 48 | 28 (58.3) | 20 (41.7) |
|
| Tumor size, cm |
|
|
| 0.0024 |
| ≥2 | 54 | 22 (40.7) | 32 (59.3) |
|
|
<2 | 26 | 20 (76.92) | 6 (23.08) |
|
| Histopathology |
|
|
| 0.2367 |
|
Ductal | 55 | 32 (58.2) | 23 (41.8) |
|
|
Lobular | 25 | 18 (72.0) | 7 (28.0) |
|
| Histological
grade |
|
|
| 0.0317 |
| 1 | 13 | 3 (23.1) | 10 (76.9) |
|
| 2 | 44 | 28 (63.6) | 16 (36.4) |
|
| 3 | 23 | 11 (47.8) | 12 (52.2) |
|
| AJCC stage |
|
|
| 0.0265 |
| I | 18 | 3 (16.7) | 15 (83.3) |
|
| II | 20 | 12 (60) | 8 (40) |
|
|
III | 42 | 27 (64.3) | 15 (35.7) |
|
| Tumor stage |
|
|
| 0.0382 |
| T1 | 11 | 3 (27.3) | 8 (72.7) |
|
| T2 | 20 | 15 (66.7) | 5 (33.3) |
|
| T3 | 49 | 39 (33.3) | 10 (66.7) |
|
| Distant
metastasis |
|
|
| 0.0147 |
|
Presence | 40 | 33 (82.5) | 7(17.5) |
|
|
Absence | 40 | 23 (57.5) | 17 (42.5) |
|
| ER status |
|
|
| 0.0150 |
|
Positive | 50 | 32 (62.7) | 19 (37.3) |
|
|
Negative | 30 | 10 (34.5) | 19 (65.5) |
|
| PR status |
|
|
| 0.0015 |
|
Positive | 54 | 35 (64.8) | 19 (35.2) |
|
|
Negative | 26 | 7 (26.9) | 19 (73.1) |
|
| HER2 status |
|
|
| 0.0257 |
|
Positive | 22 | 16 (72.7) | 6 (27.3) |
|
|
Negative | 58 | 26 (44.8) | 32 (55.2) |
|
DCXR silencing suppresses
proliferation, arrests the cell cycle and decreases glycolysis
activity in human breast cancer cells
GSEA showed that genes in the high-expression DCXR
group were primarily enriched in ‘glycolysis’ and ‘cell cycle’
(Fig. 2A and B).
