Ovary cancer (OC) is considered the most lethal
malignancy among gynecological cancers. In 2020 worldwide, OC
caused 1.6% of all new cancer-related deaths (1). Epithelial ovarian cancer (EOC) is a
clinical type of OC that is already diagnosed in 90% of women
patients with OC; patients with this OC type are typically
diagnosed in the advanced stages of the disease (75%) when cancer
has disseminated to a different abdominal tissue or metastases are
present. The majority of patients (>70%) with advanced-stage OC
do not respond to standard therapies, resulting in a resistant,
fatal disease (2). Due to this,
the identification and understanding of the molecular
characteristics associated with early disease progression,
prediction and clinical responses are necessary to improve survival
and clinical treatment in women with EOC.
In this regard, differential cancer DNA methylation,
compared with the origin tissue cells, is an early event during
carcinogenesis. That is, DNA methylation is composed of concomitant
global unmethylated DNA and local locus-methylated DNA. Methylated
DNA consists of the addition of a methyl group in the fifth carbon
of cytosine residues, forming a CpG; this addition forms
5-methylcytosine. Typical examples of DNA-methylation phenotypes
have been characterized in certain types of cancer, such as
colorectal carcinoma, breast carcinoma and glioma (3–8).
Methylated DNA molecules are more compact than unmethylated DNA
molecules (3). It has been
proposed that unmethylated DNA is a characteristic of cancer while
methylated DNA presents only a variable consequence depending on
the locus and on the specific part of the locus in cancer cells
(9,10). RNA transcription is strongly
influenced by DNA methylation. Methylated loci are silenced and
unmethylated loci are transcriptionally activated in ovarian
carcinoma tumors. The diagnosis of patients is made by transvaginal
ultrasound and detection of cancer antigen (CA)-125 levels;
however, the state of DNA methylation and RNA expression in the
tumors has been currently associated with the diagnosis of the
histological subtypes in the different types of ovarian carcinoma,
the advanced stages and, importantly, the survival of the
patients.
EOC is a heterogeneous carcinoma type and every EOC
subtype has its natural history of development. Specifically, cell
subtypes are described by histopathological and molecular
characteristics of ovarian carcinoma (11–13).
However, it is considered that serous ovarian carcinomas are
developed as a disease continuum, from low-grade to high-grade
serous ovarian carcinomas. Evidence suggests that high-grade and
low-grade serous carcinomas develop independently in their natural
course of progression and they have different prognoses (12,13).
At the molecular level, the prognosis of spontaneous
EOC type I and type II is associated with the EOC sub-type, the age
of the patient and the treatment used. Although advances in
clinical treatments have increased in the past few decades, the
pathology structure remains unaltered (14). Of patients diagnosed with ovarian
cancer, ~50% survive five years following diagnosis, including 29%
of those with metastases of ovarian carcinoma (1). In ovarian carcinomas, several factors
determine survival, including primarily histological subtype,
grade, stage, cytoreductive surgery and, secondarily, ethnicity
(15,16).
The mortalities associated with ovarian cancer
comprise 4% of all cancer-related deaths (1,14).
Germinal variations are changes in DNA locus. Germinal variants of
ovarian carcinomas are present in 5% of patients with ovarian
carcinoma. Ovarian carcinomas are mutated in the following two
ways: In germinal DNA (DNA variants of hereditary cancer origin) or
in somatic DNA (DNA variants with individual spontaneous tumor
cancer origin). Variants in the germ line DNA represent 24% of OC.
The majority of the genetic variants are present in BRCA in
hereditary breast and ovarian cancer syndrome (HBOC), whereas other
DNA repair genes are present in Lynch syndrome (LS), also called
hereditary non-polyposis colorectal cancer syndrome (17).
The loci mutations with higher hereditary penetrance
to develop ovarian carcinoma are those of BRCA1 or
BRCA2. HBOC accounts for ~80% of hereditary ovarian
carcinoma and 15% of epithelial OC cases. In HBOC, 65–85% of
cancers are due to genomic variants in BRCA1 and
BRCA2 genes, which are considered high penetrance for OC
(18). These genes encode proteins
for homologous recombination to repair DNA double-strand breaks and
maintain genomic stability. In addition, germinal carcinoma with
BRCA1/2 mutations develops a high-grade serous ovarian carcinoma
(HGSOC) subtype (15,16,19).
Other loci mutations with moderate familial penetrance involve
genes implicated in OC, such as BARD1, BRIP1, PALB2, RAD50,
RAD51C, NBN and MRE11A; these mutations are encoded in each
gene that has been involved in OC as part of the
BRCA2/Fanconi anemia signaling pathway (20). Epithelial OC deficiency in DNA
mismatch repair is the second most common cause of HBOC, accounting
for 10–15% of this condition. The lifetime risk of developing OC
with LS is ~8–12% and the mean age at presentation is ~43 years.
OC-associated genes in this pathway are the following: MLH1,
MSH2, MSH6 and PMS2. Specifically, BRIP1, MLH1, MSH2,
MSH6, PMS2 and EPCAM are moderate penetrance genes in OC
(16–19).
It is notable that each mutated locus develops
different characteristics in the phenotype of tumor cells present
in the patients (19–23). By contrast, EOCs developed from a
somatic spontaneous origin are heterogeneous. At the cellular
level, ovarian carcinoma tumors are classified into five different
types: HGSOC, low-grade serous ovarian carcinoma (LGSOC), mucinous
carcinoma (MC), clear cells carcinoma (CCC) and endometrioid
carcinoma (EC). Recently, DNA methylation and RNA transcription
have been shown to vary in a defined way to develop EOC tumors,
such as hypomethylated DNA and variation in RNA expression. It is
notable that germinal variation in the DNA locus of the large
non-coding RNA (lncRNA) HOTAIR is a risk cause of developing
ovarian carcinoma. In addition, overexpression of HOTAIR has
been found in ovarian carcinomas and it has been associated with
chemotherapy resistance (24).
Ovarian carcinoma, which is resistant to
radiotherapy and chemotherapy is another important problem. For
example, in HGSOC, the presence of the TP53 mutation and
chromosome instability (CIN) are associated with resistance to
radiotherapy and standard chemotherapy, which is a mix of
carboplatin and taxane (25). In
addition, it has been observed that DNA methylation loci induce
sensitivity to therapies. By contrast, the DNA methylation loci of
the nuclear RNA transcripts are associated with resistance to
treatment. In this regard, it has been proposed that ovarian
carcinoma cells could be sensitized to radiotherapy and
chemotherapy by the addition of DNA methylation inhibitors such as
decitabine (25).
The molecular characteristics validate the natural
history of ovarian carcinoma tumors and explain the association
between clinical characteristics of ovarian carcinoma, such as
mutations in the expression levels of DNA, RNA and proteins with
patient survival. The first characterization of ovarian carcinoma
is performed by quantifying transvaginal CA-125 levels using
ultrasonic waves (26).
Subsequently, the characterization of the macroscopic tumors in
surgery is required and finally the histological characteristics
have to be defined by microscopic observations. Finally, the
characterization of the genotype is proposed using nuclear
characteristics to improve diagnosis and prognosis, as well as to
confirm the natural history of the tumors. This is due to the
phenotype of the tumor cells being associated with DNA methylation.
Unmethylated DNA has been associated with nuclear size, aneuploidy,
carcinoma subtypes and higher proliferation of ovarian carcinoma
cells (27,28) [Table
I, (29–48)].
Currently, the following examples have been
demonstrated that indicate the DNA methylation status of ovarian
carcinomas and describe the hypomethylated nuclear locus in the
tumors compared with that of ovarian epithelial cells: The global
loci markers (satellite sequences and ALU repetitive
sequences) and the local loci markers. The assays used to
discriminate the state of methylation currently available in human
tumors are the following: Sodium bisulfite DNA treatment and
pyrosequencing, reverse transcription-quantitative PCR (RT-qPCR),
or methylation-specific PCR. High-resolution methods to determine
cell single DNA methylation are currently available, such as
droplet and digital PCR (43–50).
