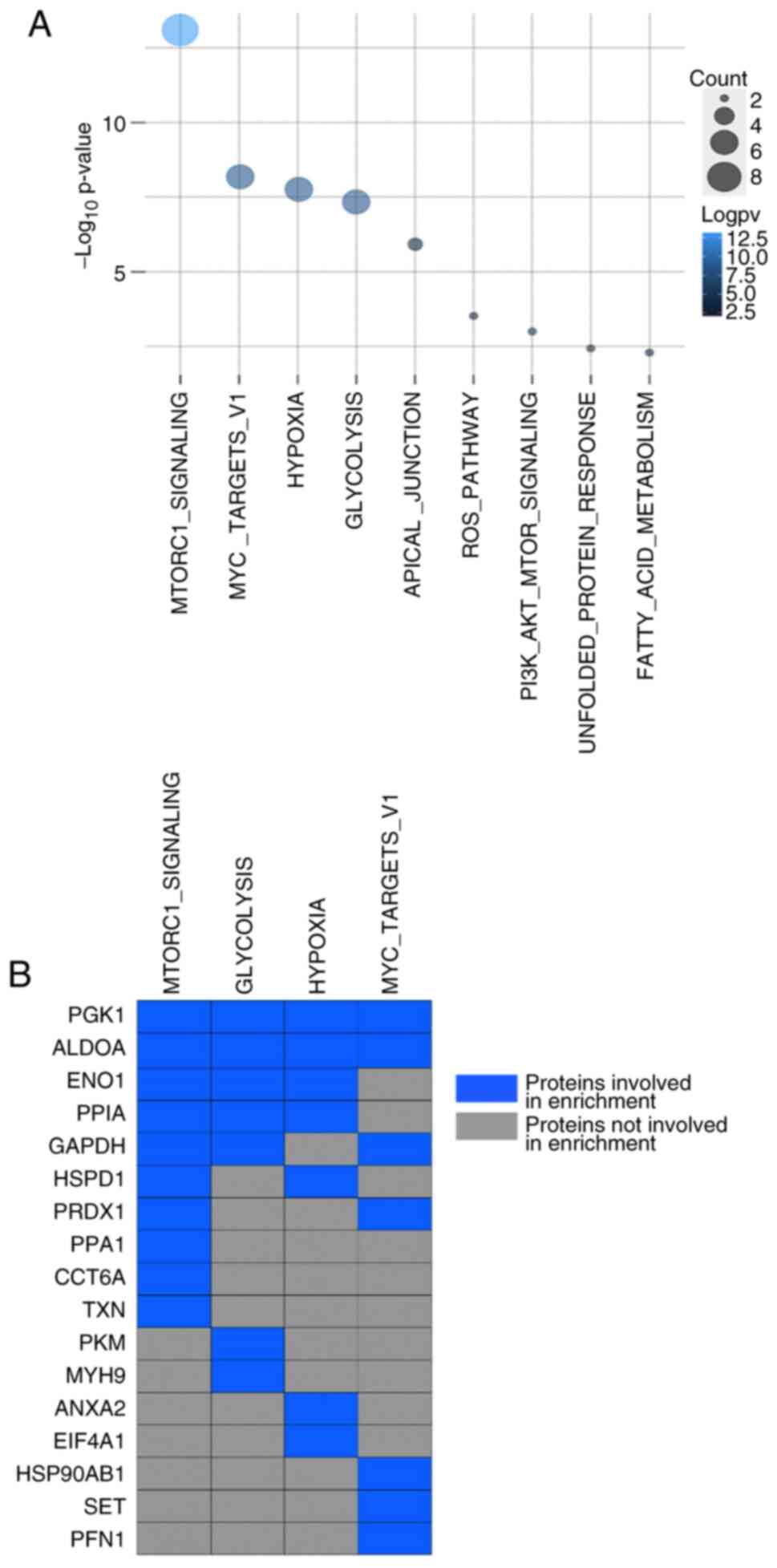
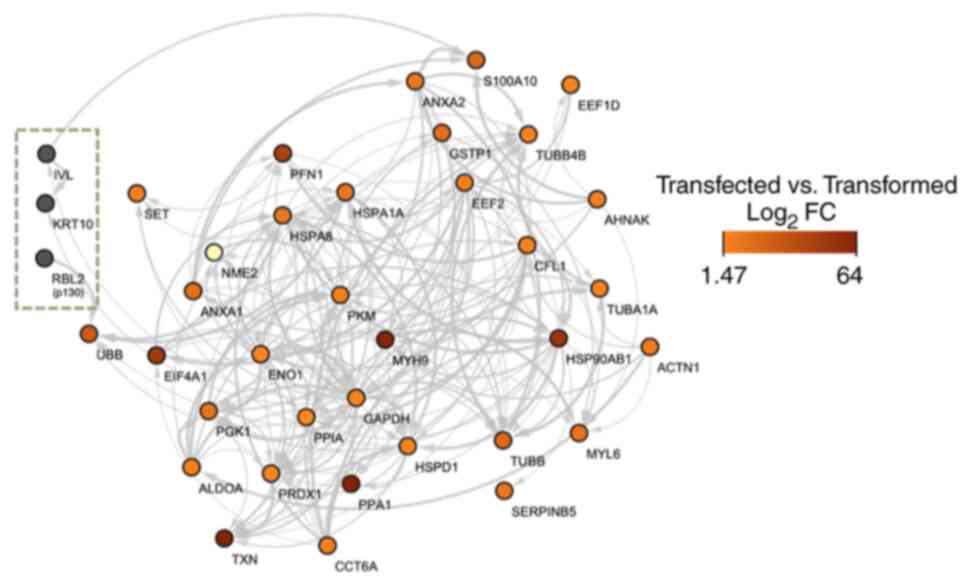
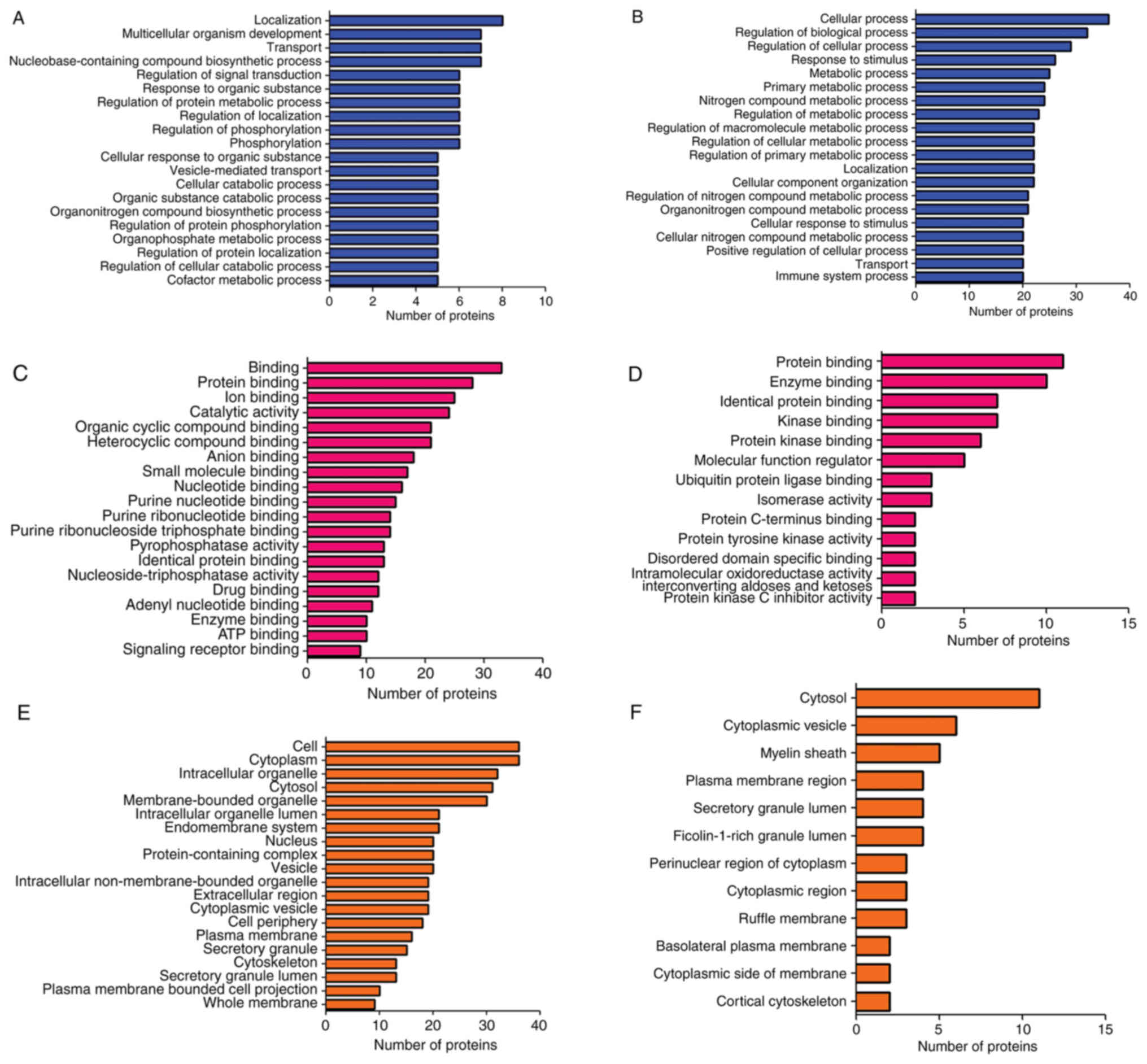
|
1
|
Berman TA and Schiller JT: Human
papillomavirus in cervical cancer and oropharyngeal cancer: One
cause, two diseases. Cancer. 123:2219–2229. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sand FL, Munk C, Jensen SM, Svahn MF,
Frederiksen K and Kjaer SK: Long-Term risk for noncervical
anogenital cancer in women with previously diagnosed high-grade
cervical intraepithelial neoplasia: A danish nationwide cohort
study. Cancer Epidemiol Biomarkers Prev. 25:1090–1097. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fuller KM and Hinyard L: Factors
associated with HPV vaccination in young males. J Community Health.
42:1127–1132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haverkos HW, Haverkos GP and O'Mara M:
