Introduction
According to the International Diabetes Federation,
there will be 592 million patients with diabetes worldwide by 2035
(1). Diabetic kidney disease (DKD)
is a complication that occurs in >20% of patients with diabetes
(2) and it is the main cause of
end-stage renal disease (ESRD) (3). The US Renal Data System 2020 Annual
Data Report demonstrated that the percentage of patients with ESRD
and diabetes mellitus increased form 58.3% in 2016 to a peak of
60.5% in 2019 before decreasing to 59.9% in 2020 (4). Furthermore, the increase in the
percentage of patients with chronic kidney disease (CKD) caused by
diabetes has shifted the spectrum of CKD in China toward its
pattern in developed countries. These findings suggested there may
be a rising incidence of diabetes-related ESRD in China (5). Tubulointerstitial fibrosis (TIF) is a
common pathway in chronic kidney diseases, which eventually leads
to ERSD. TIF is also a better predictor of kidney disease than
glomerulosclerosis (6). TIF is
characterized by deposition of the extracellular matrix, which is
mainly produced by myofibroblasts. Renal proximal tubule epithelial
cells, through epithelial-mesenchymal transition (EMT), are one of
the main sources of myofibroblasts. Mounting evidence has revealed
that interventions aimed at inhibiting EMT can significantly
prevent DKD-induced renal fibrosis (7–9).
As a signal transduction and transcription
activator, STAT is phosphorylated to form a dimer, which
translocates to the nucleus to induce the transcription of target
genes. Notably phosphorylated (p)-STAT is considered its active
form. Studies have shown that p-STAT1 and p-STAT3 are involved in
EMT and subsequent TIF under several conditions (10–13).
The role of STAT3 in EMT and fibrosis has been widely recognized
(14–18); however, other studies have
suggested that the activation of STAT1 exhibits anti-fibrotic
properties by reducing macrophage infiltration or changing the
phenotype of macrophages in renal ischemia-reperfusion injury
(19). Tumor-related studies have
also demonstrated that the activation of STAT1 mediated the effect
of interleukin-27 in reversing trophoblast cell EMT (20). In addition, STAT1 activation can
mediate the protective effect of IFN-γ on DKD glomerulosclerosis
(2,21,22).
STAT1 can be modified by small ubiquitin-related
modifier (SUMO), which affects its activity (23,24).
The human-derived SUMO family is comprised of four members: SUMO1,
SUMO2, SUMO3 and SUMO4, each with distinct distributions and
functions (25,26). However, the mechanism underlying
STAT1 SUMOylation in DKD remains unclear. Therefore, the present
study investigated the possible role of STAT1 activation and the
role of STAT1 SUMOylation in high glucose-induced tubular EMT.
Materials and methods
Materials
The HK-2 human renal proximal tubule epithelial cell
line was purchased from American Type Culture Collection. DMEM was
purchased from Gibco; Thermo Fisher Scientific, Inc. FBS was
purchased from ScienCell Research Laboratories, Inc. Mannitol was
purchased from Shanghai Aladdin Biochemical Technology Co., Ltd.
The CellTiter 96® AQueous One Solution Cell
Proliferation Assay was purchased from Promega Corporation. The BCA
Protein Assay Kit was obtained from Beijing Solarbio Science &
Technology Co., Ltd. Lipofectamine® 3000 was obtained
from Invitrogen; Thermo Fisher Scientific, Inc. RIPA lysis was
purchased from Shanghai BestBio. BSA and ECL system were obtained
from neoFroxx GmbH and Tiangen Biotech Co., Ltd., respectively.
Anti-SUMO2/3 (cat. no. ab3742), SUMO4 (cat. no. ab126606), UBC9
(cat. no. ab75854), p-STAT1 (cat. no. ab30645), vimentin (cat. no.
ab92547) and α-SMA (cat. no. ab124964) antibodies were purchased
from Abcam. Anti-E-cadherin (cat. no. 20874-1-AP), protein
inhibitor of activated STAT (PIAS)1 (cat. no. 23395-1-AP), PIAS3
(cat. no. 13486-1-AP), GFP (cat. no. 66002-1-lg) and anti-β-actin
(cat. no. 66009-1-lg) antibodies were purchased from Proteintech
Group, Ltd. Anti-SUMO1 antibody (cat. no. 4930) was purchased from
Cell Signaling Technology, Inc. Rabbit anti-STAT1 antibody (cat.
no. 10144-2-AP) was purchased from Proteintech Group, Ltd. and
mouse anti-STAT1 antibody (cat. no. sc-464) was purchased from
Santa Cruz Biotechnology, Inc. Anti-PIAS4 antibody (cat. no.
AF5329) was purchased from Affinity Biosciences, Ltd. Protein A/G
PLUS agarose (cat. no. sc-2003) and normal IgG antibodies (cat. no.
sc-2005) were obtained from Santa Cruz Biotechnology, Inc. The dual
luciferase reporter assay kit was purchased from Promega
Corporation. STAT1, PIAS4 and UBC9 small interfering RNAs (siRNAs)
and the negative control siRNA (cat. no. siN0000001-1-10) were
purchased from Guangzhou RiboBio Co., Ltd. The siRNA sequences were
as follows: UBC9 siRNA475, 5′-AGTGCGCTATCCCTGGAAA-3′, UBC9
siRNA552, 5′-GGCCAGCTATCACCATCAA-3′, UBC9 siRNA259,
5′-GGGAAGGAGGCTTGTTCAA-3′, STAT1 siRNA1, 5′-CATGCGGTTGAACCCTACA-3′,
STAT1 siRNA2, 5′-GCACGCTGCCAATGATGTT-3′, STAT1 siRNA3,
5′-CTGGATATATCAAGACTGA-3′, and PIAS4 siRNA,
5′-UUAUUGGAGGGGUAGUAGCCC-3′. The STAT1-wild type (STAT1-WT),
STAT1-K703R (STAT1-KR; SUMOylation site mutation), STAT1-Y701E
(STAT1-YE; sustained activation) plasmids and the plasmid vector
(pCMV3-C-GFPSpark) were obtained from Sino Biological, Inc. The
accession number used to design the plasmids is NM_007315.3.
STAT1-Luc plasmid was obtained from Genomeditech. The two-step
immunohistochemical detection reagent test kit (cat. no. PV-9001),
containing polymer helper and polyperoxidase-anti-rabbit IgG
antibody, was purchased from OriGene Technologies, Inc. The
fluorescent secondary antibody (cat. no. SA00013-4) and horseradish
peroxidase-conjugated secondary antibodies (cat. nos. SA00001-1 and
SA00001-2) were purchased from Proteintech Group, Ltd.
