Introduction
Human papillomavirus (HPV) infections are strongly
associated with cervical cancer (CC), and with the incidence of
oropharyngeal cancer (1) and
anogenital cancer (2). HPV
infections are also typically linked to skin or mucosal lesions,
such as warts. HPV affects both men and women; however, the disease
burden is commonly observed among women due to their high
susceptibility for cervical infections (3). Approximately 90% of CC cases are
caused by one or more HPV types, particularly HPV16 (detected in
~50% of cases) and HPV18 (detected in 10–15% of cases) (4). HPV16 and −18 are known as high-risk
HPV (HR-HPV) subtypes, alongside another 15 types, including HPV31,
−33-35, −39, −45, −51, −52, −56, −58, −59, −68, −73 and −82
(5). Usually, HR-HPV infections
persist for 1–2 years. Within this period, the virion replicates
for months in host tissue with regular transformation, along with
active suppression of both innate and adaptive immune responses
(6). HPV relies greatly on its
potential to control both viral and host gene expression (7). Viral gene transcript regulation is
directly related to keratinocyte differentiation in the host
(8). Previous studies have
reported that HPV-infected keratinocytes exhibit significantly
reduced expression of numerous inflammatory mediators (9,10).
HPV is able to promote immune evasion via the
expression of oncogenic proteins, which are responsible for the
modulation of several immune mechanisms, including antigen
presentation and inflammatory pathways (11). HPV E6 and E7 oncoproteins support
viral oncogenicity as they are expressed concurrently with whole
tumor progression and deregulate host cell gene expression
(12). Both E6 and E7 proteins
exhibit numerous functions; however, they are mainly involved in
the inactivation of p53 and retinoblastoma protein (pRb) (13). The inactivation of p53 and pRb can
result in the loss of control over DNA damage repair and cell cycle
regulation. The E7 oncoprotein binds to and induces degradation of
pocket proteins (such as pRb, p107 and p130), which leads to the
release of E2F transcription factor family proteins and the
expression of S-phase genes (14).
However, the suppression of pRb family proteins is not entirely
influenced by E7, which suggests that other mechanisms are
involved. Furthermore, the degradation of p107 and p130 by E7 is
also related to the suppression of DREAM complex-mediated genes
(13). Various proteins across
different pathways, including activator protein-1 (15), HIF1 (16), krüppel-like factor 4 (17), p73 (18) and other host cell factors have been
identified as E7 targets. However, their functional interactions
that encourage virus replication and carcinogenesis are not yet
fully understood (7).
Understanding the pathogenic mechanisms and
signaling pathways associated with HR-HPV, along with the
identification of differentially expressed protein-protein
interactions (PPIs), could aid in the design of novel therapeutic
strategies that target HPV-associated CC. Therefore, the present
study evaluated changes in protein expression levels between normal
human keratinocytes (HaCaT) transfected with recombinant HPV16/18
E7 and HPV-transformed cells (Caski and HeLa) to identify proteins
that may contribute to cancer hallmark enrichment. Deciphering
alterations in cellular protein interactions and their associated
pathways during the viral life cycle is essential to understand the
evolution of HPV E7 function in cervical carcinogenesis.
Materials and methods
Cell lines
Human HaCaT keratinocyte cells (Addexbio
Technologies), HPV16-positive CC CaSki (passage 9) and
HPV18-positive CC HeLa (passage 11) cell lines (both from the
Laboratory of Virology, Department of Medical Microbiology,
University of Malaya, Kuala Lumpur, Malaysia), were used in the
present study. The HaCaT cells were authenticated using STR
profiling. CaSki cells carry integrated viral HPV16 DNA. HeLa cells
contain integrated viral HPV18 DNA. All cell lines were maintained
in T25 flasks with 5 ml Dulbecco's Modified Eagle's Medium (Gibco;
Thermo Fisher Scientific, Inc.), 10% heat-inactivated fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
Cells were maintained under standard incubation conditions of 10%
CO2 with 95% humidity at 37°C.
Transfection of HPV16/HPV18 E7
recombinant plasmids in human keratinocytes
Transfection of HaCaT cells was performed according
to the manufacturer's protocol. Briefly, FuGENE® HD
transfection buffer (Promega Corporation) was added to 2 µg of each
plasmid [pMSCVpuro; Addgene (Fig.
S1) (control); pMSCVpuro-HPV16E7; and pMSCVpuro-HPV18E7], which
were diluted in 100 µl serum-free Opti-MEM™ Reduced Serum medium
(Gibco; Thermo Fisher Scientific, Inc.). Subsequently, 100 µl
transfection mixture containing 2 µg of the nucleic acid was added
to HaCaT cells (3×105) grown in a 6-well plate and
incubated at 37°C for 48 h. The cells were then washed and
maintained using 0.5 µg/ml puromycin (Gibco; Thermo Fisher
Scientific, Inc.) for 24 h post-transfection and incubated for a
further 48 h to establish stable transformants before harvesting.
Transfection efficiency was assessed using reverse
transcription-quantitative PCR (RT-qPCR) and the results are
presented in Fig. S2.
RNA extraction and RT-qPCR
Total RNA was extracted using the FavorPrep™
Blood/Cultured Cell Total RNA Mini Kit (Favorgen). The cDNA was
prepared using the RevertAid™ First Strand cDNA Synthesis Kit
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. qPCR was performed using Absolute SYBR Green ROX (Thermo
Fisher Scientific, Inc.) in triplicate using an ABI PRISM™ 7900HT
sequence detector (Applied Biosystems; Thermo Fisher Scientific,
Inc.) (19). The primers used for
the qPCR reactions are presented in Table SI. PCR was performed under the
following thermocycling conditions: 95°C for 15 min, followed by 40
cycles at 95°C for 15 sec, 58°C for 30 sec and 72°C for 30 sec.
Optimization was performed for each primer set and the relative
quantification was performed with normalization against β-actin
(20).
Protein extraction and
quantification
Proteins were extracted using RIPA lysis buffer
[NaCl, 150 mM; 1% Triton X-100; Tris-HCl, 50mM (pH, 8.0); 0.5%
sodium deoxycholate; 0.1% SDS; Halt™ Protease Inhibitor Cocktail
(100X; Thermo Fisher Scientific, Inc.)]. Cells were washed twice
with ice-cold PBS and mixed with ice-cold RIPA lysis buffer. The
cell lysate was centrifuged at 13,000 × g for 20 min at 4°C. The
supernatant, containing soluble proteins, was collected and stored
at −80°C until further analysis. The concentration of the extracted
proteins was determined using the Bradford assay (21). Bovine serum albumin
(MilliporeSigma) was used as a protein standard to construct the
calibration curve.
In-solution tryptic digestion and
desalting
A total of ~500 µg protein from each sample were
digested in 50 mM ammonium bicarbonate digestion buffer together
with 100 mM DTT reducing buffer and incubated at 95°C for 5 min.
The samples were cooled to ambient temperature and alkylated in 100
mM iodoacetamide buffer for 20 min in the dark. Trypsin is a serine
protease that is highly specific, digesting protein into peptides
by cutting at the carboxyl side of arginine and lysine residues.
Samples were then incubated with trypsin (Promega Corp.) at 37°C
for 3 h and another 1 µl trypsin was added for overnight incubation
at 30°C. The digested peptides were desalted, lyophilized and
stored at −80°C until further experimentation (22).
Protein identification using
nano-electrospray ionization-liquid chromatography-tandem mass
spectrometry (nano-ESI-LC-MS/MS)
Protein identification using nano-ESI-LC-MS/MS was
performed with certain modifications (23). Trypsin-digested peptides (500 µg)
were loaded onto a 300 Å, SB-C18, 160 nl enrichment column and 75
µm × 150 mm analytical column (cat. no. G4240-62010; Agilent
Technologies, Inc.) with a flow rate of 4 µl/min using a capillary
pump and 0.5 µl/min using an Agilent 1200 nano pump. The eluted
peptides were subjected to nano-ESI MS/MS using an Agilent 1200
Series HPLC-Chip/MS System Interface, coupled with the Agilent 6550
Q-TOF LC/MS system (Agilent Technologies, Inc.). Injection volume
was adjusted to 1 µl/sample. The mobile phases were 0.1% formic
acid in water (solution A) and 90% acetonitrile with 0.1% formic
acid (solution B) for 4 min and 70% solution B for 3 min, using an
Agilent 1200 Series nanoflow LC pump. Ion polarity was set to
positive ionization mode, the drying gas flow rate was 5.0 µl/min
and the temperature was fixed at 325°C. The fragmentor and
capillary voltage were set at 360 and 1,900 V, respectively. The
spectra were acquired in MS/MS mode with an MS scan range of
110–3,000 m/z and MS/MS scan range of 50–3,000 m/z. Precursor
charge selection was set as a double-, triple- or more than
triple-charged state, with the exclusion of precursors 1,221.9906
m/z (z=1) set as reference ions. Data were extracted using
MH+ (positive ion) mass range between 50–3,200 Da and
processed using the Agilent Spectrum Mill MS Proteomics Workbench
software packages version B.04.00 (Agilent Technologies, Inc.). The
raw data files obtained from the LC-MS/MS were processed using
PEAKS DB analysis to evaluate the relative protein abundance.
De novo identification of peptides
using PEAKS Studio 7.0
PEAKS DB is a proteomic software package for MS/MS
designed for peptide sequencing, protein identification and
quantification (24). Peptide
identification was performed using automated de novo
sequencing in PEAKS Studio 7.0 (Bioinformatics Solution, Inc.)
(25). Proteins were identified
from HPV-transfected and -transformed cells using the Uniprot
Homo sapiens, type Swissprot and Trembl databases, processed
with PEAKS 7.0 (Bioinformatics Solution, Inc.) and
carbamidomethylation was set as a fixed modification. Subsequently,
high-confidence proteins were identified by setting a false
discovery rate (FDR) threshold of 1%, unique peptide ≥1 and
−10lgP>20.