DNA methylation and RNA expression are associated
with ovarian carcinoma cells. This indicates that RNA transcription
could be inhibited due to the DNA methylation status of the locus,
except for certain recent paradoxical examples led by a negative
correlation in other types of cancers, where intragenic methylation
correlates with gene overexpression (Table II) (51,52).
In contrast to these observations, RNA expression is activated in
the unmethylated DNA status of the locus (9). Therefore, RNA transcription is
differentially present in ovarian carcinoma. MicroRNAs (miRs) are a
class of small RNA transcripts (19–22 nucleotides) that decrease
gene expression via translational inhibition or degradation of
target messenger RNA (mRNA). Various miRs are differentially
expressed in cancer, suggesting a link between these molecules and
the different expression levels of proteins in cancer tissues
(53). By contrast, large
non-coding RNAs (lncRNAs) function on affecting the nuclear
structure and RNA transcription. It is notable that the
hypermethylated DNA of RNA loci decrease the presence of miRs and
MEG3 (53–55), which is a lncRNA, so that the RNA
transcripts in the normal ovarian tissue, benign epithelial tumors,
benign epithelial ovarian cysts, malignant ovarian carcinoma and
serous ovarian carcinoma exhibit differential expression of RNA
transcripts (56–59).
It remains to be determined why DNA methylation in
ovarian carcinomas is heterogeneous. In 2014, the World Health
Organization classification guidelines for female reproductive
tumors defined the ovarian carcinoma type in cell-level
characterized ovarian carcinoma subtypes HGSOC, LGSOC, CCC, MC and
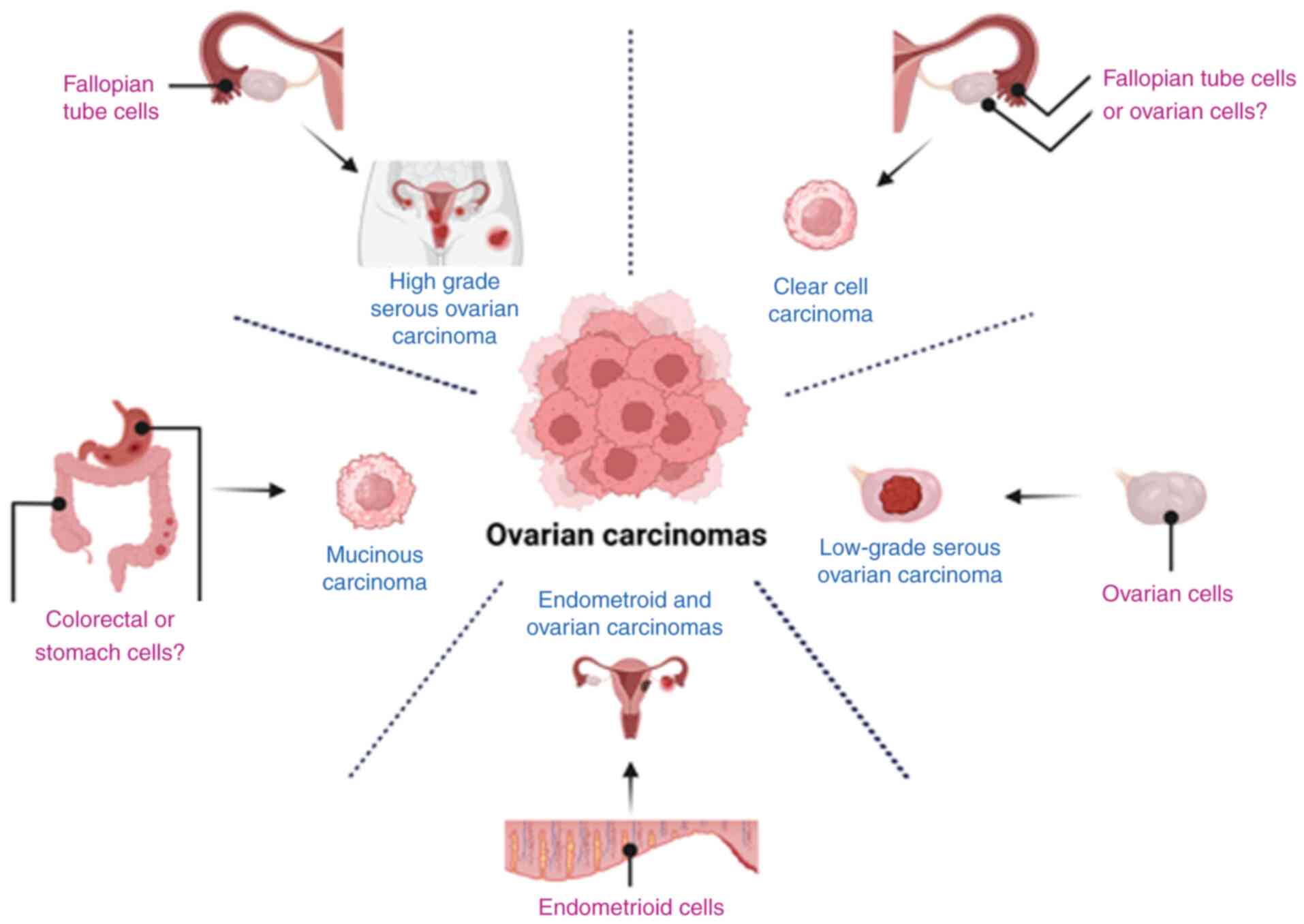
EC (11–13,21,60).
It was proposed that at the molecular level, the natural history of
ovarian carcinomas is HGSOC. A fallopian tube epithelial origin was
found when DNA methylation was compared (61,62).
HGSOC has locally DNA methylations in 6 loci that differentiate
HGSOC from the OSE DNA methylation pattern (ARMCX1, ICAM4,
LOC134466, PEG3, PYCARD and SGNE1) (41); HGSOC overexpresses miR-223,
miR-551b-3p, miR-30a-5p, miR-9 and miR-30a-5p (63–70).
Other studies using miR microarrays have described the different
expression patterns of serous ovarian carcinoma (70) and clear cell carcinoma (CCC),
specifically the SFRP1 methylated locus (41,71,72).
By contrast, DNA methylation in low-grade serous ovarian carcinoma
has not been reported to date compared with other ovarian carcinoma
subtypes or epithelial ovarian cells. By contrast, it has been
shown that endometrioid carcinoma (EC) has similarities with
endometrial and ovarian carcinomas in the promoter hypermethylated
locus (73–76). CCC has been characterized by the
HNF1 pathway to be unmethylated, whereas the RE alpha pathway is
unmethylated, similar to OSE (71). Finally, mucinous carcinomas (MUC)
exhibit 81 unmethylated genes that are different from those of
HGSOC. It is notable that MUC-DNA methylation is more similar to
colorectal and stomach carcinoma than HGSOC, providing additional
information on the MUC origins from colorectal metaplasia (12,77).
Downregulation of miR-192 and miR-2215 levels is also noted in MUC
(78) (Fig. 1).
In conclusion, DNA methylation is a characteristic
that varies early in the development of ovarian carcinoma cells.
The vestige of a methylated locus in the origin tissues of ovarian
carcinomas marks a directional methylation of every ovarian
carcinoma subtype. DNA methylation and RNA expression are
associated. Finally, DNA methylation and RNA transcripts are
associated and differentially presented by the subtype cells.
The following factors are associated with the
prognosis of patients with ovarian carcinoma: The variations in DNA
syndromes, the ethnic origin, the origin of the gynecological
pathologies, the subtypes, the advanced stages of the tumors
(metastasis to lymph node, or metastasis to distant tissues), the
size of residual tumor following cytoreductive surgery and the
resistance of cancer cells to radiotherapy and chemotherapy. It is
notable that 75% of ovarian carcinomas are of HGSOC sub-type and
the patient 5-year survival following diagnosis with ovarian
carcinoma is ~47%. In comparison, the survival of the women
diagnosed with metastasis of ovarian carcinoma is only 29%
(11,15,21–23,79,80).