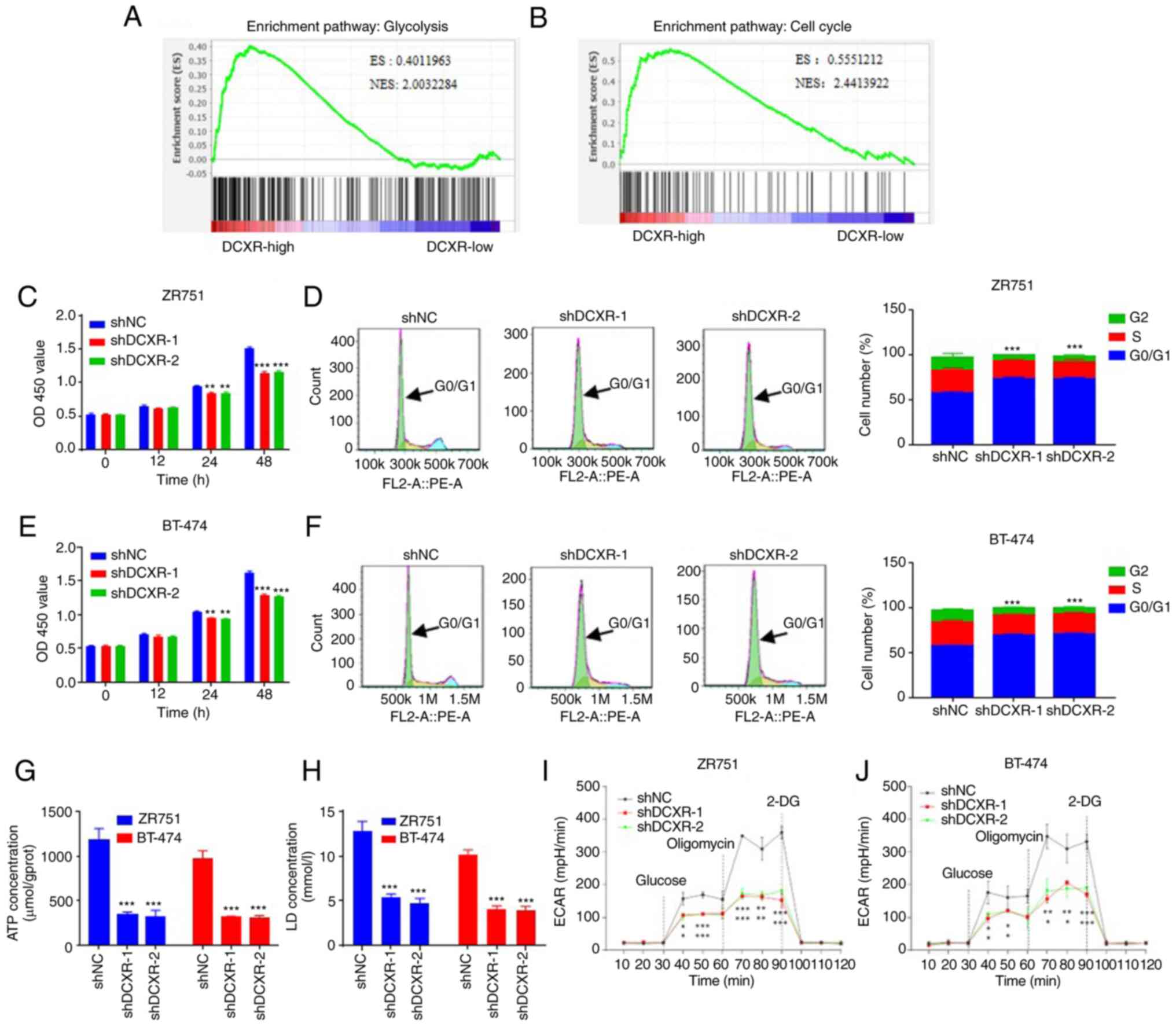 | Figure 2.DCXR silencing suppresses
proliferation, arrests the cell cycle and decreases glycolysis
activity of human breast cancer cells. Bioinformatics analysis
indicated that DCXR was enriched in (A) ‘glycolysis’ and (B) ‘cell
cycle’ in breast cancer. Proliferation of (C) ZR751 and (D) BT-474
cells was suppressed following transduction with shDCXR-1 and
shDCXR-2. Knockdown of DCXR arrested cells in G0/G1 phase in (E)
ZR751 and (F) BT-474 cells. DCXR silencing decreased production of
(G) ATP and (H) LD in ZR751 and BT-474 cells. Both shDCXR-1 and
shDCXR-2 suppressed ECAR (mpH/min) in (I) ZR751 and (J) BT-474
cells. *P<0.05, **P<0.01, ***P<0.001 vs. shNC. ES,
enrichment Score; NES, normalized enrichment score; 2-DG,
2-deoxy-D-glucose; DCXR, dicarbonyl/L-xylulose reductase; ECAR,
extracellular acidification rate; LD, lactate dehydrogenase; NC,
negative control; OD, optical density; sh, short hairpin. |
Next, in vitro experiments using breast
cancer cell lines were used to assess the function of DCXR. DCXR
mRNA and protein levels were upregulated in MDA-MB-231, BT-474,
T47D, MCF-7 and ZR751 cells compared with MCF-10A normal human
mammary epithelial cells (Fig. S1A
and B). The two cell lines with the highest expression levels
(ZR751 and BT-474) were selected for DCXR knockdown and MDA-MB-231,
with the lowest expression, was selected for DCXR overexpression.