Label-free quantification (LFQ)
Label-free mass spectrometry-based quantitative
approaches provide powerful, fast and low-cost tools for analyzing
protein changes in complex biological samples in several
large-scale biomarker discovery studies (26). However, to avoid any variation
errors in the performance of LC and MS, a carefully controlled
normalization step is required. LFQ was performed using PEAKS
studio 7.0 (Bioinformatics Solution, Inc.) the abundance of
proteins was calculated using normalized spectral protein intensity
(LFQ intensity), in which proteins were quantified by comparing the
number of identified MS/MS spectra from the same protein in each of
the multiple LC-MS/MS data sets. It is possible that an increase in
protein abundance results in a higher number of proteolytic
peptides and vice versa. In turn, a larger number of proteolytic
peptides leads to higher protein sequence coverage and increased
number of both identified unique peptides and total MS/MS spectra
(spectral count) for each protein (27). HaCaT-pMSCV puro was used as a
control to normalize the LFQ of fold changes for significant
proteins. The pheatmap package was used to present differentially
expressed proteins with hierarchical clustering.
Data acquisition and statistical
analysis
Three biological replicates for each cell line,
including the control, underwent LC-MS/MS. P-values were obtained
using the Benjamini-Hochberg FDR. Statistical analysis was
performed using GraphPad Prism version 5.0.0 for Windows (GraphPad
Software, Inc.) and the data are presented as the mean ± standard
deviation (SD). A comparison between the transfected samples and
control was conducted using Student'st-test. and P<0.05 was
considered to indicate a statistically significant difference.
Bioinformatics analysis
UpSet plot analysis
An UpSet plot is commonly applied to visualize a
dataset with more than three overlapping patterns. It is similar to
a Venn diagram but more comprehensive (28). An UpSet plot (https://upset.app/) for HaCaT control, HaCaT-16 E7,
HaCaT-18 E7, CaSki and HeLa datasets was generated to identify
unique and overlapping proteins across all five cell lines.
Volcano plot analysis
The identified proteins were further analyzed using
Perseus (version 1.5.3.0) (29)
for differential expression analysis. Firstly, the reverse,
site-only and contaminant peptides were removed from the dataset,
with missing values input using a normal distribution. This
analysis identified significant protein changes using Student's
t-test with an FDR<0.05 between HPV-transfected and
HPV-transformed cells. The data were presented using a volcano
plot, where the x-axis represented the fold change and the y-axis
presented the -logP-value.
Molecular signature database (MSigDB)
analysis
The MSigDB (30) is
one of the largest and most popular repositories of gene sets for
use with the Gene Set Enrichment Analysis (GSEA) software tool. The
GSEA tool was used to characterize protein expression levels in
HaCaT control and HPV-transfected cells with the accession no.
MSV000090470 (massive.ucsd.edu). The cancer hallmarks for
upregulated proteins in HPV-transfected cells were also identified
(31). The P-value summary was
used to determine the FDR according to the method described by
Benjamini (32). The proteins were
identified for each cancer hallmark, sorted by their P-value and
FDR values. The top-scoring protein with a summary FDR<0.01 was
considered to encompass the definitive cancer hallmark set.
PPI network
Cytoscape 3.8.2 is a freely available platform for
network visualization and analysis (Cytoscape_v3.8.2). PPI network
construction in Cytoscape requires each protein in the input file
to have the same identifiers. Therefore, UniProt ID mapping with
Cytoscape was applied to standardize the identifiers. All
upregulated proteins in HPV-transfected cells were selected to
create a PPI network. The interactors in the network were carefully
evaluated to identify potential interactions between the nodes.
Search Tools for the Retrieval of
Interacting Genes (STRING) functional enrichment analysis
Gene Ontology (GO) enrichment analysis was applied
to identify the function of differentially expressed proteins based
on three main aspects as follows: i) Molecular function (MF); ii)
cellular component (CC); and iii) biological process (BP) in which
they were mutually involved. The list of identified proteins was
evaluated using version 10 of the STRING database (http://string10.embl.de) to predict the identified
proteins. The interaction score was set to a high confidence level
of 0.700. GO terms with FDR<0.01 were defined as the enriched
terms for the differentially expressed proteins; Homo
sapiens was selected as the organism.
Results
Protein identification in
HPV-transfected and -transformed cells
Protein profiling analysis was performed using total
protein fractions from each cell type to identify differentially
expressed proteins in HPV-transfected human keratinocytes
(HaCaT-HPV16/18 E7) and HPV-transformed cells (CaSki and HeLa),
with empty vector pMSCV-puro human keratinocytes (HaCaT) used as a
control. After quality control assessment of each protein,
including average mass, number of peptides and number of uniquely
expressed peptides, high confidence proteins (−10lgP>20
considered significant) were identified. A total of 41 proteins
were identified in HaCaT cells (Table
SII), 29 proteins were identified in HaCaT-pMSCVpuro (control)
cells (Table SIII), 85 proteins
were identified in HaCaT-HPV16 E7 cells (Table SIV) and 104 proteins were
identified in HaCaT-HPV18 E7 cells (Table SV). For HPV-transformed cells, 45
and 39 proteins were identified in CaSki (Table SVI) and HeLa (Table SVII) cells, respectively. The
results of the present study demonstrated that glucose-6-phosphate
isomerase (GPI) and fructose-bisphosphate aldolase (ALDOA) were the
two common proteins among the cell lines. GAPDH, tubulin α-1A chain
(TUBA1A), tubulin α chain, heat shock 70kDa protein 8 isoform 2
variant (HSPA8), profilin (PFN1), keratin type I cytoskeletal 17,
annexin 5, transketolase, annexin 2 (ANXA2), keratin type II
cytoskeletal 8 (KRT8) and peroxiredoxin-1 (PRDX1) were identified
in CaSki and HeLa cells. Tubulin α-1B chain and elongation factor 2
(EEF2) were common proteins identified in HaCaT-HPV16/18 E7, CaSki
and HeLa cells.
LFQ and heat map clustering analysis
between HPV-transfected and -transformed cells
The high-confidence proteins (P=0.01) were
identified across HPV-transfected and -transformed cells were
subjected to LFQ to assess the differentially expressed proteins
with significant fold changes (Table
SVIII). HaCaT control cells were used for normalization. A
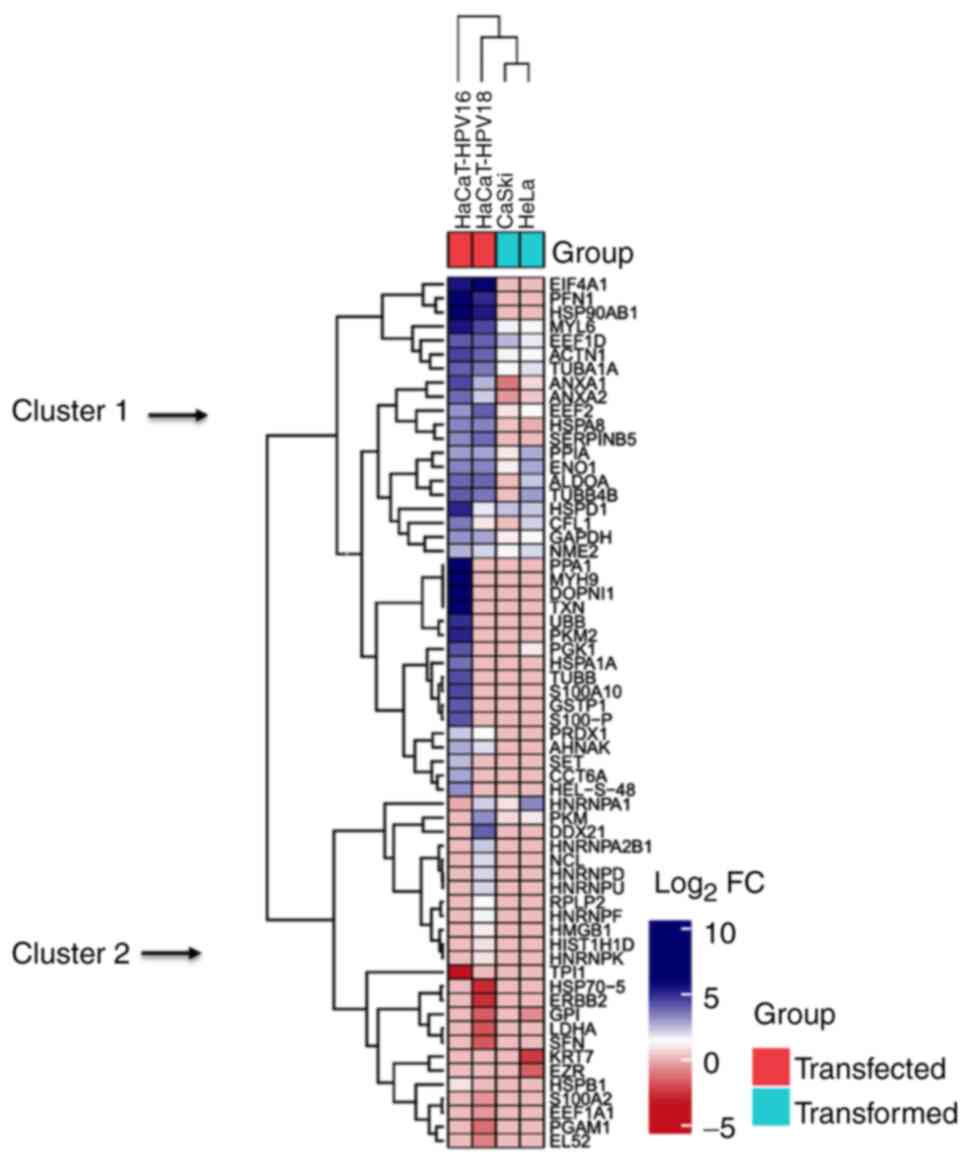
total of 62 differentially expressed proteins were identified. A
heat map of differentially expressed proteins was generated with
legend color bar, column and row annotations (Fig. 1). All proteins were subsequently
grouped based on the hierarchical clustering between
HPV-transfected and -transformed cells. The upregulated proteins
were clustered at the top of the heat map (blue), whereas the
downregulated proteins were clustered at the bottom (red). The
results of the present study identified 37 upregulated and 13
downregulated proteins. The hierarchical clustering (column) of
HPV-transfected cells demonstrated more proteins in common between
HaCaT-HPV16 E7 and HaCaT-HPV18 E7 cells compared with those
identified in both HPV-transformed cells. Furthermore, hierarchical
clustering (row) resulted in two distinct clusters, separated by
upregulated and downregulated proteins based on assessed fold
change. Most proteins in HPV-transfected cells were upregulated.