Cell culture and treatment
HK-2 cells were cultured in DMEM (normal glucose,
5.5 mmol/l) supplemented with 10% FBS at 37°C in a 5%
CO2 incubator. Cells were exposed to 30 mmol/l glucose
for 0, 12, 24 and 48 h at 37°C. Mannitol was used as an osmotic
control. Cells were treated with mannitol (30 mmol/l) in the same
manner as high glucose. An inverted fluorescence microscope was
used to observe cell morphology.
Cell viability assay
Cell viability was determined using the CellTiter 96
AQueous One Solution assay, according to the manufacturer's
protocol. Briefly, HK-2 cells were seeded in 96-well plates.
Following treatment with high glucose for 0, 24 and 48 h at 37°C,
cells were incubated with CellTiter 96 AQueous One Solution for 2 h
at 37°C. Finally, the absorbance of the reaction product was
measured at 490 nm using a microplate reader.
Cell transfection
When cells reached 70% confluence, they were
transfected using Lipofectamine® 3000 according to the
manufacturer's protocol. For siRNA transfection, Lipofectamine 3000
reagent and siRNAs (2.5 µg) were incubated for 15 min at room
temperature, and the mixture was then added to the cell medium. For
plasmid transfection, Lipofectamine 3000 reagent, P3000 reagent and
plasmids (3 µg) were incubated for 15 min. The empty plasmid was
used as a negative control. The mixture was then added to the cell
culture medium. After 8 h incubation with the mixture at 37°C, the
cells were cultured under normal condition for 40 h. The cells were
then treated with 30 mmol/l glucose for 12, 24 and 48 h at 37°C.
The transfection efficiency of plasmids and siRNAs was confirmed by
western blotting.
Western blotting
HK-2 cells were lysed with RIPA lysis buffer
containing 0.4% protease inhibitor and 1% phosphatase inhibitor for
40 min and were then quantified using a BCA assay kit. Protein
samples (30 µg per lane) were separated by SDS-PAGE on 10% gels and
were then transferred to polyvinylidene fluoride membranes.
Subsequently, the membranes were blocked with BSA for 2 h at 37°C
and incubated with primary antibodies (1:1,000) overnight at 4°C.
Thereafter, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibodies (1:5,000) for 2 h at
37°C. Protein samples were detected using an ECL system. β-actin
was used as an internal reference protein and relative protein
expression levels were semi-quantified using a Gel-Pro analyzer 4.0
(Media Cybernetics, Inc.).
Immunocytochemistry and
immunofluorescence
E-cadherin and α-SMA were detected by
immunocytochemistry and SUMOs were detected by immunofluorescence.
A sterile cover glass was placed on a six-well plate to culture the
cells. For immunocytochemistry, cells (~50% confluence) were fixed
with 4% paraformaldehyde for 30 min at room temperature. Following
permeabilization with 0.1% Triton X-100 for 30 min at 37°C, cells
were incubated with 3% H2O2 at 37°C for 30
min to remove endogenous peroxidase. Primary antibodies
(E-cadherin: 1:200, α-SMA: 1:250) were then added and incubated
overnight at 4°C. Subsequently, the cells were incubated with
polymer helper and polyperoxidase-anti-rabbit IgG antibody at 37°C
for 30 min. Finally, the cells were stained with diaminobenzidine.
A positive signal was observed using an Olympus light microscope
(Olympus Corporation). With the exception that a fluorescent
secondary antibody (1:200) was used at 37°C for 30 min and
observation was performed under a fluorescence microscope, the
immunofluorescence procedure was similar to the immunocytochemistry
procedure.
Co-immunoprecipitation (Co-IP)
HK-2 cells were lysed with RIPA lysis buffer
containing 0.4% protease inhibitor and 1% phosphatase inhibitor
mixture for 40 min and proteins were then quantified using a BCA
assay kit. The lysate (800 µl) was incubated with mouse STAT1
antibody (1:200) or IgG antibody (1:200) on a mixer for 24 h at 4°C
and then incubated with protein A/G beads (20 µl) on the mixer at
4°C overnight. Subsequently, the samples were washed four times
with IP washing buffer at 500 × g for 5 min at 4°C. After adding
bromophenol blue protein indicator buffer, the extracted protein
was boiled at 100°C for 7 min and the expression of SUMOs and PIASs
were analyzed using western blotting.
Dual luciferase reporter analysis
Lipofectamine 3000 was used for plasmid transfection
and the dual-luciferase reporter assay system was used for STAT1
activity detection. The STAT1-WT or STAT1-KR plasmids were
co-transfected into cells with the STAT1-Luc plasmid, and each
group was simultaneously transfected with the Renilla
luciferase plasmid to eliminate the difference caused by different
efficiencies. A total of 48 h after transfection, glucose (30
mmol/l) was added to the cells for 24 h. Subsequently, the dual
luciferase reporter assay system was used to detect STAT1 activity.
Briefly, the cells were lysed with PLB lysis buffer at room
temperature. First, the LARII reagent was added to the lysate to
detect firefly luciferase activity, and the Stop & Glo reagent
was then added to detect the Renilla luciferase activity.
STAT1 activity was expressed as the ratio of firefly luciferase
activity to Renilla luciferase activity.
Statistical analysis
Each experiment was repeated at least three times.
Statistical analyses were conducted using SPSS 19.0 statistical
software (IBM Corporation). The measured data are expressed as the
mean ± standard deviation. Normally distributed quantitative data
were compared between two groups using independent-samples
Student's t-test and data among multiple groups were compared using
one-way ANOVA (with LSD or Tukey's test) or two-way ANOVA (with
Bonferroni test). P<0.05 was considered to indicate a
statistically significant difference.