DCXR was knocked down by transducing three different shDCXR into
ZR751 and BT-474 cells; cells with shDCXR-1 and shDCXR-2 were
selected for follow-up study based on the efficiency of shRNA; the
results showed that all three shRNAs could effectively inhibit the
expression of DCXR, and the shRNA with the best knockdown effect
(shRNA-1) was selected for subsequent experiments (Fig. S1C and D). In shDCXR-1- and
shDCXR-2-transduced ZR751 and BT-474 cells, proliferation was
decreased at 24 and 48 h (Fig. 2C and
E), cell cycle was arrested in G1 phase (Fig. 2E and F), the production of ATP and
LD was decreased (Fig. 2G and H)
and ECAR was decreased (Fig. 2I and
J). These data suggested DCXR knockdown inhibited glycolysis
and cell cycle of breast cancer cells.
DCXR silencing suppresses
tumorigenicity of human breast cancer cells in a mouse tumor
model
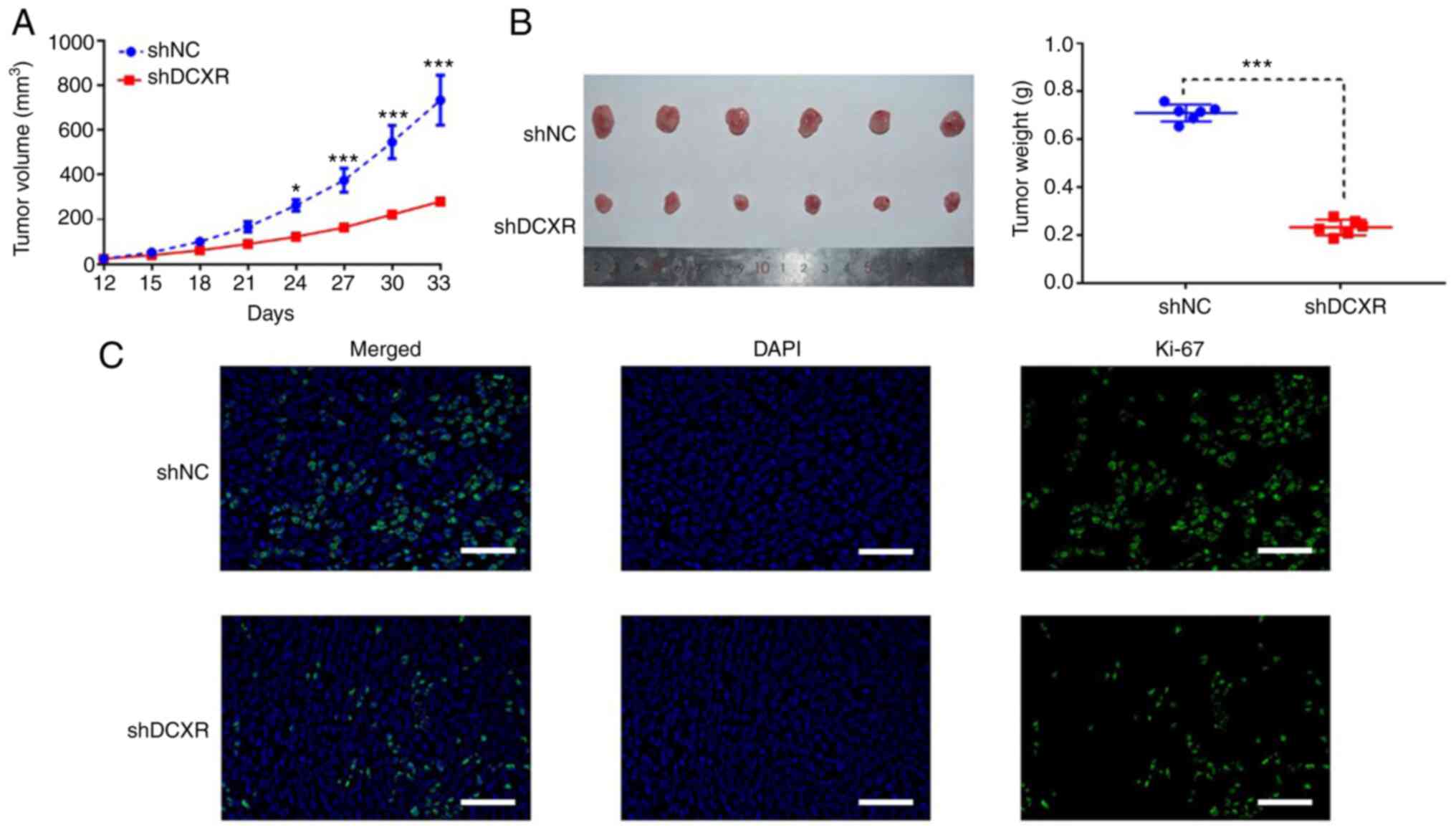
To identify the effect of DCXR on tumorigenicity of
breast cancer cells, tumor growth was observed following injection
of shDCXR- or shNC-transduced ZR751 cells into left flank of nude
mice. At 33 days post-injection, DCXR interference was found to