EIF4A1, PFN1, HSP90AB1, MYL6, EEF1D, ACTN1, TUBA1A, ANXA1, ANXA2,
EEF2, HSPA8, SERPINB5, PPIA, ENO1, ALSOA, TUBB4B, HSPD1, CFL1,
GAPDH, NME2, PPA1, MYH9, DOPNI1, TXN, UBB, PKM2, PGK1, HSPA1A,
TUBB, S100A10, GSTP1, S100-P, PRDX1, AHNAK, SET, CCT6A and HEL-S-48
were upregulated in HPV-transfected cells compared with in both
HaCaT (control) and HPV-transformed cells (Cluster 1). However,
Cluster 2 demonstrated a mixed pattern with both upregulated and
downregulated proteins. The upregulated proteins in Cluster 2 were
HNRNPA1, PKM, DDX21, HNRNPA2B1, NCL, HNRNPD, HNRNPU, RPLP2, HNRNPF,
HMGB1, HIST1H1D, HNRNPK and TPI1. The downregulated proteins were
HSP70-5, ERBB2, GPI, LDHA, SFN, KRT7, EZR, HSPB1, S100A2, EEF1A1,
PGAM1 and EL52.
Volcano plot of proteins with
significantly increased expression levels in HPV-transfected and
-transformed cells
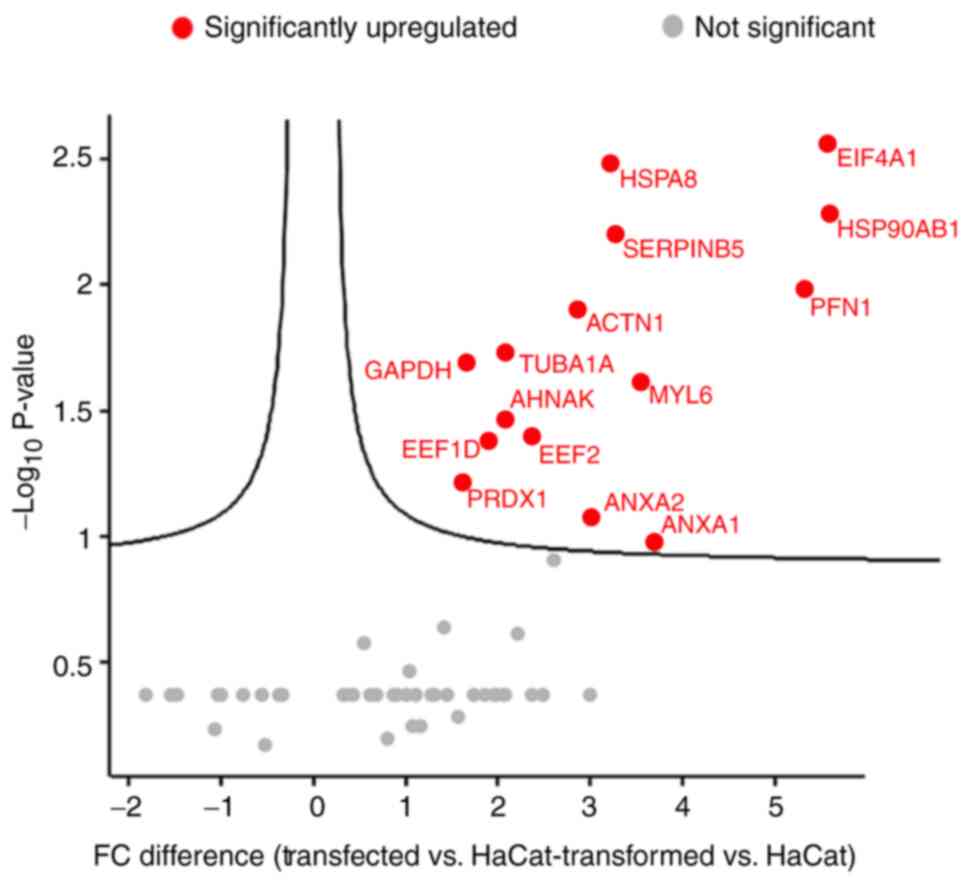
Proteins in the aforementioned heat map analysis
(Fig. 1) were further analyzed
using a volcano plot to assess the proteins with the most
significantly increased protein expression levels in
HPV-transfected and -transformed cells (Fig. 2). The volcano plot presented
fold-change differences and protein expression level distribution
in HPV-transfected cells vs. HaCaT cells, and in HPV-transformed
cells vs. HaCaT cells. Proteins were presented in graphs according
to fold change (difference) and significance (−log10 P-value). The
15 upregulated proteins mainly observed in HPV-transfected cells
were GAPDH, EEF1D, PRDX1, TUBA1A, AHNAK, EEF2, ANXA2, ANXA1, ACTN1,
MYL6, SERPINB5, HSPA8, EIF4A1, HSP90AB1 and PFN1. Among these
proteins, EIF4A1, HSP90AB1, HSPA8 and SERPINB5 were presented
toward the top right of the plot, which indicated high statistical
significance and fold change. EIF4A1 serves a crucial role in the
transformation and progression of various types of cancer as it is
part of the EIF4F complex that controls initiation rates of
pro-oncogenic mRNAs involving the PI3K/Akt/mTOR signaling pathway
(33). HSP90AB1, a chaperone
protein, is often upregulated in cancerous cells and it is able to
stabilize their protein functions with activating mutation
(34). HSPA8 is a member of the
HSP70 family, which collectively functions as a buffering system
for cellular stress which is required for cancer cell survival
(35). Fold-change differences in
the expression levels of the remaining proteins were not
statistically significant.
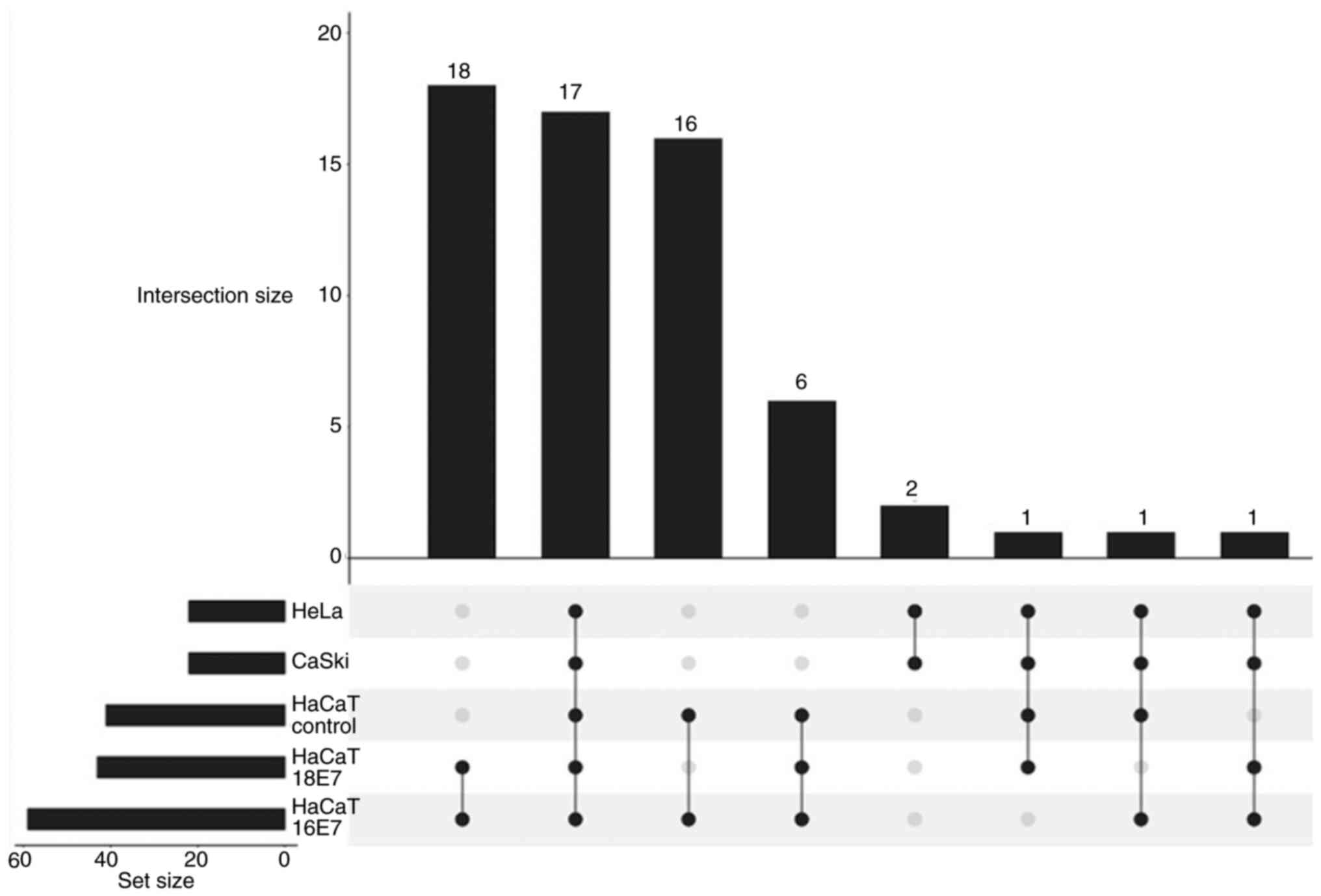
UpSet intersection plot of
HPV-transfected and -transformed cells
The total number of quantified proteins in HaCaT
control, HPV-transfected (HaCaT-HPV16 E7 and HaCaT-HPV18 E7) and
HPV-transformed (CaSki and HeLa) cells were further evaluated using
UpSet plot analysis (Fig. 3). The
UpSet intersection plot was presented as a matrix layout to
visualize overlaps and differences between qualified proteins
across all five cell lines. Dark circles in the matrix indicate
sets that are part of the intersection. A high number of linkages
indicate a marked association of proteins between the respective
cell lines (36). A total of 18
protein linkages were demonstrated between HaCaT-HPV16 E7 and
HaCaT-HPV18 E7 cells, the most in the entire analysis (GAPDH, PKM,
PPIA, HSPD1, ENO1, TUBB4B, EEF2, ANXA2, ANXA1, TUBA1A, ALDOA, NME2,
EEFID, ACTN1, MYL6, CFL1, HSPA8 and HNRNPA1). The second-highest
number of protein linkages (n=17) was demonstrated across all five
cell lines (GAPDH, PPIA, HSPD1, ENO1, TUBB4B, EEF2, ANXA2, ANXA1,
TUBA1A, ALDOA, NME2, EEFID, ACTN1, MYL6, CFL1, HSPA8 and HNRNPA1).