Co-carcinogenesis: Human papillomaviruses, coal tar derivatives,
and squamous cell cervical cancer. Front Microbiol. 8:22532017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Muñoz N, Bosch FX, De Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ;
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group, : Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Della Fera AN, Warburton A, Coursey TL,
Khurana S and McBride AA: Persistent human papillomavirus
infection. Viruses. 13:3212021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Songock WK, Kim SM and Bodily JM: The
human papillomavirus E7 oncoprotein as a regulator of
transcription. Virus Res. 231:56–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Graham SV and Faizo AAA: Control of human
papillomavirus gene expression by alternative splicing. Virus Res.
231:83–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong HS, Akhavan J, Lee SH, Kim RH, Kang
MK, Park NH and Shin KH: Proinflammatory cytokine TNFα promotes
HPV-associated oral carcinogenesis by increasing cancer stemness.
Int J Oral Sci. 12:32020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Richards KH, Wasson CW, Watherston O,
Doble R, Eric Blair G, Wittmann M and Macdonald A: The human
papillomavirus (HPV) E7 protein antagonises an Imiquimod-induced
inflammatory pathway in primary human keratinocytes. Sci Rep.
5:129222015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Freitas AC, de Oliveira THA, Barros MR
Jr and Venuti A: hrHPV E5 oncoprotein: Immune evasion and related
immunotherapies. J Exp Clin Cancer Res. 36:712017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yeo-The NSL, Ito Y and Jha S: High-risk
human papillomaviral oncogenes E6 and E7 target key cellular
pathways to achieve oncogenesis. Int J Mol Sci. 19:17062018.