suppress the tumorigenic and proliferative ability of breast cancer
cells; the volume of subcutaneous tumors was significantly smaller
in the shDCXR group compared with the shNC group at 33 days
post-injection (Fig. 3A and B).
IHC assay for Ki-67 was used to assess cancer cell proliferation
(23). The results showed that
expression of Ki-67 was downregulated in tumor tissue of mice
transplanted with DCXR-silenced cells by fluorescence staining
(Fig. 3C), suggesting that DCXR
silencing may suppress cell proliferation.
Glycolysis inhibitor 2-DG abolishes
the effect of DCXR on proliferation and glycolysis of human
MDA-MB-231 cells
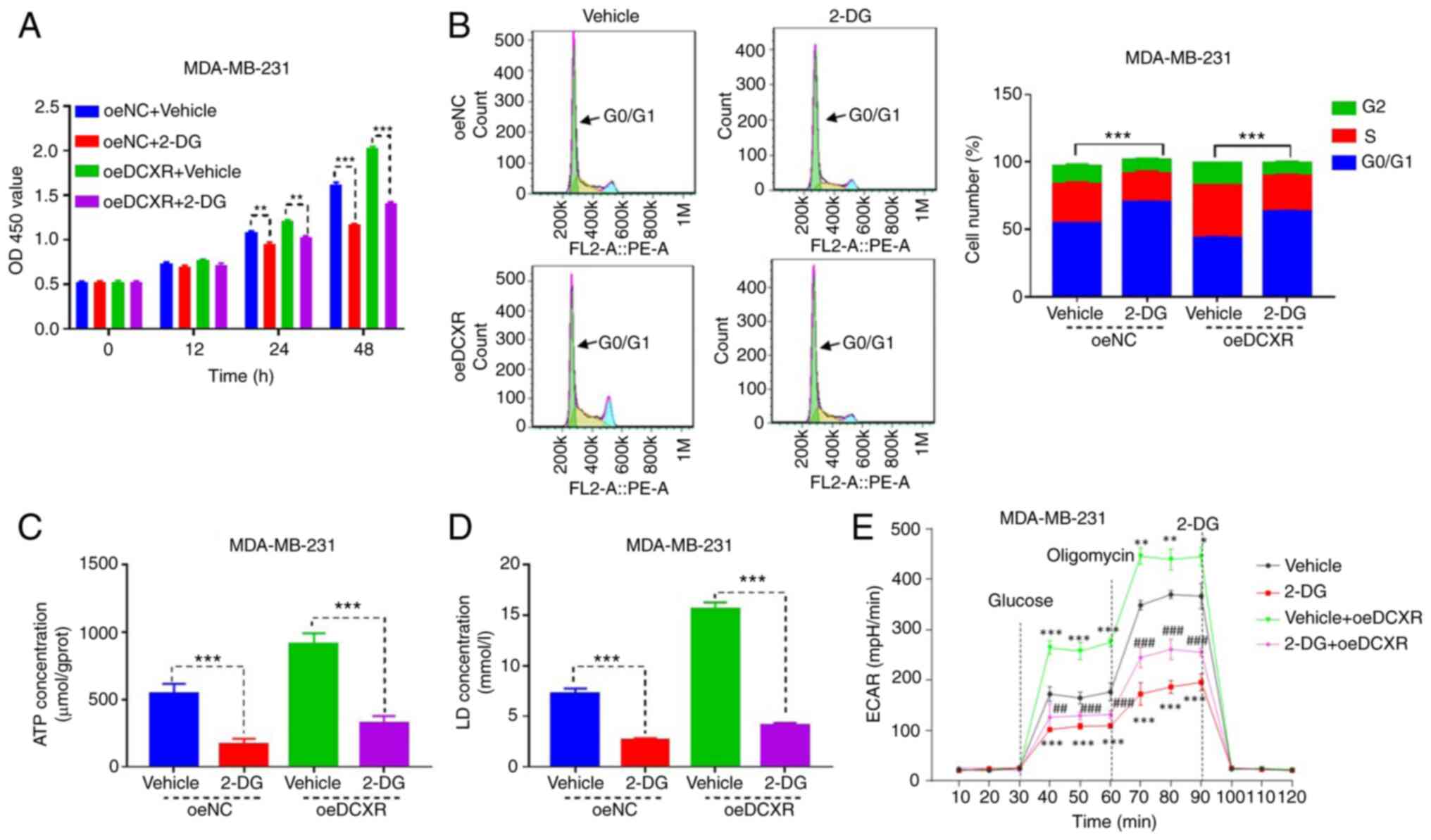
The MDA-MB-231 cell line overexpressing DCXR was
cultured to investigate the role of glycolysis inhibitor 2-DG on
DCXR function in MDA-MB-231 breast cancer cells. Overexpression and
knockdown of DCXR were detected by RT-qPCR and western blotting
(Fig. S1C-F). DCXR
overexpression promoted proliferation after 24 h (Fig. S2A), progression at S phase
(Fig. S2B) and the production of
ATP, LD and ECAR (Fig. S2C-E,
respectively). 2-DG exposure for 24 and 48 h significantly
inhibited cell proliferation, and its combination with DCXR
overexpression suppressed oe-DCXR-induced cell proliferation
(Fig. 4A). Subsequently, the role
of 2-DG on DCXR-induced effects on cell cycle and
glycolysis-associated indexes were evaluated. 2-DG exposure
increased the number of MDA-MB-231 cells in G1 phase (Fig. 4B) and decreased production of ATP
(Fig. 4C), LD (Fig. 4D) and ECAR (Fig. 4E), indicating that 2-DG suppressed
the cell cycle and glycolytic metabolism. 2-DG treatment combined
with oe-DCXR in MDA-MB-231 cells attenuated the effects of DC XR
protein overexpression on the cell cycle, and glycolysis-associated
indexes were also attenuated (Fig.
4A-E). These data indicated that the effect of DC XR on cell
cycle progression and glycolysis was abrogated when glycolysis was
inhibited.
Discussion
To the best of our knowledge, the present study is
the first to report an association between DCXR and disease
progression with respect to breast cancer. DCXR expression was
upregulated and promoted cell cycle, proliferation and glycolytic
metabolism in breast cancer.