Only two protein linkages, KRT7 and EZR, were demonstrated between
HeLa and CaSki cells. The lowest number of protein linkages (n=1)
was demonstrated in three different groups of linkages as follows:
i) HeLa, CaSki, HaCaT control and HaCaT-HPV18 E7; ii) HeLa, CaSki,
HaCaT control and HaCaT-HPV16 E7; and iii) HeLa, CaSki, HaCaT-HPV16
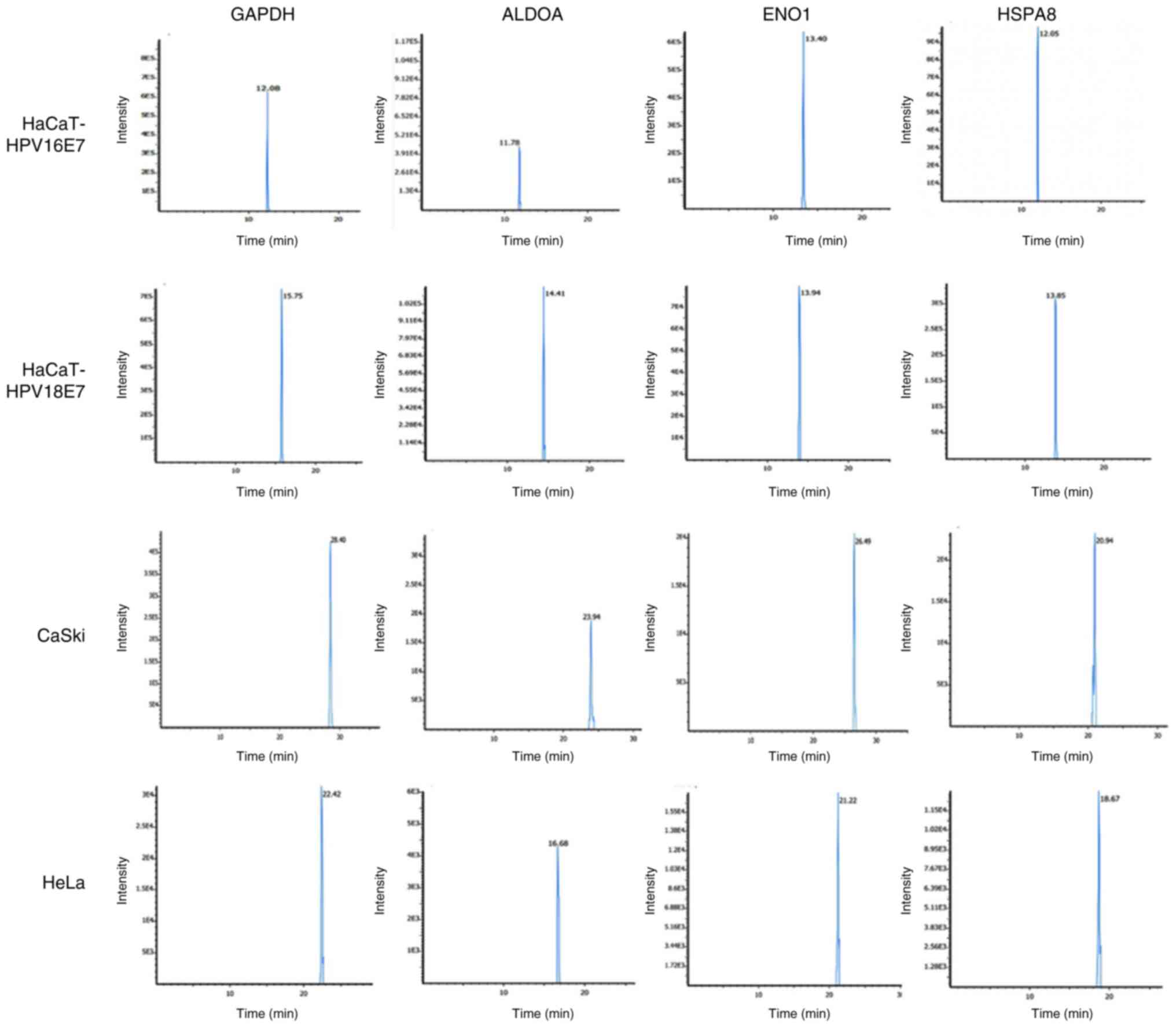
E7 and HaCaT-HPV18 E7. The representative chromatograms presented
the intensity count for GAPDH, ENO1, ALDOA and HSPA8 proteins with
the time taken to pass through the column assessed using the
LC-MS/MS analysis of the HPV-transfected (HaCaT-HPV16 E7 and
HaCaT-HPV18 E7) and HPV-transformed (CaSki and HeLa) cells
(Fig. 4).
Cancer hallmark enrichment of
upregulated proteins in HPV-transfected cells
Cancer hallmark enrichment was analyzed based on the
P-value and FDR values of the aforementioned upregulated proteins
in HaCaT-HPV16/18 E7 cells. The analysis demonstrated that these
differentially expressed proteins were significantly enriched
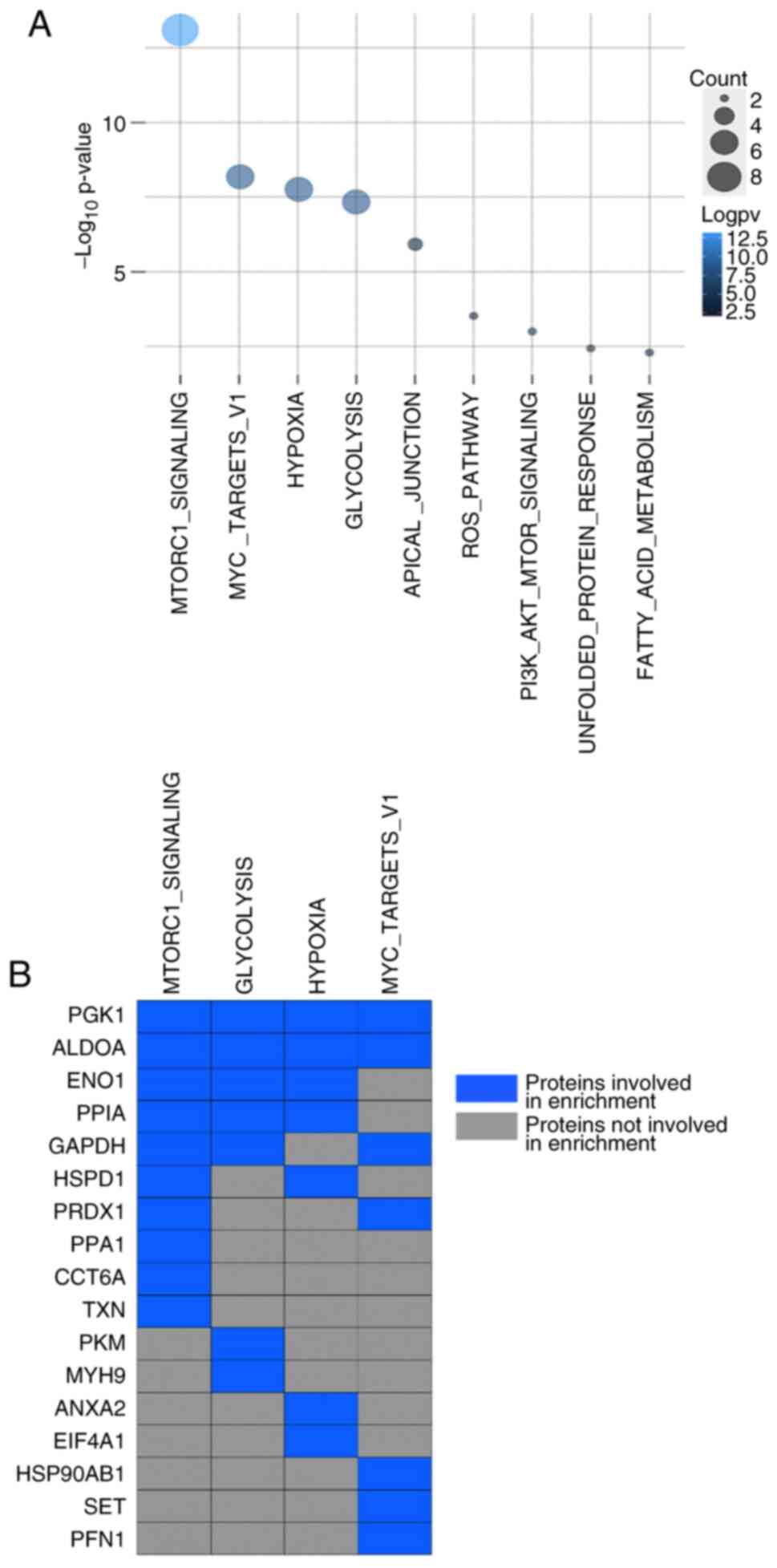
across nine pathways (Fig. 5A). A
total of 17 of these proteins were involved in the top four
enriched pathways: MTORC1 signaling, glycolysis, hypoxia and MYC
target VI (Fig. 5B). A total of 10
proteins (PGK1, ALDOA, ENO1, PPIA, GAPDH, HSPD1, PRDX1, PPA1, CCT6A
and TXN) were involved with the activation of the mTORC1 complex. A
total of seven proteins (PGK1, ALDOA, GAPDH, PRDX1, HSP90AB1, SET
and PFN1) were involved with the MYC target VI pathway. Another
seven proteins were demonstrated to be involved in cellular
response to hypoxia (PGK1, ALDOA, ENO1, PPIA, HSPD1, ANXA2 and
EIF4A1). Furthermore, PGK1, ALDOA, ENO1, PPIA, GAPDH, PKM and MYH9
proteins were involved in glycolysis. Notably, PGK1, ALDOA and ENO1
were involved in all four pathways. Other noteworthy cancer
hallmark enrichment pathways were apical junction, reactive oxygen
species (ROS) pathway, P13K/AKT/mTOR signaling pathway, unfolded
protein response and fatty acid metabolism. A total of three
proteins (PGK1, ANXA2 and ACTN1) were demonstrated to be engaged in
the apical junction pathway. PGK1, PPA1 and PKM were linked to the
ROS pathway. PGK1, ACTN1 and EEF2 were linked with the
PI3K/AKT/mTOR signaling pathway. PGK1, HSP90AB1 and S100A1 were
involved with the unfolded protein response, a cellular stress
response related to the endoplasmic reticulum. Finally, PGK1, ENO1
and HSPA1A were associated with the metabolism of fatty acids.