View Article : Google Scholar
|
|
13
|
Fischer M, Uxa S, Stanko C, Magin TM and
Engeland K: Human papilloma virus E7 oncoprotein abrogates the
p53-p21-DREAM pathway. Sci Rep. 7:26032017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bienkowska-Haba M, Luszczek W, Zwolinska
K, Scott RS and Sapp M: Genome-Wide transcriptome analysis of human
papillomavirus 16-infected primary keratinocytes reveals subtle
perturbations mostly due to E7 protein expression. J Virol.
94:e01360–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pal A and Kundu R: Human papillomavirus E6
and E7: The cervical cancer hallmarks and targets for therapy.
Front Microbiol. 10:31162020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cuninghame S, Jackson R and Zehbe I:
Hypoxia-inducible factor 1 and its role in viral carcinogenesis.
Virology. 456–457. 370–383. 2014.PubMed/NCBI
|
|
17
|
Gunasekharan VK, Li Y, Andrade J and
Laimins LA: Post-Transcriptional regulation of KLF4 by high-risk
human papillomaviruses is necessary for the
differentiation-dependent viral life cycle. PLoS Pathog.
12:e10057472016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sen P, Ganguly P and Ganguly N: Modulation
of DNA methylation by human papillomavirus E6 and E7 oncoproteins
in cervical cancer. Oncol Lett. 15:11–22. 2018.PubMed/NCBI
|
|
19
|
Gandhi S, Nor Rashid N, Mohamad Razif MF
and Othman S: Proteasomal degradation of p130 facilitate cell cycle
deregulation and impairment of cellular differentiation in
high-risk Human Papillomavirus 16 and 18 E7 transfected cells. Mol
Biol Rep. 48:5121–5133. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Olson BJSC: Assays for determination of
protein concentration. Curr Protoc Pharmacol. 73:A.3A.1–A.3A.32.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan CH, Tan KY and Tan NH: Revisiting
Notechis scutatus venom: On shotgun proteomics and neutralization
by the ‘bivalent’ Sea Snake Antivenom. J Proteomics. 144:33–38.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zainal Abidin SA, Rajadurai P, Chowdhury
ME, Ahmad Rusmili MR, Othman I and Naidu R: Proteomic
characterization and comparison of malaysian tropidolaemus wagleri
and cryptelytrops purpureomaculatus venom using shotgun-proteomics.
Toxins (Basel). 8:2992016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Xin L, Shan B, Chen W, Xie M,
Yuen D, Zhang W, Zhang Z, Lajoie GA and Ma B: PEAKS DB: de novo
sequencing assisted database search for sensitive and accurate
peptide identification. Mol Cell Proteomics. 11:M111.010587. 2012.
View Article : Google Scholar
|
|
25
|
Ma B, Zhang K, Hendrie C, Liang C, Li M,
Doherty-Kirby A and Lajoie G: PEAKS: Powerful software for peptide
de novo sequencing by tandem mass spectrometry. Rapid Commun Mass
Spectrom. 17:2337–2342. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Levin Y, Schwarz E, Wang L, Leweke FM and
Bahn S: Label-free LC-MS/MS quantitative proteomics for large-scale
biomarker discovery in complex samples. J Sep Sci. 30:2198–2203.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Washburn MP, Wolters D and Yates JR Jr:
Large-scale analysis of the yeast proteome by multidimensional
protein identification technology. Nat Biotechnol. 19:242–247.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alsallakh B, Aigner W, Miksch S and Hauser
H: Radial sets: Interactive visual analysis of large overlapping
sets. IEEE Trans Vis Comput Graph. 19:2496–2505. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tyanova S, Temu T, Sinitcyn P, Carlson A,
Hein MY, Geiger T, Mann M and Cox J: The Perseus computational
platform for comprehensive analysis of (prote)omics data. Nat
Methods. 13:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liberzon A, Birger C, Thorvaldsdottir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
33
|
Lin Y, Zhang J, Cai J, Liang R, Chen G,
Qin G, Han X, Yuan C, Liu Z, Li Y, et al: Systematic analysis of
gene expression alteration and co-expression network of eukaryotic
initiation factor 4A-3 in cancer. J Cancer. 9:4568–4577. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zeng J, He SL, Li LJ and Wang C: Hsp90
up-regulates PD-L1 to promote HPV-positive cervical cancer via
HER2/PI3K/AKT pathway. Mol Med. 27:1302021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shan N, Zhou W, Zhang S and Zhang Y:
Identification of HSPA8 as a candidate biomarker for endometrial
carcinoma by using iTRAQ-based proteomic analysis. Onco Targets
Ther. 9:2169–2179. 2016.PubMed/NCBI
|
|
36
|
Potriquet J, Laohaviroj M, Bethony JM and
Mulvenna J: A modified FASP protocol for high-throughput
preparation of protein samples for mass spectrometry. PLoS One.