The RNA expression could be driven by a random
accumulation or by a directional and defined development as
determined by the stages of International Federation of Gynecology
and Obstetrics (FIGO) (81) in
every molecular ovarian carcinoma subtype. Several pieces of
evidence have concluded that RNA transcripts are overexpressed and
downregulated in a directional way (Tables II and III). First, this has been presented in
the diagnosis biomarkers without poor prognosis. Except for the
BRCA1/2 mutation and the DNA methylation locus associated
with response to chemotherapy, the RNA transcripts are not related
to the prognosis of the patients or the single nucleotide
polymorphism in HOXA11 that protects cells from developing
HGSOC (82). This suggests that
biologically, hereditary mutations have a higher risk weight than
DNA methylation or RNA expression, which are more sensitive to
alterations of the phenotype state but not of the patient's
outcome. The nuclear characteristics determined of the advanced
FIGO stages of ovarian carcinoma are CIN, DNA methylated locus in
genes, such as BRCA1, FANCF, RASSF1A and Wnt5A, the
downregulated levels of TUBA14B and the overexpressed RNA
transcripts of the following genes and lncRNAs: MGMT, OSMR,
ESR1 and FOXL2 and long non-coding (lnc)BRM, HOTAIR,
HOXDAS-1 and lncSOX4 and CPS1-IT1 (83–87)
(Table IV).
The assays used to analyze RNAs are transcriptomics
or expression analysis of single locus transcription detected by
RT-qPCR (88,89). It is notable that the overexpressed
and downregulated transcripts associated with tumor development
classified by FIGO stages could probably result in equilibration of
their levels in the nuclear structure; in addition, the variation
in transcript levels within patients has to be taken into account;
for example, the levels of MLK7-AS1 and TUG1 were
increased 2.5- and 2.2-fold, respectively; the levels of
CASC2 were diminished 0.6-fold compared with the relative
expression noted in advanced stage ovarian carcinoma cells
(90–98). These types of variation in the RNA
expression levels are individual assessments in ovarian carcinoma
subtypes derived from a population, which are defined by comparison
of the expression levels of their corresponding counterparts.
However, in the majority of the studies, the ovarian cancer cells
are used as a point of calibration (66,67,98–162).
Molecular and cellular biology allows the improved
understanding of the microscopic and macroscopic observations of
ovarian carcinomas. The selection of the therapeutic methods, such
as surgery, radiotherapy and chemotherapy, depends on several
factors, such as the carcinoma grade, FIGO stage and patient
characteristics, namely age (81).
Surgery is performed to obtain the tumor via cytoreduction, which
involves extracting carcinoma cells from the patient. Chemotherapy
is subsequently administered following cytoreduction in cases of
advanced ovarian carcinoma. By contrast, radiation therapy uses
high-energy particles to destroy tumor cells, either directly or
indirectly, to inhibit further cell growth. Concomitantly,
radiotherapy has been commonly used as a first-line treatment for
ovarian carcinoma until the 1990s. It is now rarely used alone and
is typically used with surgery (191). However, radiotherapy can still be
beneficial in certain ways, such as reducing tumor size prior to
surgery, treating areas where cancer has spread and providing
palliative care (192,193). Ovarian carcinoma cells are
radiosensitive at the early stages of development (194), notably in low-grade carcinoma
subtypes such as EC (195).
Chemotherapy presents a challenging environment for
cells, aiming to eliminate tumor cells and improve the prognosis
for patients with cancer. The standard chemotherapy treatment for
ovarian carcinoma combines carboplatin and paclitaxel. Chemotherapy
can alter the nuclear structure, DNA methylation and RNA expression
in ovarian carcinoma cells (203–205). To date, the association of
methylation with the incidence of cancer in patients is not
directly known. However, DNA methylation is associated with
chromosomal instability of high-grade serous ovarian carcinoma.
This is the most aggressive subtype of ovarian cancer. It can be
inferred that by understanding the relationship between methylation
and CIN, the progression of ovarian cancer can be predicted. For
example, it is known that the HGSOC subtype with TP53
mutation and CIN is associated with DNA hypermethylation in
patients with poorer prognosis (206–213). HGSOC is characterized by
TP53 mutation and CIN and is linked to chemotherapy
resistance; it is also considered to be the more resistant and
heterogeneous subtype of ovarian carcinomas (214). For example, treatment with
paclitaxel in specific cell line models induces overexpression of
mdr1 and the lncRNAs UCA1 and long intergenic non-coding RNA
linc00312 (215).
Paclitaxel is a drug that inhibits depolymerization
of microtubules. It arrests cells in the G2/M phase. It
also induces CIN, leading to cell death. However, certain ovarian
carcinoma cells are resistant to paclitaxel. Certain miR
biomarkers, such as miR-134 and miR-224-5p, are associated with EOC
resistance to paclitaxel chemotherapy with 85 and 90% sensitivity,
respectively (94,102). Studies in ovarian carcinoma have
shown that RNA transcript levels are altered during paclitaxel
treatment in ovarian cancer cells. For example, treatment of A2780,
OVCAR3, SKOV3, and SW626 cells with paclitaxel induces
overexpression of mdr1 (215), UCA1 and
lincRNA00312.
Ovarian carcinoma is a type of cancer resulting from
tissue transformation and can occur both in hereditary (20%) and
sporadic (80%) forms. DNA methylations repress RNA transcription
differentially in ovarian carcinoma subtypes. This suggests that
DNA methylation of ovarian carcinoma subtypes is more similar to
their tissue of origin with regard to their nuclear
characteristics. Ovarian carcinoma presents a complex molecular
landscape where DNA methylation and RNA expression play crucial
roles in the disease development, progression and treatment
response. DNA methylation serves as both an early event in
carcinogenesis and a key regulator of gene expression, either
silencing or activating RNA transcription based on the methylation
status of specific loci.
The characteristics of nuclear vestiges vary in a
directional range of RNA transcripts. In addition, the directional
way of variation of the RNA transcripts is directed by FIGO stages
observed in every carcinoma subtype. The molecular ovarian
carcinoma subtypes are five and can be classified as follows:
High-grade serous, low-grade serous, endometrioid, mucinous and
clear cell; however, there are other subtypes to be described since
they belong in the five subtypes or other similar subtypes, such as
carcinosarcoma (analogous to malignant mixed Mullerian/mesodermal
tumors) and malignant Brenner tumors. Other cellular subtypes are
currently in discovery. This is particularly relevant in HGSOC,
where distinct methylation patterns have been identified, linking
them to tumor origin, subtype differentiation and patient
prognosis. The heterogeneity of ovarian carcinoma is further
exemplified by the differential expression of miRs and lncRNAs,
which are involved in gene regulation.
The molecular prognosis of ovarian carcinoma varies.
Firstly, hereditary ovarian carcinoma exhibits a greater effect
than sporadic ovarian carcinoma. By contrast, the survival of each
patient with spontaneous ovarian carcinoma depends on the carcinoma
development and the subtype. At a molecular level, deviations in
the DNA methylation and RNA transcription have been associated with
metastasis stages and the development of therapy-resistant cancer
cells. These findings suggest that molecular signatures, including
miR and lncRNA profiles, could serve as valuable biomarkers for
diagnosis, prognosis and therapeutic targeting in ovarian
carcinomas. Despite the advances in the understanding of the
molecular underpinnings of ovarian carcinomas, the disease
prognosis for patients, notably those diagnosed with advanced-stage
or metastatic disease remains poor, with survival rates being
markedly lower among these groups.
Current diagnostic and therapeutic strategies are
increasingly incorporating molecular data, including RNA
transcriptomics and DNA methylation analysis, to improve the
precision of treatment approaches. While hereditary mutations, such
as those noted in BRCA1/2, confer a significant risk,
epigenetic changes, notably in the later stages of the disease, are
critical in shaping the tumor phenotype and response to therapy. In
clinical practice, evidence leads to the hypothesis that certain
genes could be used as a combination to adjust cancer treatments.