DCXR was first noticed because of its association
with pentosuria, a deficiency that elevates urine levels of
L-xylulose (27). In addition to
acting as a catalytic enzyme, DCXR performs other functions through
protein.protein interactions, such as affecting sperm-zona
pellucida interaction and cell adhesion (22,28). Its role in disease, such as
infertility and cancer, has also been considered (18); however, our understanding of the
role of DCXR in cancer is limited. In the present study, DCXR was
highly expressed in breast cancer tissue, based on the results of
microarray assay from TCGA. Genome-wide gene expression analysis
based on microarray technology is a key step in elucidating the
molecular mechanisms of chronic disease, such as obesity,
cardiovascular disease and cancer, and to identifying the key genes
involved. The characteristic genes that discriminate healthy from
unhealthy samples may be used as a target for drug development or
as a molecular marker for diagnosis and prognosis (29). The present study results
demonstrated DCXR expression levels were significantly higher in 30
clinical breast cancer samples compared with paracancerous tissue,
which was consistent with results from TCGA. Thus, DCXR may be a
molecular marker for breast cancer diagnosis. DCXR was subsequently
associated with poor prognosis, tumor size, tumor stage, AJCC stage
and distant metastasis. Overall, these results suggested that DCXR
may serve as a potential clinical diagnostic/prognostic marker and
therapeutic target in breast cancer. Moreover, consistent with
previously published articles, the presented data supported DCXR as
an oncogene (18,19). The present study showed DCXR
upregulation may be associated with breast cancer tumor progression
and may be an independent risk factor for breast cancer.
The link between glycolysis and the cell cycle is
important for cancer progression (30). The cell cycle is a process of cell
division regulated by checkpoint controls; disrupted cell cycle
underlies abnormal cell division and proliferation (31). Moreover, the cell cycle is an
energy-intensive process, supported by oxidative phosphorylation
and/or glycolysis (32). Cancer
cells usually exhibit enhanced glycolysis that provides abundant
ATP when necessary (for example at G1/S transition checkpoint) to
support the proliferation and invasion of cancer cells (33,34). Certain studies have shown that
signaling molecules critical for glycolysis regulate breast cancer
cell proliferation (11,35–37). To the best of our knowledge,
however, the function of DCXR in cell events and glycolytic
metabolism during disease progression has been rarely studied. The
present study investigated the role of DCXR in regulating
glycolysis and cell cycle in breast cancer cells because the
function of DCXR was significantly enriched in ‘glycolysis’, as
indicated by GSEA bioinformatics analysis. DCXR loss- or
gain-of-function results suggested that DCXR silencing inhibited
both glycolysis and cell cycle G1/S checkpoint progression, whereas
DCXR overexpression promoted glycolysis and G1/S phase progression.
To the best of our knowledge, this is the first study demonstrating
that DCXR promotes glycolytic metabolism. Therefore, we
hypothesized that DCXR may promote breast cancer development by
regulating glycolysis and cell cycle. This phenomenon was further
supported by in vivo experiments using mice with xenograft
tumors, which showed that DCXR silencing limited tumor growth and
proliferation; in vitro human breast cancer cell assays
showed that exposure to glycolysis inhibitor 2-DG abolished the
promoting effect of DCXR overexpression on cell cycle and
glycolysis. Previous studies have shown that the function of DCXR
is enzyme catalysis and protein-protein interaction-mediated cell
adhesion in cancer cells (19,20). The present study demonstrated a
novel function of DCXR in enhancing glycolytic metabolism and
promoting proliferation of breast cancer cells. The present study
had certain limitations. First, the reason for the increase in DCXR
has not been identified. Furthermore, the molecular mechanism of
glucose metabolism in animals has not been clarified. These will be
the focus of future research.
The present findings suggested that DCXR may be an
oncogene in breast cancer cells and may be associated with tumor
progression. These results revealed that DCXR may regulate
glycolysis, cell cycle progression and proliferation in breast
cancer cells. This study investigated the biological function of
DCXR and provided a basis for the application of DCXR as a marker
or therapeutic target for breast cancer.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Scientific Research
Project of Shanghai Municipal Health Commission (grant no.
201940502) and The Science and Technology Development Fund of
Shanghai Pudong New Area (grant no. PKJ2019-Y14).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
YY and BZ contributed to the conception and design
of this study. YJ and MZ performed the experiments, collected the
data and performed statistical analysis with the help of YT and LQ.