Results
Effects of high glucose on cell
viability and the phenotypic transition of HK-2 cells
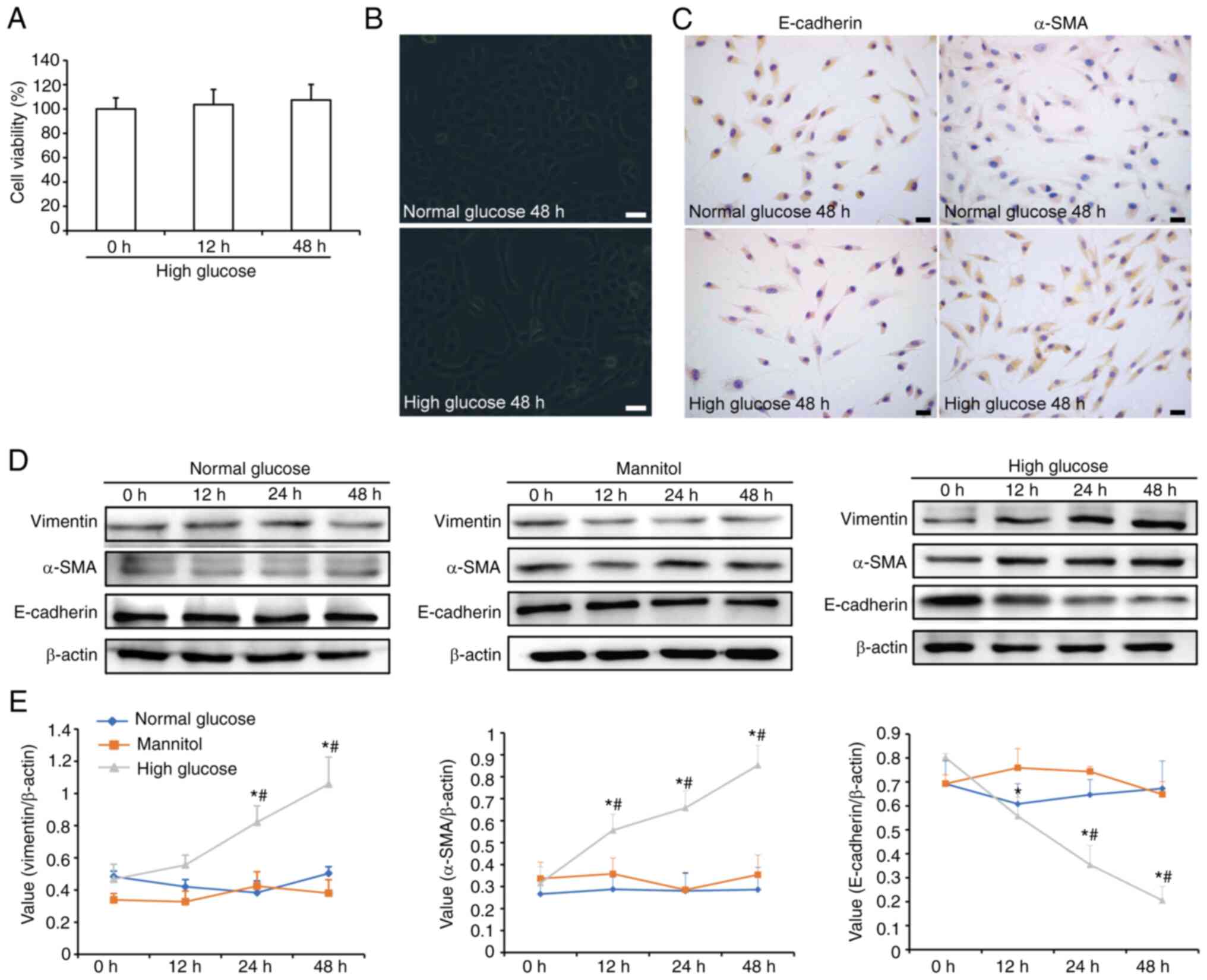
The CellTiter 96 AQueous One Solution assay was used
to determine the viability of HK-2 cells. The results revealed that
high glucose did not affect cell viability (Fig. 1A). Under an inverted fluorescence
microscope, most HK-2 cells were arranged in a typical
cobblestone-like pattern when cultured with normal glucose, whereas
the cells were elongated and exhibited a spindle-shaped morphology
after 48 h of high-glucose treatment (Fig. 1B). Immunocytochemical analysis
showed weak expression of E-cadherin and strong expression of α-SMA
in the cytoplasm of HK-2 cells following high glucose treatment for
48 h (Fig. 1C). In addition,
western blotting was used to detect the phenotypic transition of
HK-2 cells (Fig. 1D and E). Among
the three treatment groups, only high glucose increased the
expression of vimentin and α-SMA, and decreased E-cadherin
expression in a time-dependent manner. Compared with in the normal
glucose treatment group, the expression levels of vimentin were
significantly increased when cells were stimulated with high
glucose for 24 h. In addition, α-SMA expression was significantly
increased after 12 h of high-glucose stimulation. By contrast, the
expression levels of E-cadherin were significantly downregulated
after 12 h of high-glucose exposure and gradually decreased. These
results indicated that high glucose may promote the EMT of HK-2
cells.
Effects of high glucose on SUMO
expression in HK-2 cells
To investigate the effects of high glucose on the
expression levels of SUMOs in HK-2 cells, immunofluorescence
analysis (Fig. 2A) and western
blotting (Fig. 2B and C) were
performed. SUMOs were primarily expressed in the nucleus. Among the
three treatment groups, only high glucose increased the expression
levels of SUMO1 and SUMO4. As for SUMO2/3, although there was no
marked change observed by immunocytochemistry, the western blotting
results indicated that high glucose elevated the expression levels
of SUMO2/3. Compared with in cells treated with normal glucose, the
relative expression levels of SUMO1 and SUMO4 peaked 12 h after
high-glucose treatment, whereas the relative expression levels of
SUMO2/3 peaked at 24 h and then decreased.