However, a pair of primers may not provide a clinical solution.
Therefore, practical techniques, such as PCR and sequencing, are
used for validation of the next biological characterization of
chromosomes. In the present review, it was hypothesized that
chromosome instability, which is characterized by gain of
chromosomes in cancer cells and the loss of specific chromosomes or
their translocation, should be the focus of future research. For
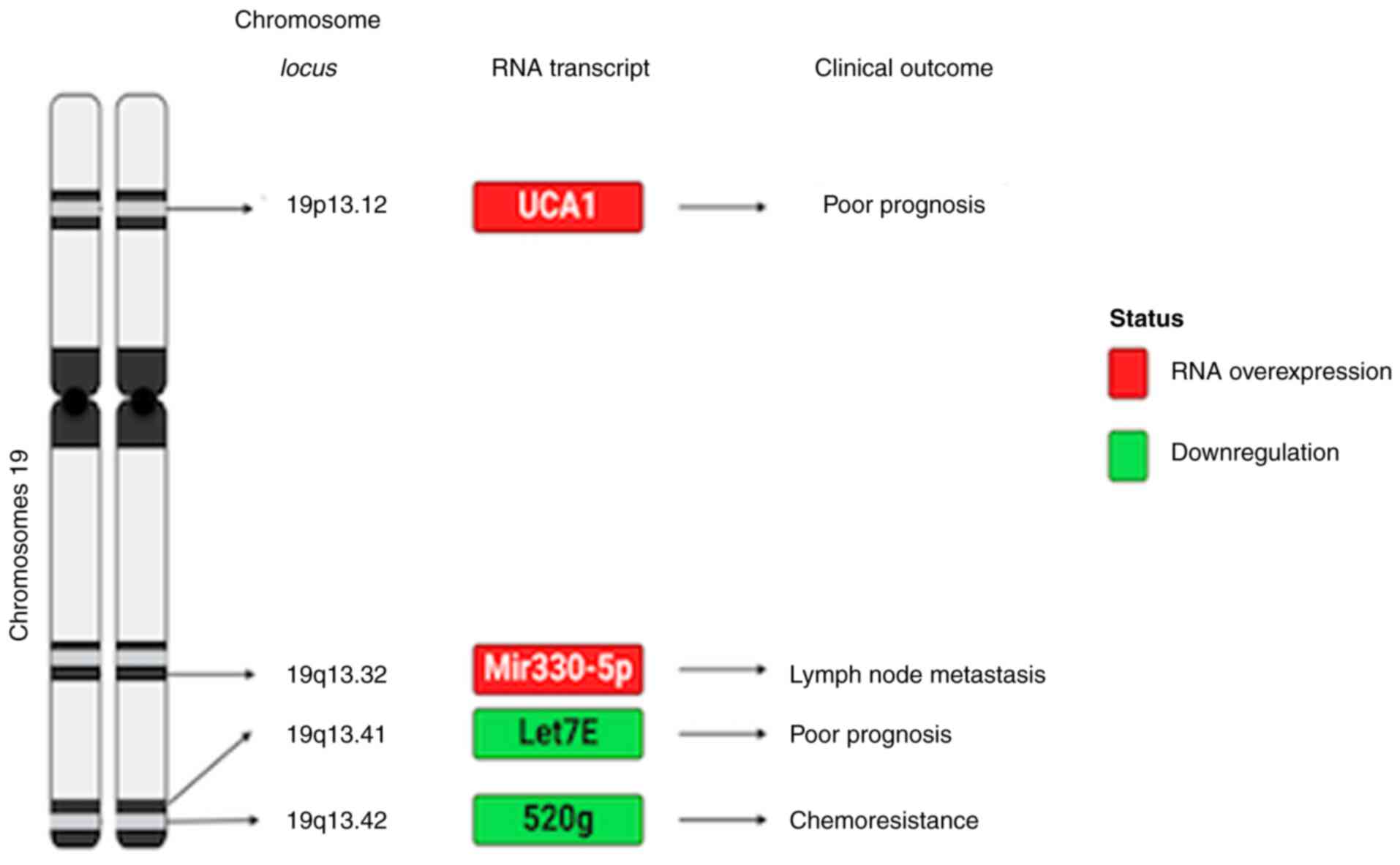
example, changes in DNA methylation of chromosomes 9 and 19
(Figs. 2 and 3) in high-grade serous ovarian carcinoma
are associated with ovarian carcinoma progression and resistance to
treatments.
Ultimately, a comprehensive understanding of the
molecular heterogeneity in ovarian carcinoma, including the roles
of DNA methylation and RNA transcription into the nucleus, is
essential to develop a vision for more effective treatments,
increase the understanding of disease progression and improve
long-term outcomes for patients. The continued exploration of these
molecular pathways holds the potential not only to revolutionize
but also to reform the clinical management of ovarian
carcinomas.
This article is part of the productivity of LCE as a
PhD student of Biological Sciences Postgrade Program, in the
Biomedicine Field.
This article was supported by the Programa de Posgrado en
Ciencias Biológicas, UNAM. This article is part of the productivity
work of the Ph.D. student LCE in the Programa de Posgrado en
Ciencias Biológicas, UNAM, and received a fellowship from CONACYT
Currículum Vitae Único (CVU)- 1003211. National Cancer Institute,
México (INCAN) and CONAHCYT supported this project (grant nos.
CMIC: 295466 and CBF 2023-2024-4004). The content is solely the
authors´s responsibility and does not necessarily represent the
official views of the National Cancer Institute of México.
Institutional Review Board Statement.
Not applicable.
Conceptualization and original draft preparation:
VMDCF, VFO, JDC, LAH and LCE. Writing and revising the manuscript
JAGR, APT, VFO, LAH and VMDCF. Supervision, project administration
and funding acquisition: JDC and LAH. All authors have read and
approved the final version of the manuscript. Data authentication
is not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Momenimovahed Z, Tiznobaik A, Taheri S and
Salehiniya H: Ovarian cancer in the world: epidemiology and risk
factors. Int J Womens Health. 11:287–299. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luo Y, Huang J, Tang Y, Luo X, Ge L, Sheng
X, Sun X, Chen Y and Zhu D: Regional methylome profiling reveals
dynamic epigenetic heterogeneity and convergent hypomethylation of
stem cell quiescence-associated genes in breast cancer following
neoadjuvant chemotherapy. Cell Biosci. 9:162019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sandhu R, Roll JD, Rivenbark AG and
Coleman WB: Dysregulation of the Epigenome in Human Breast Cancer':
Contributions of gene-specific DNA hypermethylation to breast
cancer pathobiology and targeting the breast cancer methylome for
improved therapy. Am J Pathol. 185:282–292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maire CL, Fuh MM, Kaulich K, Fita KD,
Stevic I, Heiland DH, Welsh JA, Jones JC, Görgens A, Ricklefs T, et
al: Genome-wide methylation profiling of glioblastoma cell-derived
extracellular vesicle DNA allows tumor classification. Neuro Oncol.
23:1087–1099. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Z, Cui Y, Wang F, Xu L, Yan Y, Tong X
and Yan H: DNA methylation-regulated LINC02587 inhibits ferroptosis
and promotes the progression of glioma cells through the CoQ-FSP1
pathway. BMC Cancer. 23:9892023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wielandt AM, Villarroel C, Hurtado C,
Simian D, Zamorano D, Martínez M, Castro M, Vial MT, Kronberg U and
López-Kostner F: Characterization of patients with sporadic
colorectal cancer following the new Consensus Molecular Subtypes
(CMS). Rev Méd Chile. 145:419–430. 2017.(In Spanish). View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moreno-Ortiz JM, Jiménez-García J,
Gutiérrez-Angulo M, Ayala-Madrigal MD, González-Mercado A,
González-Villaseñor CO, Flores-López BA, Alvizo-Rodríguez C,
Hernández-Sandoval JA, Fernández-Galindo MA, et al: High frequency
of MLH1 promoter methylation mediated by gender and age in
colorectal tumors from Mexican patients. GMM. 157:638–644. 2021.(In
Spanish).
|
|
9
|
Del Castillo Falconi VM, Torres-Arciga K,
Matus-Ortega G, Díaz-Chávez J and Herrera LA: DNA
methyltransferases: From evolution to clinical applications. Int J
Mol Sci. 23:89942022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li E, Bestor TH and Jaenisch R: Targeted
mutation of the DNA methyltransferase gene results in embryonic
lethality. Cell. 69:915–926. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shih IeM and Kurman RJ: Ovarian
tumorigenesis: A proposed model based on morphological and
molecular genetic analysis. Am J Pathol. 164:1511–1518. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kurman RJ and Shih IeM: Pathogenesis of
ovarian cancer: Lessons from morphology and molecular biology and
their clinical implications. Int J Gynecol Pathol. 27:151–160.