12:e01759672017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fermo E, Vercellati C, Marcello AP,
Zaninoni A, Aytac S, Cetin M, Capolsini I, Casale M, Paci S,
Zanella A, et al: Clinical and molecular spectrum of
glucose-6-phosphate isomerase deficiency. report of 12 new cases.
Front Physiol. 10:4672019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kawai K, Uemura M, Munakata K, Takahashi
H, Haraguchi N, Nishimura J, Hata T, Matsuda C, Ikenaga M, Murata
K, et al: Fructose-bisphosphate aldolase A is a key regulator of
hypoxic adaptation in colorectal cancer cells and involved in
treatment resistance and poor prognosis. Int J Oncol. 50:525–534.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ždralević M, Marchiq I, de Padua MMC,
Parks SK and Pouysségur J: Metabolic plasiticy in cancers-distinct
role of glycolytic enzymes GPI, LDHs or membrane transporters MCTs.
Front Oncol. 7:3132017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han J, Deng X, Sun R, Luo M, Liang M, Gu
B, Zhang T, Peng Z, Lu Y, Tian C, et al: GPI is a prognostic
biomarker and correlates with immune infiltrates in lung
adenocarcinoma. Front Oncol. 11:7526422021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saito Y, Takasawa A, Takasawa K, Aoyama T,
Akimoto T, Ota M, Magara K, Murata M, Hirohashi Y, Hasegawa T, et
al: Aldolase A promotes epithelial-mesenchymal transition to
increase malignant potentials of cervical adenocarcinoma. Cancer
Sci. 111:3071–3081. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Saritha V, Abdul J, Arun S and Meenakshi
SK: Analysis of differentially expressed proteins in the exfoliated
cells of normal and squamous cell carcinoma of the uterine cervix
to define candidate markers for cervical cancer. Int J Bioc
Biotechnol. 5:626–636. 2016.
|
|
43
|
Xu H, Yu S, Peng K, Gao L, Chen S, Shen Z,
Han Z, Chen M, Lin J, Chen S and Kang M: The role of EEF1D in
disease pathogenesis: A narrative review. Ann Transl Med.
9:16002021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sohn M, Shin S, Yoo JY, Goh Y, Lee IH and
Bae YS: Ahnak promotes tumor metastasis through transforming growth
factor-β-mediated epithelial-mesenchymal transition. Sci Rep.
8:143792018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Calmon MF, Sichero L, Boccardo E, Villa LL
and Rahal P: HPV16 E6 regulates annexin 1 (ANXA1) protein
expression in cervical carcinoma cell lines. Virology. 496:35–41.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Manawapat-Klopfer A, Thomsen LT, Martus P,
Munk C, Russ R, Gmuender H, Frederiksen K, Haedicke-Jarboui J,
Stubenrauch F, Kjaer SK and Iftner T: TMEM45A, SERPINB5 and
p16INK4A transcript levels are predictive for development of
high-grade cervical lesions. Am J Cancer Res. 6:1524–1536.