Overall, the results demonstrated that PGK1 expression was
upregulated and was involved in all cancer hallmark pathways.
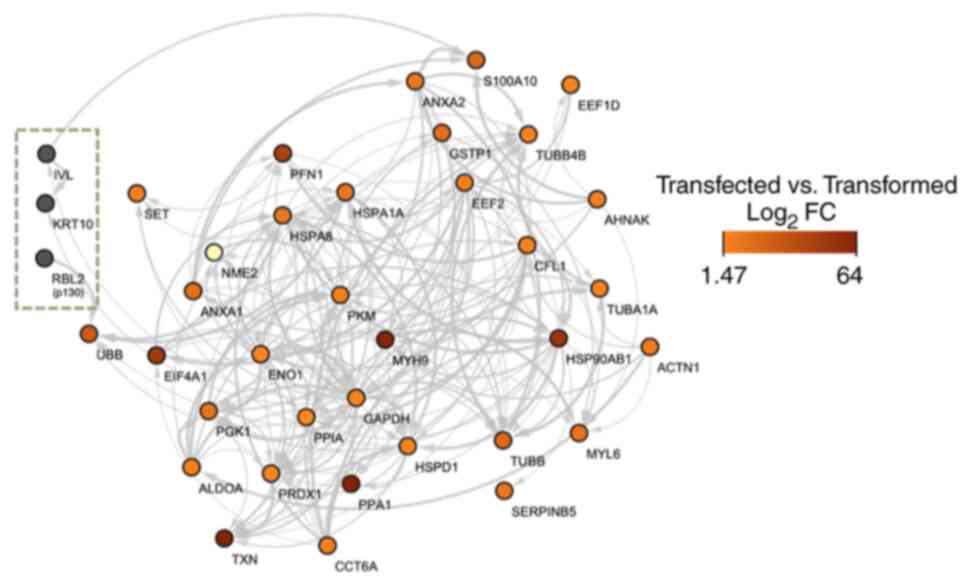
Interactions of upregulated proteins
in HPV-transfected cells and HPV-transformed cells with p130,
involucrin (IVL) and keratin 10
The upregulated proteins in HPV-transfected and
-transformed cells that interacted with p130, IVL and keratin 10
were S100A10, EEF1D, ANXA2, GSTP1, TUBB4B, EEF2, AHNAK, CFL1,
TUBA1A, HSP90AB1, ACTN1, PFN1, HSPA1A, HSPA8, PKM, MYH9, SET, NME2,
ANXA1, UBB, EIF4A1, ENO1, PGK1, ALDOA, TXN, PRDX1, PPIA, PPA1,
CCT6A, GAPDH, HSPD1, TUBB, MYL6 and SERPINB5.
The PPI network for upregulated proteins in
HaCaT-HPV16 E7 and HaCaT-HPV18 E7 cells were compared with the
target protein network, p130 (also known as RBL2), IVL and keratin
10. p130 was chosen based on its host protein localization, whereas
keratin 10 and IVL were selected as protein markers for cellular
differentiation. Based on the heat map analysis, PPIs between
HPV-transfected and -transformed cells, with their respective
log2 fold changes, were compared. The PPI network
demonstrated that p130 only interacted with polyubiquitin-B protein
(UBB), which suggested that UBB may be subjected to p130-mediated
degradation (Fig. 6). Furthermore,
the PPI network also demonstrated that IVL interacted with a
calcium-binding protein, S100A10.
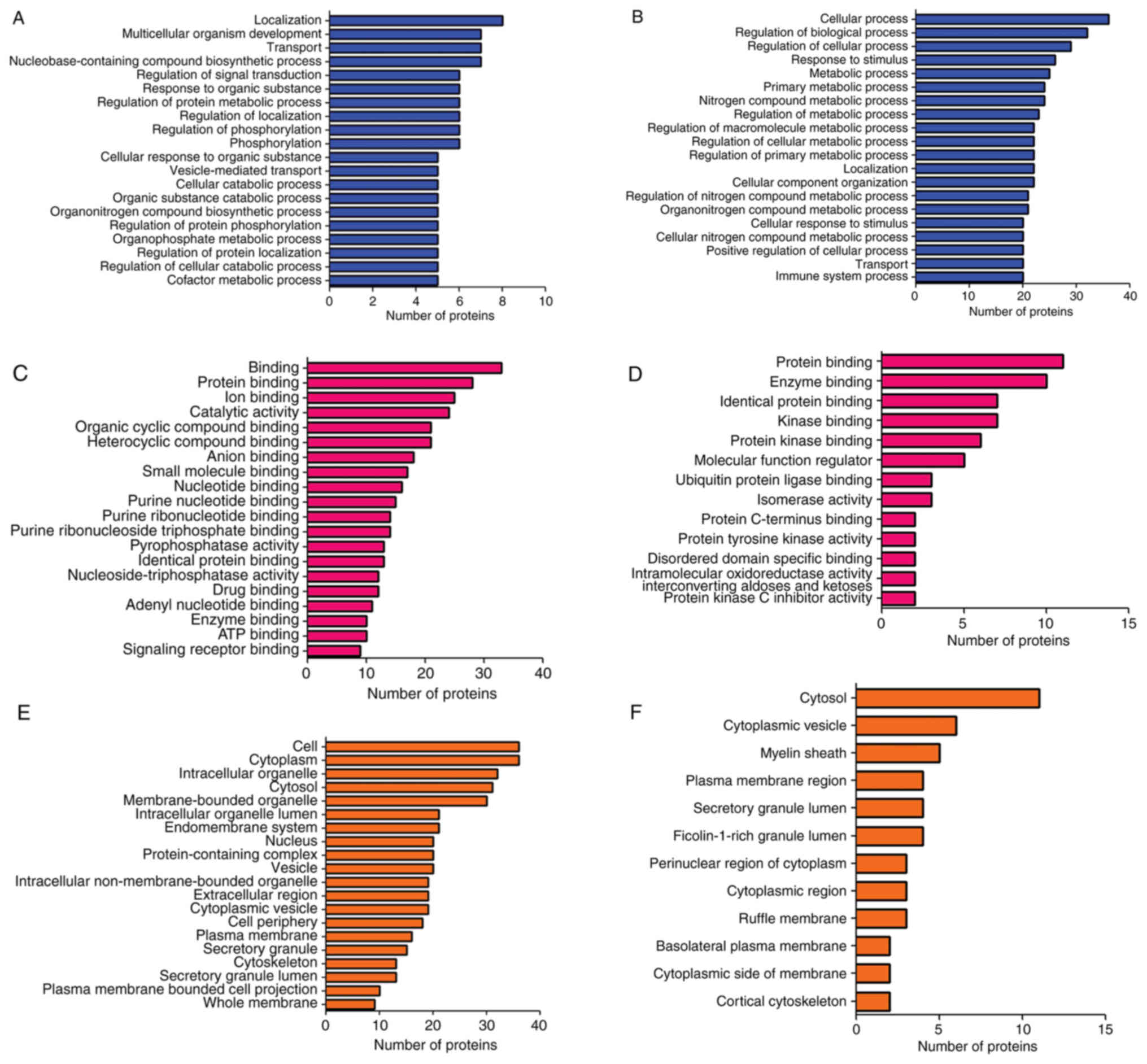
GO analysis
All 62 proteins from the LFQ analysis were subjected
to functional classification annotation. GO analysis was performed
to generate classification clusters in the categories Biological
Process (BP), Cellular Component (CC) and Molecular Function (MF).
The top upregulated and downregulated differentially expressed
proteins linked to BP (Fig. 7A and
B), MF (Fig. 7C and D) and CC
(Fig. 7E and F) were presented.
The subcategories and the number of proteins across 20 GO terms
were also labelled. For example, in the BP category, ‘cellular
process’ and ‘regulation of biological process’ both exceeded 30
upregulated proteins (Fig. 7A),
whereas the ‘localization’ subcategory had eight downregulated
proteins (Fig. 7B). In Fig. 7C, top 20 GO terms in the category
MF for the upregulated proteins are provided. A high protein count
was found in binding and protein binding categories with 34 and 28
proteins, respectively. The downregulated proteins in MF were
enriched in 13 GO terms. Protein and enzyme binding categories had
a high protein count with 12 and 11 proteins, respectively
(Fig. 7D). The top 20 GO terms
corresponding to the CC category for the upregulated proteins are
provided in Fig. 7E. Cell and
cytoplasm had the highest enrichment with 38 proteins, followed by
intracellular organelles, cytosol and membrane-bounded organelle
with 30, 31 and 32 proteins, respectively. The downregulated
proteins were enriched in 12 GO terms corresponding to CC with the
cytosol exhibiting the highest enrichment with 12 proteins,
followed by cytoplasmic vesicle with 7 proteins and myelin sheath
with 5 proteins (Fig. 7F).
The number of proteins in each subcategory was
determined; however, the value must be compared with the actual
number of occurrences and the expected number of occurrences for
each category to draw a reasonable conclusion.
Discussion
HPV E7 was the first oncogene of all the HPV
oncogenes to be identified (15).
E7 serves a crucial role in driving cells towards cancer and it may
trigger cancer characteristics during the process of viral genome
replication. Therefore, HPV E7 gene manipulation may be an
effective therapy in CC. Furthermore, transfection of keratinocytes
with the HPV E7 oncogene is a suitable model to study the oncogenic
changes induced by HPV infection.
In the present study, proteomics technology was used
to evaluate the protein expression profiles of HR-HPV E7 types
(HPV16 and 18) in HPV-transfected and -transformed cells. HaCaT
cells were transfected with recombinant HPV 16/18 E7 and
HPV-transformed cells CaSki and HeLa were used to represent native
infections with HPV16 and 18, respectively. Label-free proteomics
was performed to profile their protein contents. The results of the
present study demonstrated that GPI and ALDOA were common proteins
identified in all cell lines, with these proteins being involved in
the catalytic activity of glycolysis (37,38).