2018.PubMed/NCBI
|
|
13
|
Kurman RJ and Shih IeM: The origin and
pathogenesis of epithelial ovarian cancer: A proposed unifying
theory. Am J Surg Pathol. 34:433–443. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Samuel D, Diaz-Barbe A, Pinto A,
Schlumbrecht M and George S: Hereditary ovarian carcinoma: Cancer
pathogenesis looking beyond BRCA1 and BRCA2. Cells. 11:5392022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ramus SJ, Harrington PA, Pye C, DiCioccio
RA, Cox MJ, Garlinghouse-Jones K, Oakley-Girvan I, Jacobs IJ, Hardy
RM, Whittemore AS, et al: Contribution of BRCA1 and BRCA2 mutations
to inherited ovarian cancer. Hum Mutat. 28:1207–1215. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Menon U, Karpinskyj C and Gentry-Maharaj
A: Ovarian cancer prevention and screening. Obstet Gynecol.
131:909–927. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lavoro A, Scalisi A, Candido S, Zanghì GN,
Rizzo R, Gattuso G, Caruso G, Libra M and Falzone L: Identification
of the most common BRCA alterations through analysis of germline
mutation databases: Is droplet digital PCR an additional strategy
for the assessment of such alterations in breast and ovarian cancer
families? Int J Oncol. 60:582022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kansuttiviwat C, Lertwilaiwittaya P,
Roothumnong E, Nakthong P, Dungort P, Meesamarnpong C, Tansa-Nga W,
Pongsuktavorn K, Wiboonthanasarn S, Tititumjariya W, et al:
Germline mutations of 4567 patients with hereditary breast-ovarian
cancer spectrum in Thailand. NPJ Genom Med. 9:92024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Andrikopoulou A, Zografos E, Apostolidou
K, Kyriazoglou A, Papatheodoridi AM, Kaparelou M, Koutsoukos K,
Liontos M, Dimopoulos MA and Zagouri F: Germline and somatic
variants in ovarian carcinoma: A next-generation sequencing (NGS)
analysis. Front Oncol. 12:10307862022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ghose A, Bolina A, Mahajan I, Raza SA,
Clarke M, Pal A, Sanchez E, Rallis KS and Boussios S: Hereditary
ovarian cancer: Towards a cost-effective prevention strategy. Int J
Environ Res Public Health. 19:120572022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McCluggage WG: Morphological subtypes of
ovarian carcinoma: A review with emphasis on new developments and
pathogenesis. Pathology. 43:420–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andrews L and Mutch DG: Hereditary ovarian
cancer and risk reduction. Best Pract Res Clin Obstet Gynaecol.
41:31–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lynch HT and Lynch JF: Hereditary
nonpolyposis colorectal cancer. Semin Surg Oncol. 18:305–313. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu H, Shang X, Shi Y, Yang Z, Zhao J, Yang
M, Li Y and Xu S: Genetic variants of lncRNA HOTAIR and risk of
epithelial ovarian cancer among Chinese women. Oncotarget.
7:41047–41052. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bronder D, Tighe A, Wangsa D, Zong D,
Meyer TJ, Wardenaar R, Minshall P, Hirsch D, Heselmeyer-Haddad K,
Nelson L, et al: TP53 loss initiates chromosomal instability in
fallopian tube epithelial cells. Dis Model Mech. 14:dmm0490012021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goff BA, Mandel L, Muntz HG and Melancon
CH: Ovarian carcinoma diagnosis. Cancer. 89:2068–2075. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zeimet AG, Fiegl H, Goebel G, Kopp F,
Allasia C, Reimer D, Steppan I, Mueller-Holzner E, Ehrlich M and
Marth C: DNA ploidy, nuclear size, proliferation index and
DNA-hypomethylation in ovarian cancer. Gynecol Oncol. 121:24–31.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Widschwendter M, Jiang G, Woods C, Müller
HM, Fiegl H, Goebel G, Marth C, Müller-Holzner E, Zeimet AG, Laird
PW and Ehrlich M: DNA hypomethylation and ovarian cancer biology.
Cancer Res. 64:4472–4480. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng W, Marquez RT, Lu Z, Liu J, Lu KH,
Issa JP, Fishman DM, Yu Y and Bast RC Jr: Imprinted tumor
suppressor genesARHI andPEG3 are the most frequently down-regulated
in human ovarian cancers by loss of heterozygosity and promoter
methylation. Cancer. 112:1489–1502. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Link PA, Zhang W, Odunsi K and Karpf AR:
BORIS/CTCFL mRNA isoform expression and epigenetic regulation in
epithelial ovarian cancer. Cancer Immun. 13:62013.PubMed/NCBI
|
|
31
|
Wang YQ, Yan Q, Zhang JR, Li SD, Yang YX
and Wan XP: Epigenetic inactivation of BRCA1 through promoter
hypermethylation in ovarian cancer progression. J Obstet Gynaecol
Res. 39:549–554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Abou-Zeid AA, Azzam AZ and Kamel NA:
Methylation status of the gene promoter of cyclin-dependent kinase
inhibitor 2A (CDKN2A) in ovarian cancer. Scand J Clin Lab Invest.
71:542–547. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bhagat R, Kumar SS, Vaderhobli S,
Premalata CS, Pallavi VR, Ramesh G and Krishnamoorthy L: Epigenetic
alteration of p16 and retinoic acid receptor beta genes in the
development of epithelial ovarian carcinoma. Tumour Biol.
35:9069–9078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang G, Zhang H, Liu Y, Zhou J, He W,
Quick CM, Xie D, Smoller BR and Fan CY: Epigenetic and
immunohistochemical characterization of the Clusterin gene in
ovarian tumors. Arch Gynecol Obstet. 287:989–995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang W, Barger CJ, Link PA,
Mhawech-Fauceglia P, Miller A, Akers SN, Odunsi K and Karpf AR: DNA
hypomethylation-mediated activation of Cancer/Testis Antigen 45
(CT45) genes is associated with disease progression and reduced
survival in epithelial ovarian cancer. Epigenetics. 10:736–748.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang B, Yu L, Luo X, Huang L, Li QS, Shao
XS, Liu Y, Fan Y and Yang GZ: Detection of OPCML methylation, a
possible epigenetic marker, from free serum circulating DNA to
improve the diagnosis of early-stage ovarian epithelial cancer.