Effects of STAT1 activation on the
phenotypic transition in HK-2 cells
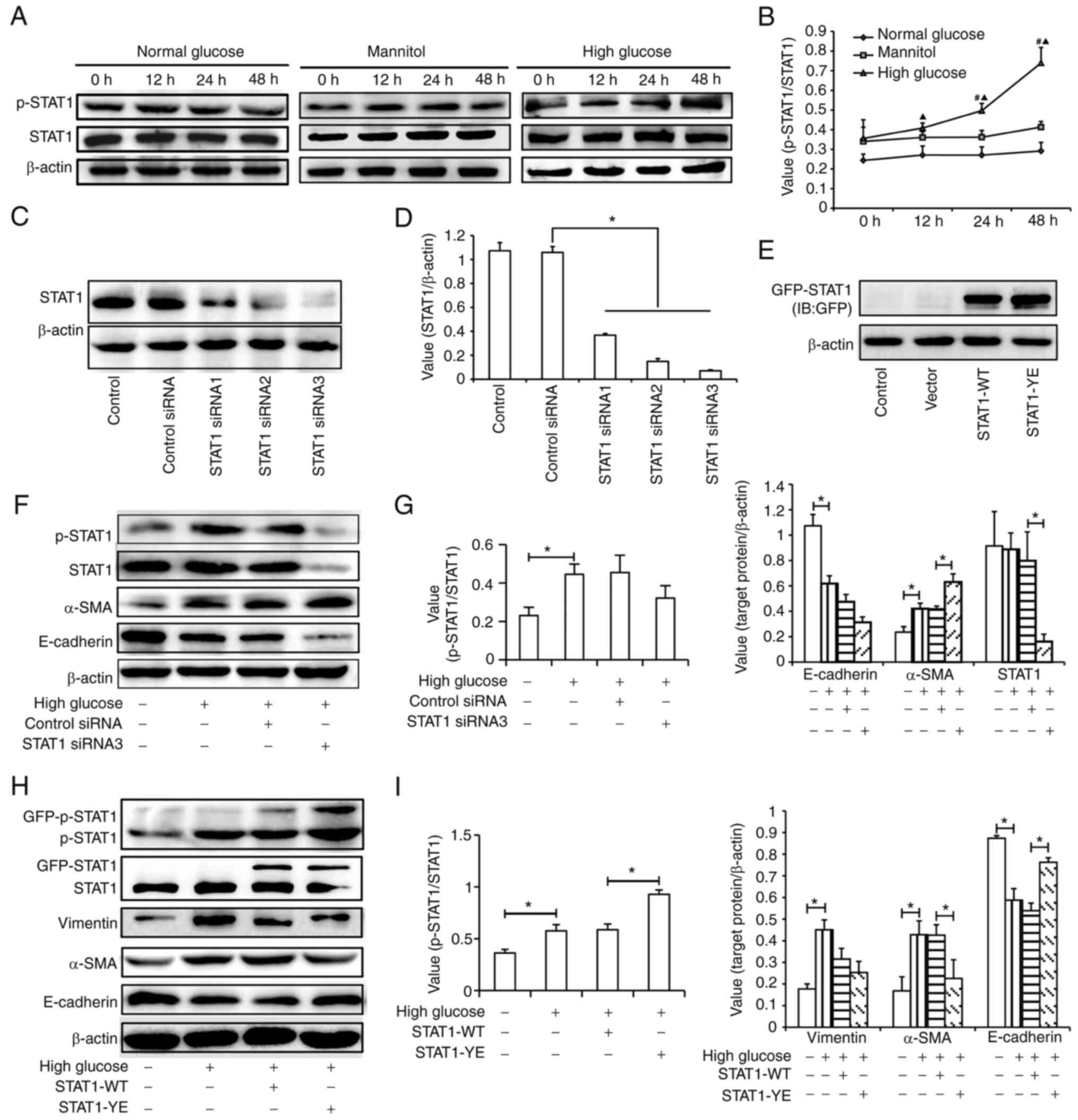
The present study observed the effect of high
glucose on the phosphorylation of STAT1 (Fig. 3A and B). The results suggested that
high glucose, but not normal glucose or mannitol, increased the
relative expression levels of p-STAT1 (Tyr701) in a time-dependent
manner.
To determine the role of STAT1 activation in tubular
EMT induced by high glucose, STAT1 siRNA or STAT1-YE plasmids were
transfected into the HK-2 cells. The transfection efficiency of
siRNA and plasmids are presented in Fig. 3C-E. All three siRNAs inhibited
STAT1 expression and STAT1 siRNA3, which had the best inhibitory
effect, was selected for subsequent experiments (Fig. 3C and D). The fusion expression of
GPF and STAT1 resulted in the molecular weight of exogenous STAT1
being higher than that of endogenous STAT1. The efficiency of
transfection with the STAT1 plasmid was confirmed by detecting the
expression of GFP-STAT1 (Fig. 3E).
As shown in Fig. 3F and G, STAT1
siRNA transfection did not significantly affect the level of
p-STAT1/STAT1 compared with control siRNA transfection; however,
STAT1 siRNA transfection aggravated the effect of high glucose on
the expression level of α-SMA. This may be due to the decreased
level of STAT1. In order to verify that p-STAT1 affected high
glucose-induced EMT, STAT1-YE plasmid was transfected into cells.
The results showed that the expression levels of p-STAT1 were
higher in the STAT1-YE group compared with those in the STAT1-WT
group. The upregulation of STAT1 phosphorylation in the STAT1-YE
group may alleviate the high glucose-induced changes in the
expression levels of E-cadherin and α-SMA (Fig. 3H and 3I). These results suggested
that STAT1 activation may inhibit EMT in HK-2 cells.