Glycolysis in tumor cells provides energy to support both rapid
proliferation and increased metabolic requirements for
macromolecule synthesis. GPI is a dimeric enzyme that acts in the
second step of glycolysis, where it catalyzes the conversion of
glucose-6-phosphate to fructose-6-phosphate (37). Furthermore, it also functions as a
cytokine/growth factor induced by c-Myc and HIF-1 (39), and is markedly expressed in
numerous types of cancer, such as bladder, colon, stomach, kidney,
lung and ovarian cancer, and lymphoma (40). ALDOA functions as a key enzyme
catalyzing the reversible reaction of fructose-1,6-bisphosphate to
glyceraldehyde-3-phosphate and dihydroxyacetone phosphate in
glycolysis. It also serves an essential role in ATP biosynthesis,
is ubiquitous in all organs or cells and is upregulated in numerous
types of cancer, including cervical adenocarcinoma (41).
The heat map generated based on the LFQ analysis
performed in the present study demonstrated that 37 proteins
(Cluster 1) were upregulated in HPV-transfected cells compared with
in control and HPV-transformed cells. These cluster 1 proteins were
further analyzed using volcano plot analysis, which demonstrated
that GAPDH, EEF1D, PRDX1, TUBA1A, AHNAK, EEF2, ANXA2, ANXA1, ACTN1,
MYL6, SERPINB5, HSPA8, EIF4A1, HSP90AB1 and PFN1 proteins were
significantly upregulated in transfected cells. A previous study
reported that TUBA1A, PFN1 and ANXA2 proteins were upregulated in
cervical squamous cell carcinoma, and HSPA8 and KRT8 were
downregulated (42). EEF1D has
previously been reported to be upregulated in various types of
cancer, such as colorectal carcinoma, esophageal carcinoma,
glioblastoma, glioma, liver, lymphoma, medulloblastoma, melanoma,
oral squamous cell carcinoma, osteosarcoma, prostate and papillary
renal cell carcinoma, thus indicating that the protein may act as a
promoter for cell proliferation and tumor growth (43). AHNAK acts as a tumor suppressor via
activation of the TGFβ/Smad3 signaling cascade, which arrests the
cell cycle in the G0/G1 phase and
downregulates c-Myc expression during cell growth (44). It is closely associated with
metastasis of aggressive tumors. A previous study reported
unregulated ANXA1 protein expression in cells transfected with
HPV16 E6/E7, which suggested its involvement in HPV-mediated
carcinogenesis (45). Furthermore,
SERPINB5 is differentially expressed in different cancer types. It
is upregulated in gastric adenocarcinoma, and breast, colon,
gallbladder, ovarian and pancreatic cancer, but downregulated in
prostate and gastric cancer (46).
SERPINB5 exhibits tumor-suppressive properties due to its nuclear
localization, binding to chromatin and inhibiting cancer cell
metastasis (47). Overexpression
of EIF4A1 has been reported to promote CC progression due to its
ATP-dependent RNA helicase activity in mRNA translation of
oncoproteins involved in cell apoptosis and proliferation (48). HSP90AB1 is required for cancer cell
invasion and migration, and its protein expression levels have been
reported to be upregulated in HPV-transfected cells (49).
There were four significant cancer hallmark pathways
identified through cancer hallmark enrichment analysis, the mTORC1
signaling pathway, MYC target VI, hypoxia and glycolysis. The
mTORC1 signaling pathway was the prominent hallmark in the present
study. In cancer, cells often use the mTOR signaling pathway as a
mechanism to enhance their proliferation. mTOR is a Ser/Thr kinase
that performs various functions, and is associated with growth
(increase in cell mass and size), proliferation, survival,
autophagy, metabolism and cytoskeletal organization (50). mTOR activity has been reported to
be dysregulated in numerous types of cancer as it serves a vital
role in the autophagy of tumor cells. Notably, the inhibitory
effect of rapamycin on mTOR activity may increase cell autophagic
flux and result in decreased tumor growth. Furthermore, previous
studies have demonstrated that rapamycin may promote the formation
of autophagosomes and induce autophagosome-lysosome fusion
(51,52). Ji and Zheng (53) reported that the mTOR signaling
pathway activated cervical carcinoma, and mTOR-specific small
interfering RNA was revealed to effectively suppress HeLa cell
proliferation via inhibiting the cell cycle and increasing
apoptosis, which is similar to the mechanism of action of
rapamycin. Rapamycin is a highly specific inhibitor of mTOR that
has been used to impede cell proliferation (54). HPV-transfected cells exhibit highly
reduced pRB protein expression levels as a consequence of
functional E6 and E7 expression (55). It was previously reported that
rapamycin resistance in the proliferation of human keratinocytes
expressing HPV16 was associated with the ability of E7 to induce
pRb degradation. HPV16 E7 was also reported to have conferred
resistance to the anti-proliferative effect of rapamycin. This was
also associated with the integrity of the LxCxE motif, which has
been reported to affect rapamycin resistance (56).
The second significant hallmark was the MYC target
VI pathway. MYC is a transcription factor that regulates multiple
human genes that promote cell proliferation (57). It also affects apoptosis via
alterations to the pro-and anti-apoptotic members of the BCL-2
family, activates telomerase and regulates the expression of
vascular endothelial growth factor, which is associated with
angiogenesis (58). These
downstream targets make MYC one of the most influential oncogenes.
Both mTORC1 signaling and MYC target VI (59) are involved in cell
proliferation.
The third cancer hallmark was the hypoxia signaling
pathway, which is governed by HIF stabilization. While adapting to
hypoxia, tumor cells can become more aggressive and become
resistant to therapeutics. Hypoxia induces changes to gene
expression and the subsequent proteome changes can have notable
effects on various functions, which may negatively affect patient
prognosis (60). Notably, slowly
dividing cells in hypoxic regions are able to escape most cytotoxic
drugs, as these treatments target rapidly dividing cells. Cancer
stem cells may also be present in poorly hypoxic regions, thus
ensuring epithelial-to-mesenchymal transition (61). Tumor cell survival under hypoxia or
substances which block HIF-1α and HIF-2α-linked signaling pathways
makes tumor cells adapt to hypoxia. They do so by inducing
metabolic reprogramming, improving the survival of tumor cells, and
supporting both angiogenesis and metastasis (62,63).
Previous studies have reported that high glucose concentrations (25
mM), which are a common occurrence in the blood of patients with
uncontrolled diabetes, may efficiently counteract hypoxic E6/E7
repression. The basis of glucose-linked effects on gene expression
is complex, and may involve epigenetic mechanisms and specific
transcription factors, such as MondoA/ChREBP-Mlx, NF-κB, c-Myc and
SP1 (64). It is necessary to
study how E6/E7 repression under hypoxia influences viral antigen
presentation on HPV-positive cancer cells, thereby assisting their
escape from host immune defense mechanisms.
Glycolysis was the final cancer hallmark addressed
in the present study. Viral proteins regulate the cell cycle via
interacting with the tumor suppressor proteins p53 and pRB. HPV
E6/E7, as well as E5 and E2, favor the Warburg effect and can
contribute to radioresistance and chemoresistance, supporting
glycolytic enzyme activities, Krebs cycle and respiratory chain
inhibition (65). These processes
lead to the accelerated production of ATP, which may satisfy the
energy demands of cancer cells during proliferation. In this
manner, HPV proteins may promote cancer hallmarks; however, it is
also possible that during early HPV infection, the Warburg effect
may aid efficient viral replication (66).
The present study demonstrated that PGK1 was present
in all cancer hallmark enrichment pathways, which indicated it as a
potential biomarker. Previous studies reported that the protein
expression levels of PGK1 were elevated in breast cancer (67), astrocytoma (68), metastatic colon cancer (69) and pancreatic ductal adenocarcinoma
(70). Furthermore, its mRNA
expression levels have been shown to be elevated in gastric cancer
(71). PGK1 is an essential enzyme
in aerobic glycolysis, which catalyzes the reversible transfer of a
phosphate group from 1,3-bisphosphoglycerate to ADP, thus producing
3-phosphoglycerate and ATP. PGK1 can affect the function of some
transcription factors, such as β-catenin, a tumor-associated
oncoprotein (72). PGK1 is the
upstream regulator of β-catenin, which affects tumor growth,
proliferation, invasion, metastasis, angiogenesis and drug
resistance (69,73,74).
Furthermore, PGK1 serves an important role in the tumor occurrence
and progression not only as a metabolic enzyme, but also as a
protein kinase. Mitochondrial PGK1 activates pyruvate dehydrogenase
kinase isoenzyme 1, through which tumor cells are able to inhibit
mitochondrial pyruvate metabolism and promote the Warburg effect
(75). Abnormal expression of PGK1
has been detected in tumor tissues, and also in peripheral blood
and saliva samples of patients (76). Therefore, PGK1 may be considered a
potential target for tumor therapy and may become a popular
molecule in tumor therapy research. However, the role of PGK1 in
different tumors may vary according to its tissue specificity and
associated level of expression. Furthermore, the development of
therapeutic drugs targeting PGK1 according to its function is also
an important consideration. Therefore, PGK1 has a broad research
potential in cancer, especially as a therapeutic target for
cervical cancer.
The PPI network analysis performed in the present
study demonstrated that p130 interacted exclusively with UBB, which
is subjected to p130-mediated degradation via proteasomal
degradation (77). The UBB protein
is part of the ubiquitin-proteasome system (UPS) that is associated
with the degradation of various intracellular proteins in
eukaryotic cells (78). It is also
involved in cellular signaling pathways, such as cell cycle
control, cell survival, proliferation, transcription, DNA repair,
apoptosis, cellular metabolism, membrane trafficking and
ubiquitination, which are vital for the immune response (79). E7 inactivates most cellular
substrates via the interaction of UPS components, leading to their
degradation at the proteasome (80). The present study demonstrated that
IVL interacted with a calcium-binding protein, S100A10. This
protein appears as a small dimeric helix-loop-helix tightly
associated with ANXA2 and S100A10 tends to be degraded in the
absence of ANXA2. It is involved in the intracellular post-entry
trafficking of several membrane-bound proteins (81). It has also been reported to mediate
the migration of macrophages to the tumor site (82).