Oncol Lett. 14:217–223. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kaur M, Singh A, Singh K, Gupta S and
Sachan M: Development of a multiplex MethyLight assay for the
detection of DAPK1 and SOX1 methylation in epithelial ovarian
cancer in a north Indian population. Genes Genet Syst. 91:175–181.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rattanapan Y, Korkiatsakul V, Kongruang A,
Chareonsirisuthigul T, Rerkamnuaychoke B, Wongkularb A and Wilailak
S: EGFL7 and RASSF1 promoter hypermethylation in epithelial ovarian
cancer. Cancer Genet. 224–225. 37–40. 2018.PubMed/NCBI
|
|
39
|
da Conceição Braga C, Silva LM, Piedade
JB, Traiman P and da Silva Filho AL: Epigenetic and expression
analysis of TRAIL-R2 and BCL2: On the TRAIL to knowledge of
apoptosis in ovarian tumors. Arch Gynecol Obstet. 289:1061–1069.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bonito NA, Borley J, Wilhelm-Benartzi CS,
Ghaem-Maghami S and Brown R: Epigenetic regulation of the homeobox
gene MSX1 associates with platinum-resistant disease in high-grade
serous epithelial ovarian cancer. Clin Cancer Res. 22:3097–3104.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kardum V, Karin V, Glibo M, Skrtic A,
Martic TN, Ibisevic N, Skenderi F, Vranic S and Serman L:
Methylation-associated silencing of SFRP1 gene in high-grade serous
ovarian carcinomas. Ann Diagn Pathol. 31:45–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Suzuki F, Akahira J, Miura I, Suzuki T,
Ito K, Hayashi S, Sasano H and Yaegashi N: Loss of estrogen
receptor beta isoform expression and its correlation with aberrant
DNA methylation of the 5′-untranslated region in human epithelial
ovarian carcinoma. Cancer Sci. 99:2365–2372. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Baranova I, Kovarikova H, Laco J, Dvorak
O, Sedlakova I, Palicka V and Chmelarova M: Aberrant methylation of
PCDH17 gene in high-grade serous ovarian carcinoma. Cancer Biomark.
23:125–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ding JJ, Wang G, Shi WX, Zhou HH and Zhao
EF: Promoter hypermethylation of FANCF and susceptibility and
prognosis of epithelial ovarian cancer. Reprod Sci. 23:24–30. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gozzi G, Chelbi ST, Manni P, Alberti L,
Fonda S, Saponaro S, Fabbiani L, Rivasi F, Benhattar J and Losi L:
Promoter methylation and downregulated expression of the TBX15 gene
in ovarian carcinoma. Oncol Lett. 12:2811–2819. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Choi YL, Kang SY, Shin YK, Choi JS, Kim
SH, Lee SJ, Bae DS and Ahn G: Aberrant hypermethylation of RASSF1A
promoter in ovarian borderline tumors and carcinomas. Virchows
Archiv. 448:331–336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Häfner N, Steinbach D, Jansen L, Diebolder
H, Dürst M and Runnebaum IB: RUNX3 and CAMK2N1 hypermethylation as
prognostic marker for epithelial ovarian cancer. Int J Cancer.
138:217–228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jin P, Song Y and Yu G: The role of
abnormal methylation of Wnt5a gene promoter regions in human
epithelial ovarian cancer: A clinical and experimental study. Anal
Cell Pathol (Amst). 2018:65670812018.PubMed/NCBI
|
|
49
|
Khodadadi E, Fahmideh L, Khodadadi E, Dao
S, Yousefi M, Taghizadeh S, Asgharzadeh M, Yousefi B and Kafil HS:
Current advances in DNA methylation analysis methods. Biomed Res
Int. 2021:88275162021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gattuso G, Lavoro A, Caltabiano R, Madonna
G, Capone M, Ascierto PA, Falzone L, Libra M and Candido S:
Methylation-sensitive restriction enzyme-droplet digital PCR assay
for the one-step highly sensitive analysis of DNA methylation
hotspots. Int J Mol Med. 53:422024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Falzone L, Salemi R, Travali S, Scalisi A,
McCubrey JA, Candido S and Libra M: MMP-9 overexpression is
associated with intragenic hypermethylation of MMP9 gene in
melanoma. Aging (Albany NY). 8:933–944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Singer M, Kosti I, Pachter L and
Mandel-Gutfreund Y: A diverse epigenetic landscape at human exons
with implication for expression. Nucleic Acids Res. 43:3498–3508.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Davidson B, Tropé CG and Reich R: The
clinical and diagnostic role of microRNAs in ovarian carcinoma.
Gynecol Oncol. 133:640–646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sheng X and Li J, Yang L, Chen Z, Zhao Q,
Tan L, Zhou Y and Li J: Promoter hypermethylation influences the
suppressive role of maternally expressed 3, a long non-coding RNA,
in the development of epithelial ovarian cancer. Oncol Rep.
32:277–285. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Loginov VI, Pronina IV, Burdennyy AM,
Filippova EA, Kazubskaya TP, Kushlinsky DN, Utkin DO, Khodyrev DS,
Kushlinskii NE, Dmitriev AA and Braga EA: Novel miRNA genes
deregulated by aberrant methylation in ovarian carcinoma are
involved in metastasis. Gene. 662:28–36. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Filippov-Levy N, Cohen-Schussheim H, Tropé
CG, Hetland Falkenthal TE, Smith Y, Davidson B and Reich R:
Expression and clinical role of long non-coding RNA in high-grade
serous carcinoma. Gynecol Oncol. 148:559–566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu X, Dai C, Jia G, Xu S, Fu Z, Xu J, Li
Q, Ruan H and Xu P: Microarray analysis reveals differentially
expressed lncRNAs in benign epithelial ovarian cysts and normal
ovaries. Oncol Rep. 38:799–808. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lu YM, Wang Y, Liu SQ, Zhou MY and Guo YR:
Profile and validation of dysregulated long non-coding RNAs and
mRNAs in ovarian cancer. Oncol Rep. 40:2964–2976. 2018.PubMed/NCBI
|
|
59
|
Wang H, Fu Z, Dai C, Cao J, Liu X, Xu J,
Lv M, Gu Y, Zhang J, Hua X, et al: LncRNAs expression profiling in
normal ovary, benign ovarian cyst and malignant epithelial ovarian
cancer. Sci Rep. 6:389832016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Boyd C and McCluggage WG: Low-grade
ovarian serous neoplasms (low-grade serous carcinoma and serous
borderline tumor) associated with high-grade serous carcinoma or
undifferentiated carcinoma: Report of a series of cases of an
unusual phenomenon. Am J Surg Pathol. 36:368–375. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pisanic TR II, Cope LM, Lin SF, Yen TT,
Athamanolap P, Asaka R, Nakayama K, Fader AN, Wang TH, Shih IM and
Wang TL: Methylomic analysis of ovarian cancers identifies
tumor-specific alterations readily detectable in early precursor
lesions. Clin Cancer Res. 24:6536–6547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Klinkebiel D, Zhang W, Akers SN, Odunsi K
and Karpf AR: DNA Methylome analyses implicate fallopian tube
epithelia as the origin for high-grade serous ovarian cancer. Mol
Cancer Res. 14:787–794. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Givel AM, Kieffer Y, Scholer-Dahirel A,
Sirven P, Cardon M, Pelon F, Magagna I, Gentric G, Costa A, Bonneau
C, Mieulet V, et al: miR200-regulated CXCL12β promotes fibroblast
heterogeneity and immunosuppression in ovarian cancers. Nat Commun.
9:10562018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang Y, Qiu C, Lu N, Liu Z, Jin C, Sun C,
Bu H, Yu H, Dongol S and Kong B: FOXD1 is targeted by miR-30a-5p
and miR-200a-5p and suppresses the proliferation of human ovarian
carcinoma cells by promoting p21 expression in a p53-independent
manner. Int J Oncol. 52:2130–2142. 2018.PubMed/NCBI
|
|
65
|
Ma H, Tian T, Liang S, Liu X, Shen H, Xia
M, Liu X, Zhang W, Wang L, Chen S and Yu L: Estrogen
receptor-mediated miR-486-5p regulation of OLFM4 expression in
ovarian cancer. Oncotarget. 7:10594–1605. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Nymoen DA, Slipicevic A, Holth A, Emilsen
E, Hetland Falkenthal TE, Tropé CG, Reich R, Flørenes VA and
Davidson B: MiR-29a is a candidate biomarker of better survival in
metastatic high-grade serous carcinoma. Hum Pathol. 54:74–81. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Arts FA, Keogh L, Smyth P, O'Toole S, Ta
R, Gleeson N, O'Leary JJ, Flavin R and Sheils O: miR-223
potentially targets SWI/SNF complex protein SMARCD1 in atypical
proliferative serous tumor and high-grade ovarian serous carcinoma.