SUMOylation of STAT1 in HK-2
cells
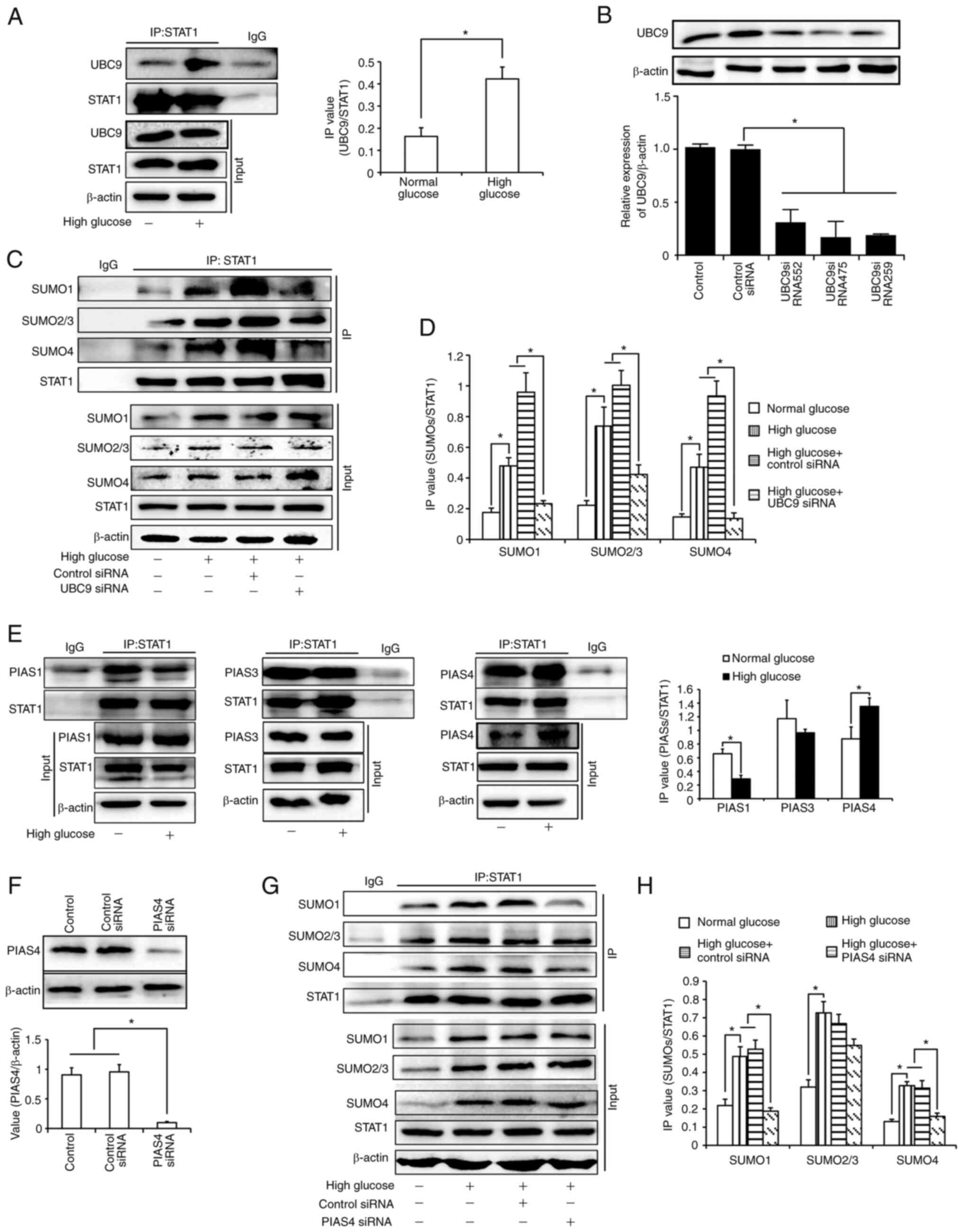
It has been reported that STAT1 can be modified
using SUMOs. As UBC9 is the only E2 enzyme involved in SUMOylation,
the present study first verified the conjunction of STAT1 and UBC9.
As shown in Fig. 4A, there was
enhanced conjunction of STAT1 and UBC9 under high glucose
conditions. Subsequently, the binding of STAT1 and SUMOs in UBC9
siRNA-transfected cells was detected. All three siRNAs inhibited
UBC9 expression (Fig. 4B);
UBC9-si475 exhibited the best inhibitory effect and was selected
for subsequent experiments. As shown in Fig. 4C and D, high glucose levels
upregulated the binding of STAT1 with all three SUMOs, which could
be inhibited by UBC9 knockdown. This indicated that STAT1 could be
modified by SUMOs in HK-2 cells. SUMOylation of STAT1 was enhanced
under high glucose conditions. Subsequently, it was confirmed by
co-IP experiments that the binding of PIAS4, instead of PIAS1 or
PIAS3, to STAT1 was increased under high glucose conditions. These
findings suggested that PIAS4 may be the E3 ligase which mediated
the upregulation of STAT1 SUMOylation (Fig. 4E). Subsequently, HK-2 cells were
transfected with PIAS4 siRNA (Fig.
4F). The results revealed that the binding of SUMO1 and SUMO4
with STAT1 was inhibited after PIAS4 was knocked down (Fig 4G and H). The slight bands seen in
Fig. 4A and E may be caused by the
light or heavy chains of IgG, respectively. In addition, the slight
bands seen in Fig. 4G may be due
to insufficient cleaning in the experiments.
Effects of STAT1 SUMOylation on its
activity
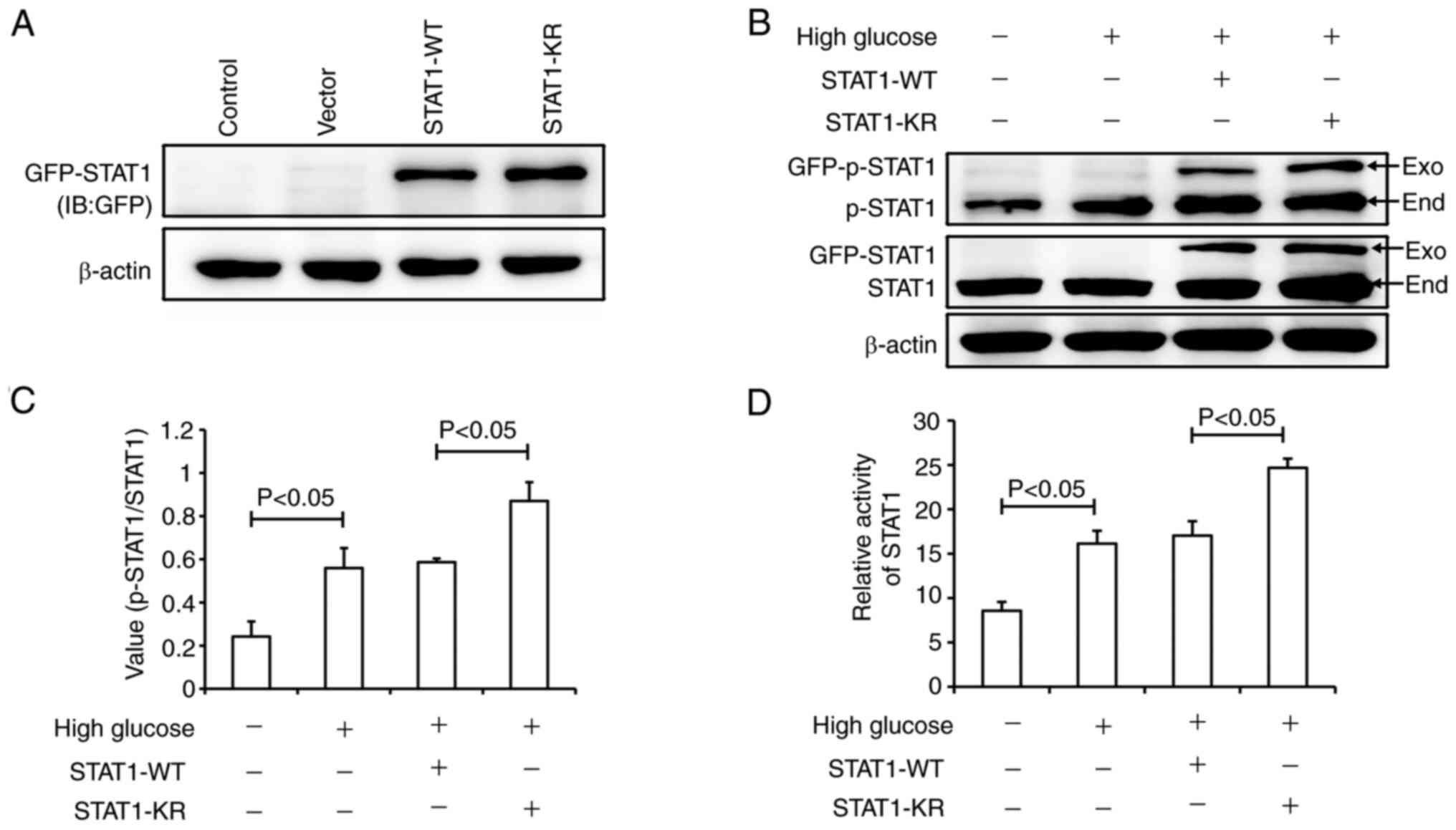
In order to verify the effect of STAT1 SUMOylation
on its activity, the STAT1-KR plasmid was generated. In STAT1-KR,
Lys 703 was mutated to Arg; this mutation can inhibit the
SUMOylation of STAT1 at Lys 703. It has previously been suggested
that the SUMOylation of STAT1 at Lys 703 affects the
phosphorylation of STAT1 at Tyr701 (27). Fig.