2016.PubMed/NCBI
|
|
47
|
Chang IW, Liu KW, Ragunanan M, He HL,
Shiue YL and Yu SC: SERPINB5 Expression: Association with CCRT
Response and Prognostic Value in Rectal Cancer. Int J Med Sci.
15:376–384. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liang S, Ju X, Zhou Y, Chen Y, Ke G, Wen H
and Wu X: Downregulation of eukaryotic initiation factor 4A1
improves radiosensitivity by delaying DNA double strand break
repair in cervical cancer. Oncol Lett. 14:6976–6982.
2017.PubMed/NCBI
|
|
49
|
Wang H, Deng G, Ai M, Xu Z, Mou T, Yu J,
Liu H, Wang S and Li G: Hsp90ab1 stabilizes LRP5 to promote
epithelial-mesenchymal transition via activating of AKT and
Wnt/β-catenin signaling pathways in gastric cancer progression.
Oncogene. 38:1489–1507. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Saxton RA and Sabatini DM: mTOR signaling
in growth, metabolism, and disease. Cell. 168:960–976. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rezazadeh D, Norooznezhad AH, Mansouri K,
Jahani M, Mostafaie A, Mohammadi MH and Modarressi MH: Rapamycin
reduces cervical cancer cells viability in hypoxic condition:
Investigation of the role of autophagy and apoptosis. Onco Targets
Ther. 13:4239–4247. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dossou AS and Basu A: The emerging roles
of mTORC1 in macromanaging autophagy. Cancers (Basel). 11:14222019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ji J and Zheng PS: Activation of mTOR
signaling pathway contributes to survival of cervical cancer cells.
Gynecol Oncol. 117:103–108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lamming DW: Inhibition of the mechanistic
target of rapamycin (mTOR)-rapamycin and beyond. Cold Spring Harb
Perspect Med. 6:a0259242016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yim EK and Park JS: The role of HPV E6 and
E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer
Res Treat. 37:319–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rabachini T, Boccardo E, Andrade R, Perez
KR, Nonogaki S, Cuccovia IM and Villa LL: HPV-16 E7 expression
up-regulates phospholipase D activity and promotes rapamycin
resistance in a pRB-dependent manner. BMC Cancer. 18:4852018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chanvorachote P, Sriratanasak N and
Nonpanya N: C-myc contributes to malignancy of lung Cancer: A
potential anticancer drug target. Anticancer Res. 40:609–618. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu J, Chen Y and Olopade OI: MYC and
breast cancer. Genes Cancer. 1:629–640. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Meyer N and Penn LZ: Reflecting on 25
years with MYC. Nat Rev Cancer. 8:976–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20:8402019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Balamurugan K: HIF-1 at the crossroads of
hypoxia, inflammation, and cancer. Int J Cancer. 138:1058–1066.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Semenza GL: Hypoxia-inducible factors:
Mediators of cancer progression and targets for cancer therapy.
Trends Pharmacol Sci. 33:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Havula E and Hietakangas V: Glucose
sensing by ChREBP/MondoA-Mlx transcription factors. Semin Cell Dev
Biol. 23:640–647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Martínez-Ramírez I, Carrillo-García A,
Contreras-Paredes A, Ortiz-Sánchez E, Cruz-Gregorio A and Lizano M:
Regulation of cellular metabolism by high-risk human
papillomaviruses. Int J Mol Sci. 19:18392018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Medda A, Duca D and Chiocca S: Human
papillomavirus and cellular pathways: Hits and targets. Pathogens.