Hum Pathol. 70:98–104. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Chaluvally-Raghavan P, Jeong KJ, Pradeep
S, Silva AM, Yu S, Liu W, Moss T, Rodriguez-Aguayo C, Zhang D, Ram
P, et al: Direct upregulation of STAT3 by MicroRNA-551b-3p
deregulates growth and metastasis of ovarian cancer. Cell Rep.
15:1493–1504. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhao H, Liu S, Wang G, Wu X, Ding Y, Guo
G, Jiang J and Cui S: Expression of miR-136 is associated with the
primary cisplatin resistance of human epithelial ovarian cancer.
Oncol Rep. 33:591–598. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kuznetsov VA, Tang Z and Ivshina AV:
Identification of common oncogenic and early developmental pathways
in the ovarian carcinomas controlling by distinct prognostically
significant microRNA subsets. BMC Genomics. 18 (Suppl 6):6922017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang X, Guo G, Wang G, Zhao J, Wang B, Yu
X and Ding Y: Profile of differentially expressed miRNAs in
high-grade serous carcinoma and clear cell ovarian carcinoma, and
the expression of miR-510 in ovarian carcinoma. Mol Med Rep.
12:8021–8031. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yanaihara N, Noguchi Y, Saito M, Takenaka
M, Takakura S, Yamada K and Okamoto A: MicroRNA gene expression
signature driven by miR-9 overexpression in ovarian clear cell
carcinoma. PLoS One. 11:e01625842016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Furlan D, Carnevali I, Marcomini B,
Cerutti R, Dainese E, Capella C and Riva C: The high frequency of
de novo promoter methylation in synchronous primary endometrial and
ovarian carcinomas. Clin Cancer Res. 12:3329–3336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Niskakoski A, Pasanen A, Porkka N, Eldfors
S, Lassus H, Renkonen-Sinisalo L, Kaur S, Mecklin JP, Bützow R and
Peltomäki P: Converging endometrial and ovarian tumorigenesis in
Lynch syndrome: Shared origin of synchronous carcinomas. Gynecol
Oncol. 150:92–98. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kolbe DL, DeLoia JA, Porter-Gill P,
Strange M, Petrykowska HM, Guirguis A, Krivak TC, Brody LC and
Elnitski L: Differential analysis of ovarian and endometrial
cancers identifies a methylator phenotype. PLoS One. 7:e329412012.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Guo C, Ren F, Wang D, Li Y, Liu K, Liu S
and Chen P: RUNX3 is inactivated by promoter hypermethylation in
malignant transformation of ovarian endometriosis. Oncol Rep.
32:2580–2588. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liew PL, Huang RL, Weng YC, Fang CL,
Hui-Ming Huang T and Lai HC: Distinct methylation profile of
mucinous ovarian carcinoma reveals susceptibility to proteasome
inhibitors: Methylation profile of MuOC and PSMB8. Int J Cancer.
143:355–367. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Agostini A, Brunetti M, Davidson B, Tropé
CG, Eriksson AGZ, Heim S, Panagopoulos I and Micci F: The microRNA
miR-192/215 family is upregulated in mucinous ovarian carcinomas.
Sci Rep. 8:110692018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Vang R, Shih IeM and Kurman RJ: Ovarian
low-grade and high-grade serous carcinoma: Pathogenesis,
clinicopathologic and molecular biologic features, and diagnostic
problems. Adv Anat Pathol. 16:267–282. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Bowtell DD: The genesis and evolution of
high-grade serous ovarian cancer. Nat Rev Cancer. 10:803–808. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
O'Shea AS: Clinical staging of ovarian
cancer. Methods Mol Biol. 2424:3–10. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Richards EJ, Permuth-Wey J, Li Y, Chen YA,
Coppola D, Reid BM, Lin HY, Teer JK, Berchuck A, Birrer MJ, et al:
A functional variant in HOXA11-AS, a novel long non-coding RNA,
inhibits the oncogenic phenotype of epithelial ovarian cancer.
Oncotarget. 6:34745–34757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zhang T, Wu D, Deng S, Han R, Liu T, Li J
and Xu Y: Integrated analysis reveals that long non-coding RNA
TUBA4B can be used as a prognostic biomarker in various cancers.
Cell Physiol Biochem. 49:530–544. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Meryet-Figuière M, Lambert B, Gauduchon P,
Vigneron N, Brotin E, Poulain L and Denoyelle C: An overview of
long non-coding RNAs in ovarian cancers. Oncotarget. 7:44719–44734.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhong Y, Gao D, He S, Shuai C and Peng S:
Dysregulated expression of long noncoding RNAs in ovarian cancer.
Int J Gynecol Cancer. 26:1564–1570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ma Y, Lu Y and Lu B: MicroRNA and Long
Non-Coding RNA in ovarian carcinoma: Translational insights and
potential clinical applications. Cancer Invest. 34:465–476. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Lin X, Qiu J and Hua K: Long non-coding
RNAs as emerging regulators of epithelial to mesenchymal transition
in gynecologic cancers. Biosci Trends. 12:342–353. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Micheel J, Safrastyan A and Wollny D:
Advances in non-coding RNA sequencing. Noncoding RNA.
7:702021.PubMed/NCBI
|
|
89
|
Zhang N, Hu G, Myers TG and Williamson PR:
Protocols for the analysis of microRNA expression, biogenesis, and
function in immune cells. Curr Protoc Immunol. 126:e782019.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhang S, Leng T, Zhang Q, Zhao Q, Nie X
and Yang L: Sanguinarine inhibits epithelial ovarian cancer
development via regulating long non-coding RNA CASC2-EIF4A3 axis
and/or inhibiting NF-κB signaling or PI3K/AKT/mTOR pathway. Biomed
Pharmacother. 102:302–308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW,
Jin HY, Zhang Y, Li Q and Hua KQ: Overexpression of long non-coding
RNA HOTAIR predicts poor patient prognosis and promotes tumor
metastasis in epithelial ovarian cancer. Gynecol Oncol.
134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Xi J, Feng J and Zeng S: Long noncoding
RNA lncBRM facilitates the proliferation, migration and invasion of
ovarian cancer cells via upregulation of Sox4. Am J Cancer Res.
7:2180–2189. 2017.PubMed/NCBI
|
|
93
|
Zhang Y, Dun Y, Zhou S and Huang XH:
LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells
proliferation and invasion by targeting miR-133a-3p and activating
Wnt/β-catenin signaling pathway. Biomed Pharmacother. 96:1216–1221.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Liu Y, Wang Y, Yao D and Cui D: LncSOX4
serves an oncogenic role in the tumorigenesis of epithelial ovarian
cancer by promoting cell proliferation and inhibiting apoptosis.
Mol Med Rep. 17:8282–8288. 2018.PubMed/NCBI
|
|
95
|
Yan H, Li H, Li P, Li X, Lin J, Zhu L,
Silva MA, Wang X, Wang P and Zhang Z: Long noncoding RNA MLK7-AS1
promotes ovarian cancer cells progression by modulating
miR-375/YAP1 axis. J Exp Clin Cancer Res. 37:2372018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li T, Chen Y, Zhang J and Liu S: LncRNA
TUG1 promotes cells proliferation and inhibits cells apoptosis
through regulating AURKA in epithelial ovarian cancer cells.
Medicine (Baltimore). 97:e121312018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang YS, Ma LN, Sun JX, Liu N and Wang H:
Long non-coding CPS1-IT1 is a positive prognostic factor and
inhibits epithelial ovarian cancer tumorigenesis. Eur Rev Med
Pharmacol Sci. 21:3169–3175. 2017.PubMed/NCBI
|
|
98
|
Zhu FF, Zheng FY, Wang HO, Zheng JJ and
Zhang Q: Downregulation of lncRNA TUBA4B is associated with poor
prognosis for epithelial ovarian cancer. Pathol Oncol Res.