5A shows the expression of GFP-STAT1, which confirmed the
transfection efficiency of the STAT1 plasmids. As shown in Fig. 5B, high glucose induced the
phosphorylation of exogenous STAT1 both in STAT1-WT and STAT1-KR
groups. Compared with the STAT1-WT plasmid group, the level of
total p-STAT1 was higher in the STAT1-KR plasmid group (Fig. 5B and C). Dual-luciferase reporter
gene analysis suggested that high glucose significantly increased
STAT1 activity; however, STAT1 was more active in the STAT1-KR
plasmid group than that in the STAT1-WT plasmid group (Fig. 5D). These results suggested that
STAT1-KR transfection increased p-STAT1 expression and STAT1
activity.
Role of STAT1 SUMOylation in the
phenotypic transition of HK-2 cells
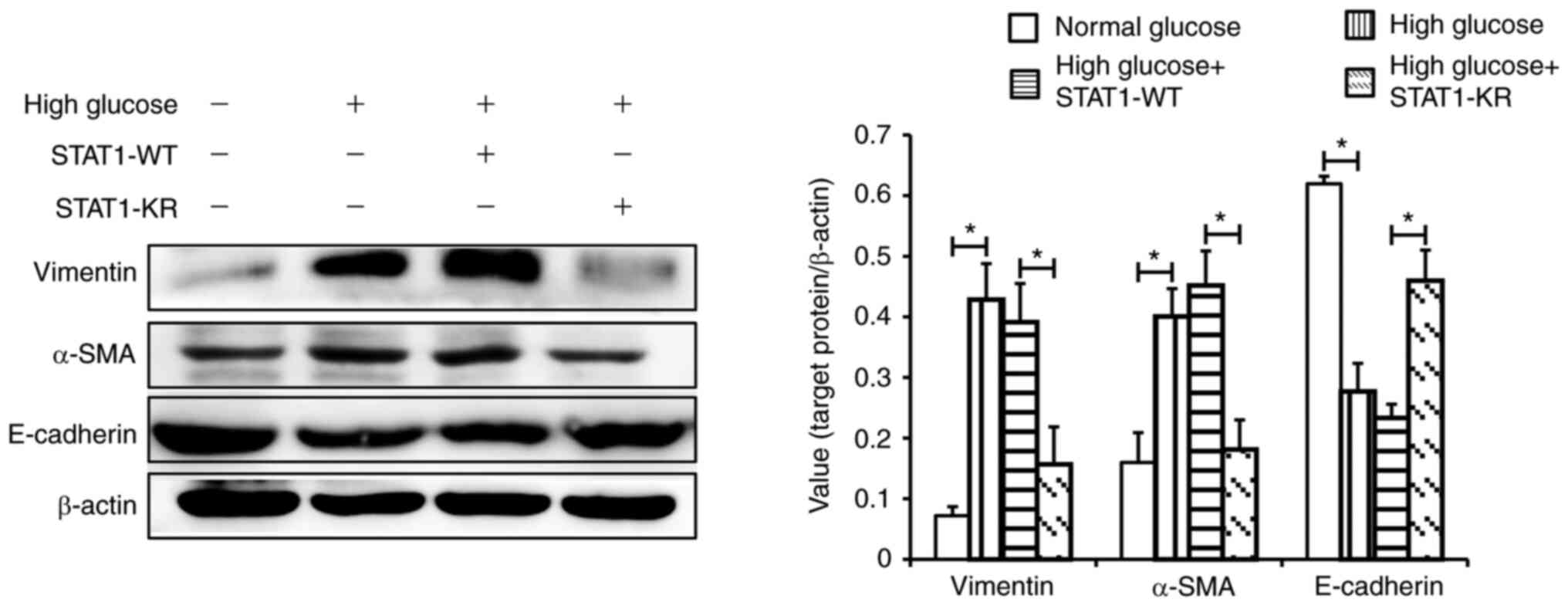
As shown in Fig. 6,
under high glucose, E-cadherin expression was higher, and α-SMA and
vimentin expression was lower in the STAT1-KR plasmid group
compared with that in the STAT1-WT plasmid group. These results
suggested that activated STAT1 may suppress the high
glucose-induced phenotypic transition of HK-2 cells.
Discussion
DKD is one of the most common microvascular
complications associated with diabetes. The major pathological
changes in DKD include glomerulosclerosis and TIF. Although
glomerulopathy was originally considered the main pathological
change in DKD, there is increasing evidence that TIF is more
closely related to renal function. TIF can be detected in the early
stages of DKD and directly contributes to decreased renal function,
independent of glomerular dysfunction (10,28).
Myofibroblasts are the main cells that produce the extracellular
matrix, leading to TIF, and EMT in renal tubular epithelial cells
is a key source of myofibroblasts (29,30).
EMT is characterized by the loss of epithelial adhesion molecules
and the expression of myofibroblast markers in the renal tubular
epithelial cells (31,32). In the present study, HK-2 cells
were stimulated with 30 mmol/l glucose. The subsequent decrease in
E-cadherin, and the increase in α-SMA and vimentin confirmed that
high glucose induced tubular EMT.