YJ and MZ drafted the manuscript, which was corrected and revised
by YY and BZ. YY, BZ, YJ and MZ confirm the authenticity of all the
raw data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients and research protocols were approved by the Ethics
Committee of Shanghai Seventh's People's Hospital (approval no.
2020-7th-HIRB-031). All experimental procedures involving animals
were approved by The Animal Care and Use Committee of Shanghai
Seventh's People's Hospital (approval no. 2021-AR-011).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Solanki M and Visscher D: Pathology of
breast cancer in the last half century. Hum Pathol. 95:137–148.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang HX and Gires O: Tumor-derived
extracellular vesicles in breast cancer: From bench to bedside.
Cancer Lett. 460:54–64. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsang JYS and Tse GM: Molecular
classification of breast cancer. Adv Anat Pathol. 27:27–35. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kandel A, Dhillon SK, Prabaharan CB,
Hisham SFB, Rajamanickam K, Napper S, Chidambaram SB, Essa MM, Yang
J and Sakharkar MK: Identifying kinase targets of PPARgamma in
human breast cancer. J Drug Target. 29:660–668. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rossi FA, Steinberg JH, Roitberg EH, Joshi
MU, Pandey A, Abba MC, Dufrusine B, Buglioni S, Laurenzi VD, Sala
G, et al: USP19 modulates cancer cell migration and invasion and
acts as a novel prognostic marker in patients with early breast
cancer. Oncogenesis. 10:282021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang YP and Lei QY: Perspectives of
reprogramming breast cancer metabolism. Adv Exp Med Biol.
1026:217–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu Z, Wu J, Zhao Q, Fu S and Jin J:
Emerging roles of aerobic glycolysis in breast cancer. Clin Transl
Oncol. 22:631–646. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heiden MG, Cantley LC and Thompson CB:
Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vaupel P, Schmidberger H and Mayer A: The
Warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Enzo E, Santinon G, Pocaterra A, Aragona
M, Bresolin S, Forcato M, Grifoni D, Pession A, Zanconato F, Guzzo
G, et al: Aerobic glycolysis tunes YAP/TAZ transcriptional
activity. EMBO J. 34:1349–1370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Y, He J, Yang L, Luo Q and Liu Z:
Histone deacetylase-3 modification of MicroRNA-31 promotes cell
proliferation and aerobic glycolysis in breast cancer and is
predictive of poor prognosis. J Breast Cancer. 21:112–123. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang HS, Du GY, Zhang ZG, Zhou Z, Sun HL,
Yu XY, Shi YT, Xiong DN, Li H and Huang YH: NRF2 facilitates breast
cancer cell growth via HIF1a-mediated metabolic reprogramming. Int
J Biochem Cell Biol. 95:85–92. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
van Weverwijk A, Koundouros N, Iravani M,
Ashenden M, Gao Q, Poulogiannis G, Jungwirth U and Isacke CM:
Metabolic adaptability in metastatic breast cancer by
AKR1B10-dependent balancing of glycolysis and fatty acid oxidation.
Nat Commun. 10:26982019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Perco P, Ju W, Kerschbaum J, Leierer J,
Menon R, Zhu C, Kretzler M, Mayer G and Rudnicki M; Nephrotic
Syndrome Study Network (NEPTUNE), : Identification of dicarbonyl
and L-xylulose reductase as a therapeutic target in human chronic
kidney disease. JCI Insight. 4:e1281202019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu XH, Ding LY, Huang WX, Yang XM, Xie F,
Xu M and Yu L: (−)-Epigallocatechin-3-gallate, a potential
inhibitor to human dicarbonyl/L-xylulose reductase. J Biochem.
154:167–175. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang S, Jan YH, Mishin V, Heck DE, Laskin
DL and Laskin JD: Diacetyl/l-xylulose reductase mediates chemical
redox cycling in lung epithelial cells. Chem Res Toxicol.