The present study also investigated the functional
classification of differentially expressed proteins identified
using LFQ analysis to create three distinct clusters: BP, CC and
MF. The present study was able to distinguish proteins according to
the main clusters affected by HPV infection. Due to its
interactions with a wide range of proteins, the effects of E7 were
demonstrated across numerous cellular processes, including viral
replication, transformation, cell cycle and cell death (83,84).
The active site for binding of tumor suppressors with HPV E7
oncoprotein is within conserved region 2, encompassing the LxCxE
motif. This motif is responsible for binding with cellular targets.
Conserved regions 2 and 3 of HPV are responsible for the
degradation of tumor suppressors that ultimately lead to inhibition
of cell cycle arrest (85).
In conclusion, developments in HPV research have
continued to identify numerous mechanisms that can be exploited by
the virus to overcome cellular growth controls. At present,
development of accurate therapeutic targets for CC treatment still
poses a challenge. The results of the present study demonstrated
the importance of elucidating the involvement of multiple pathways
perturbed by HPV infection. Furthermore, it was demonstrated that
PGK1 was present across all cancer hallmark enrichment pathways.
Further investigations are required to identify other PGK1
functions, which could further evaluate its potential as a
therapeutic target in CC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Malaysian Ministry of
Higher Education through the Fundamental Research Grant Scheme
[grant no. FRGS/1/2016/SKK08/UM/02/16 (FP015-2016)].
Availability of data and materials
The data generated in the present study maybe
obtained from the MassIVE database (massive.ucsd.edu) under
accession no. MSV000079070. The direct URL is
ftp://massive.ucsd.edu/MSV000090470/. The datasets were summarized
in Table SI, Table SII, Table SIII, Table SIV, Table SV, Table SVI. The datasets used and/or
analyzed during the current study are available from the
corresponding author on reasonable request.
Authors' contributions
SG, NFMR, SO, SC and NNR contributed to the
conception and design of the present study. Material preparation,
data collection and analysis were performed by SG. NNR and MFMR
confirm the authenticity of all the raw data. The first draft of
the manuscript was written by SG. NNR, MFMR and SO contributed to
acquisition of data. SC assisted with data analysis and
interpretation. NNR, MFMR, SO and SC critically reviewed the
manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Berman TA and Schiller JT: Human
papillomavirus in cervical cancer and oropharyngeal cancer: One
cause, two diseases. Cancer. 123:2219–2229. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sand FL, Munk C, Jensen SM, Svahn MF,
Frederiksen K and Kjaer SK: Long-Term risk for noncervical
anogenital cancer in women with previously diagnosed high-grade
cervical intraepithelial neoplasia: A danish nationwide cohort
study. Cancer Epidemiol Biomarkers Prev. 25:1090–1097. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fuller KM and Hinyard L: Factors
associated with HPV vaccination in young males. J Community Health.
42:1127–1132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haverkos HW, Haverkos GP and O'Mara M:
Co-carcinogenesis: Human papillomaviruses, coal tar derivatives,
and squamous cell cervical cancer. Front Microbiol. 8:22532017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Muñoz N, Bosch FX, De Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ;
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group, : Epidemiologic classification of human
papillomavirus types associated with cervical cancer. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Della Fera AN, Warburton A, Coursey TL,
Khurana S and McBride AA: Persistent human papillomavirus
infection. Viruses. 13:3212021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Songock WK, Kim SM and Bodily JM: The
human papillomavirus E7 oncoprotein as a regulator of
transcription. Virus Res. 231:56–75. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Graham SV and Faizo AAA: Control of human
papillomavirus gene expression by alternative splicing. Virus Res.
231:83–95. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong HS, Akhavan J, Lee SH, Kim RH, Kang
MK, Park NH and Shin KH: Proinflammatory cytokine TNFα promotes
HPV-associated oral carcinogenesis by increasing cancer stemness.
Int J Oral Sci. 12:32020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Richards KH, Wasson CW, Watherston O,
Doble R, Eric Blair G, Wittmann M and Macdonald A: The human
papillomavirus (HPV) E7 protein antagonises an Imiquimod-induced
inflammatory pathway in primary human keratinocytes. Sci Rep.
5:129222015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Freitas AC, de Oliveira THA, Barros MR
Jr and Venuti A: hrHPV E5 oncoprotein: Immune evasion and related
immunotherapies. J Exp Clin Cancer Res. 36:712017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yeo-The NSL, Ito Y and Jha S: High-risk
human papillomaviral oncogenes E6 and E7 target key cellular
pathways to achieve oncogenesis. Int J Mol Sci. 19:17062018.
View Article : Google Scholar
|
|
13
|
Fischer M, Uxa S, Stanko C, Magin TM and
Engeland K: Human papilloma virus E7 oncoprotein abrogates the
p53-p21-DREAM pathway. Sci Rep. 7:26032017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bienkowska-Haba M, Luszczek W, Zwolinska
K, Scott RS and Sapp M: Genome-Wide transcriptome analysis of human
papillomavirus 16-infected primary keratinocytes reveals subtle
perturbations mostly due to E7 protein expression. J Virol.
94:e01360–19. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pal A and Kundu R: Human papillomavirus E6
and E7: The cervical cancer hallmarks and targets for therapy.
Front Microbiol. 10:31162020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cuninghame S, Jackson R and Zehbe I:
Hypoxia-inducible factor 1 and its role in viral carcinogenesis.
Virology. 456–457. 370–383. 2014.PubMed/NCBI
|
|
17
|
Gunasekharan VK, Li Y, Andrade J and
Laimins LA: Post-Transcriptional regulation of KLF4 by high-risk
human papillomaviruses is necessary for the
differentiation-dependent viral life cycle. PLoS Pathog.
12:e10057472016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sen P, Ganguly P and Ganguly N: Modulation
of DNA methylation by human papillomavirus E6 and E7 oncoproteins
in cervical cancer. Oncol Lett. 15:11–22. 2018.PubMed/NCBI
|
|
19
|
Gandhi S, Nor Rashid N, Mohamad Razif MF
and Othman S: Proteasomal degradation of p130 facilitate cell cycle
deregulation and impairment of cellular differentiation in
high-risk Human Papillomavirus 16 and 18 E7 transfected cells. Mol
Biol Rep. 48:5121–5133. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Olson BJSC: Assays for determination of
protein concentration. Curr Protoc Pharmacol. 73:A.3A.1–A.3A.32.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan CH, Tan KY and Tan NH: Revisiting
Notechis scutatus venom: On shotgun proteomics and neutralization
by the ‘bivalent’ Sea Snake Antivenom. J Proteomics. 144:33–38.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zainal Abidin SA, Rajadurai P, Chowdhury
ME, Ahmad Rusmili MR, Othman I and Naidu R: Proteomic
characterization and comparison of malaysian tropidolaemus wagleri
and cryptelytrops purpureomaculatus venom using shotgun-proteomics.
Toxins (Basel). 8:2992016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Xin L, Shan B, Chen W, Xie M,
Yuen D, Zhang W, Zhang Z, Lajoie GA and Ma B: PEAKS DB: de novo
sequencing assisted database search for sensitive and accurate
peptide identification. Mol Cell Proteomics. 11:M111.010587. 2012.
View Article : Google Scholar
|
|
25
|
Ma B, Zhang K, Hendrie C, Liang C, Li M,
Doherty-Kirby A and Lajoie G: PEAKS: Powerful software for peptide
de novo sequencing by tandem mass spectrometry. Rapid Commun Mass
Spectrom. 17:2337–2342. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Levin Y, Schwarz E, Wang L, Leweke FM and
Bahn S: Label-free LC-MS/MS quantitative proteomics for large-scale
biomarker discovery in complex samples. J Sep Sci. 30:2198–2203.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Washburn MP, Wolters D and Yates JR Jr:
Large-scale analysis of the yeast proteome by multidimensional
protein identification technology. Nat Biotechnol. 19:242–247.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alsallakh B, Aigner W, Miksch S and Hauser
H: Radial sets: Interactive visual analysis of large overlapping
sets. IEEE Trans Vis Comput Graph. 19:2496–2505. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tyanova S, Temu T, Sinitcyn P, Carlson A,
Hein MY, Geiger T, Mann M and Cox J: The Perseus computational
platform for comprehensive analysis of (prote)omics data. Nat
Methods. 13:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liberzon A, Birger C, Thorvaldsdottir H,
Ghandi M, Mesirov JP and Tamayo P: The molecular signatures
database (MSigDB) hallmark gene set collection. Cell Syst.
1:417–425. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Statist Soc B. 57:289–300. 1995.
|
|
33
|
Lin Y, Zhang J, Cai J, Liang R, Chen G,
Qin G, Han X, Yuan C, Liu Z, Li Y, et al: Systematic analysis of
gene expression alteration and co-expression network of eukaryotic
initiation factor 4A-3 in cancer. J Cancer. 9:4568–4577. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zeng J, He SL, Li LJ and Wang C: Hsp90
up-regulates PD-L1 to promote HPV-positive cervical cancer via
HER2/PI3K/AKT pathway. Mol Med. 27:1302021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shan N, Zhou W, Zhang S and Zhang Y:
Identification of HSPA8 as a candidate biomarker for endometrial
carcinoma by using iTRAQ-based proteomic analysis. Onco Targets
Ther. 9:2169–2179. 2016.PubMed/NCBI
|
|
36
|
Potriquet J, Laohaviroj M, Bethony JM and
Mulvenna J: A modified FASP protocol for high-throughput
preparation of protein samples for mass spectrometry. PLoS One.
12:e01759672017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fermo E, Vercellati C, Marcello AP,
Zaninoni A, Aytac S, Cetin M, Capolsini I, Casale M, Paci S,
Zanella A, et al: Clinical and molecular spectrum of
glucose-6-phosphate isomerase deficiency. report of 12 new cases.
Front Physiol. 10:4672019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kawai K, Uemura M, Munakata K, Takahashi
H, Haraguchi N, Nishimura J, Hata T, Matsuda C, Ikenaga M, Murata
K, et al: Fructose-bisphosphate aldolase A is a key regulator of
hypoxic adaptation in colorectal cancer cells and involved in
treatment resistance and poor prognosis. Int J Oncol. 50:525–534.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ždralević M, Marchiq I, de Padua MMC,
Parks SK and Pouysségur J: Metabolic plasiticy in cancers-distinct
role of glycolytic enzymes GPI, LDHs or membrane transporters MCTs.