Given the uncertainty regarding the role of STAT1
activation in EMT and fibrosis, STAT1 siRNA was transfected into
HK-2 cells to explore the possible role of STAT1 in tubular EMT.
The results revealed that STAT1 knockdown aggravated the effects of
high glucose on the expression levels of α-SMA in HK-2 cells. As
unphosphorylated STAT1 also induces the expression of some genes
(33), and STAT1 siRNA reduced the
expression levels of STAT1 rather than p-STAT1/STAT1, it may be
suggested that the effect of STAT1 siRNA transfection on the
expression of α-SMA could be due to the decreased levels of STAT1.
The present study transfected the cells with the STAT1-YE plasmid
to verify the role of STAT1 activation in tubular EMT. The levels
of p-STAT1 were higher in the STAT1-YE group than in the STAT1-WT
group and the increased STAT1 phosphorylation exhibited an
inhibitory effect on EMT. Therefore, it was suggested that
activated STAT1 may play a protective role in EMT. Similar results
demonstrated a protective effect of STAT1 against renal fibrosis.
For example, in a previous study, STAT1−/− mice
exhibited higher amounts of α-SMA, glomerular density, and Masson's
trichrome, silver, and collagen staining (19). IFN-γ can activate STAT1 to suppress
the overexpression of TGF-β, collagen IV and connective tissue
growth factor in diabetic mice and in high glucose-stimulated
mesangial cells (22). In
addition, p-STAT1 mediates the inhibitory role of IL-27 in EMT of
trophoblast cells (20). These
results suggest a protective role for STAT1 in EMT or fibrosis.
The activity of STAT1 is regulated by several
factors, and SUMOylation can affect its phosphorylation (24). STAT1 harbors a functional
SUMOylation consensus sequence, ΨKXE, where Ψ represents a large
hydrophobic residue, K represents lysine, X represents any amino
acid and E represents glutamate (34). The present study demonstrated that
STAT1 could be modified by SUMOs in HK-2 cells and that SUMOylation
of STAT1 was enhanced under high-glucose conditions. To identify
the effect and mechanism of STAT1 SUMOylation on high
glucose-induced EMT, STAT1-WT and STAT1-KR plasmids were
transfected into HK-2 cells. The results revealed that high glucose
weakly induced E-cadherin, vimentin and α-SMA expression in
STAT1-KR plasmid-transfected cells compared with in STAT1-WT
plasmid-transfected cells. In other words, inhibition of STAT1
SUMOylation reduced EMT. In addition, the p-STAT1 expression levels
in cells transfected with STAT1-KR plasmids were higher than those
in the STAT1-WT plasmid-transfected cells. The results of
dual-luciferase reporter gene analysis showed that the
transcriptional activity of STAT1 was significantly upregulated in
cells transfected with STAT1-KR plasmid compared with in those
transfected with STAT1-WT plasmid. Although p-STAT1 has been
reported to perform specific functions in the cytoplasm (35), the present results suggested that
the upregulation of STAT1 SUMOylation may affect tubular EMT by
inhibiting the activity of STAT1 under high-glucose conditions.
In conclusion, the upregulation of STAT1 SUMOylation
caused by high glucose levels may explain why STAT1 could not be
effectively activated to exert its protective effect. However, how
SUMOylation affects the activity of STAT1 and the genes that
mediate the effect of STAT1 activity on EMT requires further
study.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science
Foundation (grant nos. 81600564, 81800623 and 21605035), the Hebei
Natural Science Foundation (grant nos. H2020206262 and
H2019206045), and the Department of Health of Hebei Province (grant
no. 20180044).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QL and SL are responsible for conceiving and
designing the research. CG, LK, SZ, JL and YX are responsible for
performing the experiments. FG, WZ and QW are responsible for
analyzing data. XF, YD and YT are responsible for interpreting the
results of experiments and editing manuscript QL and CG confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Shi Y and Hu FB: The global implications
of diabetes and cancer. Lancet. 383:1947–1948. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Murphy D, McCulloch CE, Lin F, Banerjee T,
Bragg-Gresham JL, Eberhardt MS, Morgenstern H, Pavkov ME, Saran R,
Powe NR, et al: Trends in prevalence of chronic kidney disease in
the United States. Ann Intern Med. 165:473–481. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rossing P: Diabetic nephropathy: Worldwide
epidemic and effects of current treatment on natural history. Curr
Diab Rep. 6:479–483. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johansen KL, Chertow GM, Foley RN,
Gilbertson DT, Herzog CA, Ishani A, Israni AK, Ku E, Tamura MK, Li
S, et al: US renal data system 2020 annual data report:
Epidemiology of kidney disease in the United States. Am J kidney
Dis. 77:A7–A8. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang C, Wang H, Zhao X, Matsushita K,
Coresh J, Zhang L and Zhao MH: CKD in China: Evolving spectrum and
public health implications. Am J Kidney Dis. 76:258–264. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nauta FL, Boertien WE, Bakker SJ, van Goor
H, van Oeveren W, de Jong PE, Bilo H and Gansevoort RT: Glomerular
and tubular damage markers are elevated in patients with diabetes.
Diabetes Care. 34:975–981. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du L, Qian X, Li Y, Li XZ, He LL, Xu L,
Liu YQ, Li CC, Ma P, Shu FL, et al: Sirt1 inhibits renal tubular
cell epithelial-mesenchymal transition through YY1 deacetylation in
diabetic nephropathy. Acta Pharmacol Sin. 42:242–251. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang T, Shu F, Yang H, Heng C, Zhou Y,
Chen Y, Qian X, Du L, Zhu X, Lu Q and Yin X: YY1: A novel
therapeutic target for diabetic nephropathy orchestrated renal
fibrosis. Metabolism. 96:33–45. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang T, Heng C, Zhou Y, Hu Y, Chen S, Wang
H, Yang H, Jiang Z, Qian S, Wang Y, et al: Targeting mammalian
serine/threonine-protein kinase 4 through Yes-associated
protein/TEA domain transcription factor-mediated
epithelial-mesenchymal transition ameliorates diabetic nephropathy
orchestrated renal fibrosis. Metabolism. 108:1542582020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang F, Zhao Y, Wang Q, Hillebrands JL,
van den Born J, Ji L, An T and Qin G: Dapagliflozin attenuates
renal tubulointerstitial fibrosis associated with type 1 diabetes
by regulating STAT1/TGFβ1 signaling. Front Endocrinol (Lausanne).