10:2622021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang D, Tai LK, Wong LL, Chiu LL, Sethi
SK and Koay ES: Proteomic study reveals that proteins involved in
metabolic and detoxification pathways are highly expressed in
HER-2/neu-positive breast cancer. Mol Cell Proteomics. 4:1686–1696.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yan H, Yang K, Xiao H, Zou YJ, Zhang WB
and Liu HY: Over-expression of cofilin-1 and phosphoglycerate
kinase 1 in astrocytomas involved in pathogenesis of
radioresistance. CNS Neurosci Ther. 18:729–736. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ahmad SS, Glatzle J, Bajaeifer K, Bühler
S, Lehmann T, Königsrainer I, Vollmer JP, Sipos B, Ahmad SS,
Northoff H, et al: Phosphoglycerate kinase 1 as a promoter of
metastasis in colon cancer. Int J Oncol. 43:586–590. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hwang TL, Liang Y, Chien KY and Yu JS:
Overexpression and elevated serum levels of phosphoglycerate kinase
1 in pancreatic ductal adenocarcinoma. Proteomics. 6:2259–2272.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zieker D, Königsrainer I, Tritschler I,
Löffler M, Beckert S, Traub F, Nieselt K, Bühler S, Weller M,
Gaedcke J, et al: Phosphoglycerate kinase 1 a promoting enzyme for
peritoneal dissemination in gastric cancer. Int J Cancer.
126:1513–1520. 2010.PubMed/NCBI
|
|
72
|
Rojas-Pirela M, Andrade-Alviárez D, Rojas
V, Kemmerling U, Cáceres AJ, Michels PA, Concepción JL and Quiñones
W: Phosphoglycerate kinase: Structural aspects and functions, with
special emphasis on the enzyme from Kinetoplastea. Open Biol.
10:2003022020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lowy AM, Clements WM, Bishop J, Kong L,
Bonney T, Sisco K, Aronow B, Fenoglio-Preiser C and Groden J:
β-Catenin/Wnt signaling regulates expression of the membrane type 3
matrix metalloproteinase in gastric cancer. Cancer Res.
66:4734–4741. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yamada T, Takaoka AS, Naishiro Y, Hayashi
R, Maruyama K, Maesawa C, Ochiai A and Hirohashi S: Transactivation
of the multidrug resistance 1 gene by T-cell factor 4/beta-catenin
complex in early colorectal carcinogenesis. Cancer Res.
60:4761–4766. 2000.PubMed/NCBI
|
|
75
|
Li X, Jiang Y, Meisenhelder J, Yang W,
Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, et al:
Mitochondria-translocated PGK1 functions as a protein kinase to
coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell.
61:705–719. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yu X and Li S: Non-metabolic functions of
glycolytic enzymes in tumorigenesis. Oncogene. 36:2629–2636. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kalejta RF and Shenk T:
Proteasome-dependent, ubiquitin-independent degradation of the Rb
family of tumor suppressors by the human cytomegalovirus pp71
protein. Proc Natl Acad Sci USA. 100:3263–3268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Schwartz AL and Ciechanover A: Targeting
proteins for destruction by the ubiquitin system: Implications for
human pathobiology. Annu Rev Pharmacol Toxicol. 49:73–96. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Đukić A, Lulić L, Thomas M, Skelin J,
Bennett Saidu NE, Grce M, Banks L and Tomaić V: HPV oncoproteins
and the ubiquitin proteasome system: A signature of malignancy?
Pathogens. 9:1332020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gupta I, Singh K, Varshney NK and Khan S:
Delineating crosstalk mechanisms of the ubiquitin proteasome system
that regulate apoptosis. Front Cell Dev Biol. 6:112018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Taylor JR, Skeate JG and Kast WM: Annexin
A2 in virus infection. Front Microbiol. 9:29542018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Xia C, Braunstein Z, Toomey AC, Zhong J
and Rao X: S100 proteins as an important regulator of macrophage
inflammation. Front Immunol. 8:19082018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
McLaughlin-Drubin ME, Huh KW and Munger K:
Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J
Virol. 82:8695–8705. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
McLaughlin-Drubin ME, Meyers J and Munger
K: Cancer associated human papillomaviruses. Curr Opin Virol.
2:459–466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Aarthy M and Singh SK: Interpretations on
the interaction between protein tyrosine phosphatase and E7
oncoproteins of high and low-risk HPV: A computational perception.
ACS Omega. 6:16472–16487. 2021. View Article : Google Scholar : PubMed/NCBI
|