Front Oncol. 7:3132017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han J, Deng X, Sun R, Luo M, Liang M, Gu
B, Zhang T, Peng Z, Lu Y, Tian C, et al: GPI is a prognostic
biomarker and correlates with immune infiltrates in lung
adenocarcinoma. Front Oncol. 11:7526422021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Saito Y, Takasawa A, Takasawa K, Aoyama T,
Akimoto T, Ota M, Magara K, Murata M, Hirohashi Y, Hasegawa T, et
al: Aldolase A promotes epithelial-mesenchymal transition to
increase malignant potentials of cervical adenocarcinoma. Cancer
Sci. 111:3071–3081. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Saritha V, Abdul J, Arun S and Meenakshi
SK: Analysis of differentially expressed proteins in the exfoliated
cells of normal and squamous cell carcinoma of the uterine cervix
to define candidate markers for cervical cancer. Int J Bioc
Biotechnol. 5:626–636. 2016.
|
|
43
|
Xu H, Yu S, Peng K, Gao L, Chen S, Shen Z,
Han Z, Chen M, Lin J, Chen S and Kang M: The role of EEF1D in
disease pathogenesis: A narrative review. Ann Transl Med.
9:16002021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sohn M, Shin S, Yoo JY, Goh Y, Lee IH and
Bae YS: Ahnak promotes tumor metastasis through transforming growth
factor-β-mediated epithelial-mesenchymal transition. Sci Rep.
8:143792018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Calmon MF, Sichero L, Boccardo E, Villa LL
and Rahal P: HPV16 E6 regulates annexin 1 (ANXA1) protein
expression in cervical carcinoma cell lines. Virology. 496:35–41.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Manawapat-Klopfer A, Thomsen LT, Martus P,
Munk C, Russ R, Gmuender H, Frederiksen K, Haedicke-Jarboui J,
Stubenrauch F, Kjaer SK and Iftner T: TMEM45A, SERPINB5 and
p16INK4A transcript levels are predictive for development of
high-grade cervical lesions. Am J Cancer Res. 6:1524–1536.
2016.PubMed/NCBI
|
|
47
|
Chang IW, Liu KW, Ragunanan M, He HL,
Shiue YL and Yu SC: SERPINB5 Expression: Association with CCRT
Response and Prognostic Value in Rectal Cancer. Int J Med Sci.
15:376–384. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liang S, Ju X, Zhou Y, Chen Y, Ke G, Wen H
and Wu X: Downregulation of eukaryotic initiation factor 4A1
improves radiosensitivity by delaying DNA double strand break
repair in cervical cancer. Oncol Lett. 14:6976–6982.
2017.PubMed/NCBI
|
|
49
|
Wang H, Deng G, Ai M, Xu Z, Mou T, Yu J,
Liu H, Wang S and Li G: Hsp90ab1 stabilizes LRP5 to promote
epithelial-mesenchymal transition via activating of AKT and
Wnt/β-catenin signaling pathways in gastric cancer progression.
Oncogene. 38:1489–1507. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Saxton RA and Sabatini DM: mTOR signaling
in growth, metabolism, and disease. Cell. 168:960–976. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rezazadeh D, Norooznezhad AH, Mansouri K,
Jahani M, Mostafaie A, Mohammadi MH and Modarressi MH: Rapamycin
reduces cervical cancer cells viability in hypoxic condition:
Investigation of the role of autophagy and apoptosis. Onco Targets
Ther. 13:4239–4247. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dossou AS and Basu A: The emerging roles
of mTORC1 in macromanaging autophagy. Cancers (Basel). 11:14222019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ji J and Zheng PS: Activation of mTOR
signaling pathway contributes to survival of cervical cancer cells.
Gynecol Oncol. 117:103–108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lamming DW: Inhibition of the mechanistic
target of rapamycin (mTOR)-rapamycin and beyond. Cold Spring Harb
Perspect Med. 6:a0259242016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yim EK and Park JS: The role of HPV E6 and
E7 oncoproteins in HPV-associated cervical carcinogenesis. Cancer
Res Treat. 37:319–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rabachini T, Boccardo E, Andrade R, Perez
KR, Nonogaki S, Cuccovia IM and Villa LL: HPV-16 E7 expression
up-regulates phospholipase D activity and promotes rapamycin
resistance in a pRB-dependent manner. BMC Cancer. 18:4852018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chanvorachote P, Sriratanasak N and
Nonpanya N: C-myc contributes to malignancy of lung Cancer: A
potential anticancer drug target. Anticancer Res. 40:609–618. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xu J, Chen Y and Olopade OI: MYC and
breast cancer. Genes Cancer. 1:629–640. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Meyer N and Penn LZ: Reflecting on 25
years with MYC. Nat Rev Cancer. 8:976–990. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Roma-Rodrigues C, Mendes R, Baptista PV
and Fernandes AR: Targeting tumor microenvironment for cancer
therapy. Int J Mol Sci. 20:8402019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shibue T and Weinberg RA: EMT, CSCs, and
drug resistance: The mechanistic link and clinical implications.
Nat Rev Clin Oncol. 14:611–629. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Balamurugan K: HIF-1 at the crossroads of
hypoxia, inflammation, and cancer. Int J Cancer. 138:1058–1066.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Semenza GL: Hypoxia-inducible factors:
Mediators of cancer progression and targets for cancer therapy.
Trends Pharmacol Sci. 33:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Havula E and Hietakangas V: Glucose
sensing by ChREBP/MondoA-Mlx transcription factors. Semin Cell Dev
Biol. 23:640–647. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Martínez-Ramírez I, Carrillo-García A,
Contreras-Paredes A, Ortiz-Sánchez E, Cruz-Gregorio A and Lizano M:
Regulation of cellular metabolism by high-risk human
papillomaviruses. Int J Mol Sci. 19:18392018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Medda A, Duca D and Chiocca S: Human
papillomavirus and cellular pathways: Hits and targets. Pathogens.
10:2622021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang D, Tai LK, Wong LL, Chiu LL, Sethi
SK and Koay ES: Proteomic study reveals that proteins involved in
metabolic and detoxification pathways are highly expressed in
HER-2/neu-positive breast cancer. Mol Cell Proteomics. 4:1686–1696.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yan H, Yang K, Xiao H, Zou YJ, Zhang WB
and Liu HY: Over-expression of cofilin-1 and phosphoglycerate
kinase 1 in astrocytomas involved in pathogenesis of
radioresistance. CNS Neurosci Ther. 18:729–736. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ahmad SS, Glatzle J, Bajaeifer K, Bühler
S, Lehmann T, Königsrainer I, Vollmer JP, Sipos B, Ahmad SS,
Northoff H, et al: Phosphoglycerate kinase 1 as a promoter of
metastasis in colon cancer. Int J Oncol. 43:586–590. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Hwang TL, Liang Y, Chien KY and Yu JS:
Overexpression and elevated serum levels of phosphoglycerate kinase
1 in pancreatic ductal adenocarcinoma. Proteomics. 6:2259–2272.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zieker D, Königsrainer I, Tritschler I,
Löffler M, Beckert S, Traub F, Nieselt K, Bühler S, Weller M,
Gaedcke J, et al: Phosphoglycerate kinase 1 a promoting enzyme for
peritoneal dissemination in gastric cancer. Int J Cancer.
126:1513–1520. 2010.PubMed/NCBI
|
|
72
|
Rojas-Pirela M, Andrade-Alviárez D, Rojas
V, Kemmerling U, Cáceres AJ, Michels PA, Concepción JL and Quiñones
W: Phosphoglycerate kinase: Structural aspects and functions, with
special emphasis on the enzyme from Kinetoplastea. Open Biol.
10:2003022020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lowy AM, Clements WM, Bishop J, Kong L,
Bonney T, Sisco K, Aronow B, Fenoglio-Preiser C and Groden J:
β-Catenin/Wnt signaling regulates expression of the membrane type 3
matrix metalloproteinase in gastric cancer. Cancer Res.
66:4734–4741. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yamada T, Takaoka AS, Naishiro Y, Hayashi
R, Maruyama K, Maesawa C, Ochiai A and Hirohashi S: Transactivation
of the multidrug resistance 1 gene by T-cell factor 4/beta-catenin
complex in early colorectal carcinogenesis. Cancer Res.
60:4761–4766. 2000.PubMed/NCBI
|
|
75
|
Li X, Jiang Y, Meisenhelder J, Yang W,
Hawke DH, Zheng Y, Xia Y, Aldape K, He J, Hunter T, et al:
Mitochondria-translocated PGK1 functions as a protein kinase to
coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell.
61:705–719. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Yu X and Li S: Non-metabolic functions of
glycolytic enzymes in tumorigenesis. Oncogene. 36:2629–2636. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Kalejta RF and Shenk T:
Proteasome-dependent, ubiquitin-independent degradation of the Rb
family of tumor suppressors by the human cytomegalovirus pp71
protein. Proc Natl Acad Sci USA. 100:3263–3268. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Schwartz AL and Ciechanover A: Targeting
proteins for destruction by the ubiquitin system: Implications for
human pathobiology. Annu Rev Pharmacol Toxicol. 49:73–96. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Đukić A, Lulić L, Thomas M, Skelin J,
Bennett Saidu NE, Grce M, Banks L and Tomaić V: HPV oncoproteins
and the ubiquitin proteasome system: A signature of malignancy?
Pathogens. 9:1332020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gupta I, Singh K, Varshney NK and Khan S:
Delineating crosstalk mechanisms of the ubiquitin proteasome system
that regulate apoptosis. Front Cell Dev Biol. 6:112018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Taylor JR, Skeate JG and Kast WM: Annexin
A2 in virus infection. Front Microbiol. 9:29542018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Xia C, Braunstein Z, Toomey AC, Zhong J
and Rao X: S100 proteins as an important regulator of macrophage
inflammation. Front Immunol. 8:19082018. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
McLaughlin-Drubin ME, Huh KW and Munger K:
Human papillomavirus type 16 E7 oncoprotein associates with E2F6. J
Virol. 82:8695–8705. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
McLaughlin-Drubin ME, Meyers J and Munger
K: Cancer associated human papillomaviruses. Curr Opin Virol.
2:459–466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Aarthy M and Singh SK: Interpretations on
the interaction between protein tyrosine phosphatase and E7
oncoproteins of high and low-risk HPV: A computational perception.
ACS Omega. 6:16472–16487. 2021. View Article : Google Scholar : PubMed/NCBI
|