10:4412019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Zhou H, Li Y, Han L, Song M, Chen F,
Shang G, Wang D, Wang Z, Zhang W and Zhong M: PTPN2 improved renal
injury and fibrosis by suppressing STAT-induced inflammation in
early diabetic nephropathy. J Cell Mol Med. 23:4179–4195. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luan J, Fu J, Wang D, Jiao C, Cui X, Chen
C, Liu D, Zhang Y, Wang Y, Yuen PST, et al: MiR-150-based RNA
interference attenuates tubulointerstitial fibrosis through the
SOCS1/JAK/STAT pathway in vivo and in vitro. Mol Ther Nucleic
Acids. 22:871–884. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao SQ, Shen ZC, Gao BF and Han P:
microRNA-206 overexpression inhibits epithelial-mesenchymal
transition and glomerulosclerosis in rats with chronic kidney
disease by inhibiting JAK/STAT signaling pathway. J Cell Biochem.
120:14604–14617. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Lu H, Xie S, Wu C, Guo Y, Xiao Y,
Zheng S, Zhu H, Zhang Y and Bai Y: Resveratrol suppresses the
myofibroblastic phenotype and fibrosis formation in kidneys via
proliferation-related signalling pathways. Br J Pharmacol.
176:4745–4759. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Zhang F, Qu L, Xie Y, Ruan Y, Guo Z,
Mao Y, Zou Q, Shi M, Xiao Y, et al: Identification of YAP1 as a
novel downstream effector of the FGF2/STAT3 pathway in the
pathogenesis of renal tubulointerstitial fibrosis. J Cell Physiol.
236:7655–7671. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu J, Zhong Y, Liu G, Zhang X, Xiao B,
Huang S, Liu H and He L: Role of Stat3 signaling in control of EMT
of tubular epithelial cells during renal fibrosis. Cell Physiol
Biochem. 42:2552–2558. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Zou J, Tolbert E, Zhao TC,
Bayliss G and Zhuang S: Identification of histone deacetylase 8 as
a novel therapeutic target for renal fibrosis. FASEB J.
34:7295–7310. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi Y, Tao M, Ma X, Hu Y, Huang G, Qiu A,
Zhuang S and Liu N: Delayed treatment with an autophagy inhibitor
3-MA alleviates the progression of hyperuricemic nephropathy. Cell
Death Dis. 11:4672020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kemmner S, Bachmann Q, Steiger S, Lorenz
G, Honarpisheh M, Foresto-Neto O, Wang S, Carbajo-Lozoya J, Alt V,
Schulte C, et al: STAT1 regulates macrophage number and phenotype
and prevents renal fibrosis after ischemia-reperfusion injury. Am J
Renal Physiol. 316:F277–F291. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ge H, Yin N, Han TL, Huang D, Chen X, Xu
P, He C, Tong C and Qi H: Interleukin-27 inhibits trophoblast cell
invasion and migration by affecting the epithelial-mesenchymal
transition in preeclampsia. Reprod Sci. 26:928–938. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Du J, Dong W, Li H, Li B, Liu X, Kong Q,
Sun W, Sun T, Ma P, Cui Y and Kang P: Protective effects of IFN-γ
on the kidney of type-2 diabetic KKAy mice. Pharmacol Rep.
70:607–613. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du J, Wang L, Liu L, Fan Q, Yao L, Cui Y,
Kang P, Zhao H, Feng X and Gao H: IFN-γ suppresses the high
glucose-induced increase in TGF-β1 and CTGF synthesis in mesangial
cells. Pharmacol Rep. 63:1137–1144. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maarifi G, Maroui MA, Dutrieux J, Dianoux
L, Nisole S and Chelbi-Alix MK: Small ubiquitin-like modifier
alters IFN response. J Immunol. 195:2312–2324. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sampaio EP, Ding L, Rose SR, Cruz P, Hsu
AP, Kashyap A, Rosen LB, Smelkinson M, Tavella TA, Ferre EMN, et
al: Novel signal transducer and activator of transcription 1
mutation disrupts small ubiquitin-related modifier conjugation
causing gain of function. J Allergy Clin Immunol. 141:1844–1853.
e18422018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Johnson ES: Protein modification by SUMO.
Annu Rev Biochem. 73:355–382. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chang HM and Yeh ETH: SUMO: From bench to
bedside. Physiol Rev. 100:1599–1619. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Begitt A, Droescher M, Knobeloch KP and
Vinkemeier U: SUMO conjugation of STAT1 protects cells from
hyperresponsiveness to IFNγ. Blood. 118:1002–1007. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma Y, Yan R, Wan Q, Lv B, Yang Y, Lv T and
Xin W: Inhibitor of growth 2 regulates the high glucose-induced
cell cycle arrest and epithelial-to-mesenchymal transition in renal
proximal tubular cells. J Physiol Biochem. 76:373–382. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sun YB, Qu X, Caruana G and Li J: The
origin of renal fibroblasts/myofibroblasts and the signals that
trigger fibrosis. Differentiation. 92:102–107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang B, Ding W, Zhang M, Li H and Gu Y:
Rapamycin attenuates aldosterone induced tubulointerstitial
inflammation and fibrosis. Cell Physiol Biochem. 35:116–125. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hu J, Zhu Q, Li PL, Wang W, Yi F and Li N:
Stem cell conditioned culture media attenuated albumin-induced
epithelial-mesenchymal transition in renal tubular cells. Cell
Physion Biochem. 35:1719–1728. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Quaggin SE and Kapus A: Scar wars: Mapping
the fate of epithelial-mesenchymal-myofibroblast transition. Kidney
Int. 80:41–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chatterjee-Kishore M, Wright KL, Ting JP
and Stark GR: How Stat1 mediates constitutive gene expression: A
complex of unphosphorylated Stat1 and IRF1 supports transcription
of the LMP2 gene. EMBO J. 19:4111–4122. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rodriguez MS, Dargemont C and Hay RT:
SUMO-1 conjugation in vivo requires both a consensus modification
motif and nuclear targeting. J Biol Chem. 276:12654–12659. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang Y, Yu M, Hu X, Han L, Yang K, Ba H,
Zhang Z, Yin B, Yang XP, Li Z and Wang J: STAT1 mediates
transmembrane TNF-alpha-induced formation of death-inducing
signaling complex and apoptotic signaling via TNFR1. Cell Death
Differ. 24:660–671. 2017. View Article : Google Scholar : PubMed/NCBI
|