30:1406–1418. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ebert B, Kisiela M and Maser E: Human
DCXR-another ‘moonlighting protein’ involved in sugar metabolism,
carbonyl detoxification, cell adhesion and male fertility? Biol Rev
Camb Philos Soc. 90:254–278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jeffery CJ: Moonlighting proteins: Old
proteins learning new tricks. Trends Genet. 19:415–417. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cho-Vega JH, Tsavachidis S, Do KA,
Nakagawa J, Medeiros LJ and McDonnell TJ: Dicarbonyl/L-xylulose
reductase: A potential biomarker identified by laser-capture
microdissection-micro serial analysis of gene expression of human
prostate adenocarcinoma. Cancer Epidemiol Biomarkers Prev.
16:2615–2622. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee SK, Son LT, Choi HJ and Ahnn J:
Dicarbonyl/l-xylulose reductase (DCXR): The multifunctional
pentosuria enzyme. Int J Biochem Cell Biol. 45:2563–2567. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cho-Vega JH, Vega F, Schwartz MR and
Prieto VG: Expression of dicarbonyl/L-xylulose reductase (DCXR) in
human skin and melanocytic lesions: morphological studies
supporting cell adhesion function of DCXR. J Cutan Pathol.
34:535–542. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hang X, Wu Z, Chu K, Yu G, Peng H, Xin H,
Miao X, Wang J and Xu W: Low expression of DCXR protein indicates a
poor prognosis for hepatocellular carcinoma patients. Tumour Biol.
37:15079–15085. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu J, Qin S, Yi Y, Gao H, Liu X, Ma F and
Guan M: Delving into the heterogeneity of different breast cancer
subtypes and the prognostic models utilizing scRNA-Seq and Bulk
RNA-Seq. Int J Mol Sci. 23:99362022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi B, Xue M, Wang Y, Wang Y, Li D, Zhao X
and Li X: An improved method for increasing the efficiency of gene
transfection and transduction. Int J Physiol Pathophysiol
Pharmacol. 10:95–104. 2013.PubMed/NCBI
|
|
27
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals, . Guide for the Care and Use of Laboratory Animals. 8th
ed. Washington (DC): National Academies Press (US); 2011,
PubMed/NCBI
|
|
28
|
Penault-Llorca F and Radosevic-Robin N:
Ki67 assessment in breast cancer: An update. Pathology. 49:166–171.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim YN, Kim SH, Son LT, Ahnn J and Lee SK:
Dicarbonyl/L-xylulose reductase (DCXR) producing xylitol regulates
egg retention through osmolality control in Caenorhabditis elegans.
Anim Cells Syst (Seoul). 26:223–231. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Moskovtsev SI, Jarvi K, Legare C, Sullivan
R and Mullen JB: Epididymal P34H protein deficiency in men
evaluated for infertility. Fertil Steril. 88:1455–1457. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim K, Zakharkin SO and Allison DB:
Expectations, validity, and reality in gene expression profiling. J
Clin Epidemiol. 63:950–959. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Icard P, Fournel L, Wu Z, Alifano M and
Lincet H: Interconnection between metabolism and cell cycle in
cancer. Trends Biochem Sci. 44:490–501. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Williams GH and Stoeber K: The cell cycle
and cancer. J Pathol. 226:352–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Salazar-Roa M and Malumbres M: Fueling the
cell division cycle. Trends Cell Biol. 27:69–81. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Abbaszadeh Z, Cesmeli S and Avci CB:
Crucial players in glycolysis: Cancer progress. Gene.
726:1441582020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li L, Liang Y, Kang L, Liu Y, Gao S, Chen
S, Li Y, You W, Dong Q, Hong T, et al: Transcriptional regulation
of the warburg effect in cancer by SIX1. Cancer Cell. 33:368–385.
e3672018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qiao L, Dong C and Ma B: UBE2T promotes
proliferation, invasion and glycolysis of breast cancer cells by
regualting the PI3K/AKT signaling pathway. J Recept Signal
Transduct Res. 42:151–159. 2021. View Article : Google Scholar : PubMed/NCBI
|