24:419–425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Ying X, Wei K, Lin Z, Cui Y, Ding J, Chen
Y and Xu B: MicroRNA-125b suppresses ovarian cancer progression via
suppression of the epithelial-mesenchymal transition pathway by
targeting the SET protein. Cell Physiol Biochem. 39:501–510. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhu T, Gao W, Chen X, Zhang Y, Wu M, Zhang
P and Wang S: A pilot study of circulating MicroRNA-125b as a
diagnostic and prognostic biomarker for epithelial ovarian cancer.
Int J Gynecol Cancer. 27:3–10. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Teng Y, Zhang Y, Qu K, Yang X, Fu J, Chen
W and Li X: MicroRNA-29B (mir-29b) regulates the Warburg effect in
ovarian cancer by targeting AKT2 and AKT3. Oncotarget.
6:40799–40814. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Cao Q, Lu K, Dai S, Hu Y and Fan W:
Clinicopathological and prognostic implications of the miR-200
family in patients with epithelial ovarian cancer. Int J Clin Exp
Pathol. 7:2392–2401. 2014.PubMed/NCBI
|
|
103
|
Kapetanakis NI, Uzan C, Jimenez-Pailhes
AS, Gouy S, Bentivegna E, Morice P, Caron O, Gourzones-Dmitriev C,
Le Teuff G and Busson P: Plasma miR-200b in ovarian carcinoma
patients: Distinct pattern of pre/post-treatment variation compared
to CA-125 and potential for prediction of progression-free
survival. Oncotarget. 6:36815–36824. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Meng X, Müller V, Milde-Langosch K,
Trillsch F, Pantel K and Schwarzenbach H: Diagnostic and prognostic
relevance of circulating exosomal miR-373, miR-200a, miR-200b and
miR-200c in patients with epithelial ovarian cancer. Oncotarget.
7:16923–16935. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Du Z and Sha X: Demethoxycurcumin
inhibited human epithelia ovarian cancer cells' growth via
up-regulating miR-551a. Tumour Biol. 39:10104283176943022017.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Chen S, Chen X, Xiu YL, Sun KX and Zhao Y:
MicroRNA-490-3P targets CDK1 and inhibits ovarian epithelial
carcinoma tumorigenesis and progression. Cancer Lett. 362:122–130.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Shuang T, Wang M, Shi C, Zhou Y and Wang
D: Down-regulated expression of miR-134 contributes to paclitaxel
resistance in human ovarian cancer cells. FEBS Lett. 589:3154–3164.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zou YT, Gao JY, Wang HL, Wang Y, Wang H
and Li PL: Downregulation of microRNA-630 inhibits cell
proliferation and invasion and enhances chemosensitivity in human
ovarian carcinoma. Genet Mol Res. 14:8766–8777. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Zhang H and Li W: Dysregulation of
micro-143-3p and BALBP1 contributes to the pathogenesis of the
development of ovarian carcinoma. Oncol Rep. 36:3605–3610. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zhang W, Zeng Q, Ban Z, Cao J, Chu T, Lei
D, Liu C, Guo W and Zeng X: Effects of let-7c on the proliferation
of ovarian carcinoma cells by targeted regulation of CDC25a gene
expression. Oncol Lett. 16:5543–5550. 2018.PubMed/NCBI
|
|
111
|
Liu MX, Siu MK, Liu SS, Yam JW, Ngan HY
and Chan DW: Epigenetic silencing of microRNA-199b-5p is associated
with acquired chemoresistance via activation of JAG1-Notch1
signaling in ovarian cancer. Oncotarget. 5:944–958. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Kobayashi M, Sawada K, Nakamura K,
Yoshimura A, Miyamoto M, Shimizu A, Ishida K, Nakatsuka E, Kodama
M, Hashimoto K, et al: Exosomal miR-1290 is a potential biomarker
of high-grade serous ovarian carcinoma and can discriminate
patients from those with malignancies of other histological types.
J Ovarian Res. 11:812018. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Zhao H, Bi T, Qu Z, Jiang J, Cui S and
Wang Y: Expression of miR-224-5p is associated with the original
cisplatin resistance of ovarian papillary serous carcinoma. Oncol
Rep. 32:1003–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Chen Y, Chen Q, Liu Q and Gao F: Human
epididymis protein 4 expression positively correlated with miR-21
and served as a prognostic indicator in ovarian cancer. Tumour
Biol. 37:8359–8365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Li L, Huang K, You Y, Fu X, Hu L, Song L
and Meng Y: Hypoxia-induced miR-210 in epithelial ovarian cancer
enhances cancer cell viability via promoting proliferation and
inhibiting apoptosis. Int J Oncol. 44:2111–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Zhu X, Shen H, Yin X, Long L, Chen X, Feng
F, Liu Y, Zhao P, Xu Y, Li M, et al: IL-6R/STAT3/miR-204 feedback
loop contributes to cisplatin resistance of epithelial ovarian
cancer cells. Oncotarget. 8:39154–39166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Fan Y, Fan J, Huang L, Ye M, Huang Z, Wang
Y, Li Q and Huang J: Increased expression of microRNA-196a predicts
poor prognosis in human ovarian carcinoma. Int J Clin Exp Pathol.
8:4132–4137. 2015.PubMed/NCBI
|
|
118
|
Koukourakis MI, Kontomanolis E,
Sotiropoulou M, Mitrakas A, Dafa E, Pouliliou S, Sivridis E and
Giatromanolaki A: Increased soluble PD-L1 levels in the plasma of
patients with epithelial ovarian cancer correlate with plasma
levels of miR34a and miR200. Anticancer Res. 38:5739–5745. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Liu J, Dou Y and Sheng M: Inhibition of
microRNA-383 has tumor suppressive effect in human epithelial
ovarian cancer through the action on caspase-2 gene. Biomed
Pharmacother. 83:1286–1294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Dai F, Zhang Y and Chen Y: Involvement of
miR-29b signaling in the sensitivity to chemotherapy in patients
with ovarian carcinoma. Hum Pathol. 45:1285–1293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Xiao M, Cai J, Cai L, Jia J, Xie L, Zhu Y,
Huang B, Jin D and Wang Z: Let-7e sensitizes epithelial ovarian
cancer to cisplatin through repressing DNA double strand break
repair. J Ovarian Res. 10:242017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Li X, Pan Q, Wan X, Mao Y, Lu W, Xie X and
Cheng X: Methylation-associated Has-miR-9 deregulation in
paclitaxel-resistant epithelial ovarian carcinoma. BMC Cancer.
15:5092015. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Paudel D, Zhou W, Ouyang Y, Dong S, Huang
Q, Giri R, Wang J and Tong X: MicroRNA-130b functions as a tumor
suppressor by regulating RUNX3 in epithelial ovarian cancer. Gene.
586:48–55. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Duan S, Dong X, Hai J, Jiang J, Wang W,
Yang J, Zhang W and Chen C: MicroRNA-135a-3p is downregulated and
serves as a tumour suppressor in ovarian cancer by targeting CCR2.
Biomed Pharmacother. 107:712–720. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Chen X, Dong C, Law PT, Chan MT, Su Z,
Wang S, Wu WK and Xu H: MicroRNA-145 targets TRIM2 and exerts
tumor-suppressing functions in epithelial ovarian cancer. Gynecol
Oncol. 139:513–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Qin CZ, Lou XY, Lv QL, Cheng L, Wu NY, Hu
L and Zhou HH: MicroRNA-184 acts as a potential diagnostic and
prognostic marker in epithelial ovarian cancer and regualtes cell
proliferation, apoptosis and inflammation. Pharmazie. 70:668–673.
2015.PubMed/NCBI
|
|
127
|
Liang T, Li L, Cheng Y, Ren C and Zhang G:
MicroRNA-194 promotes the growth, migration, and invasion of
ovarian carcinoma cells by targeting protein tyrosine phosphatase
nonreceptor type 12. Onco Targets Ther. 9:4307–4315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Wei C, Zhang X, He S, Liu B, Han H and Sun
X: MicroRNA-219-5p inhibits the proliferation, migration, and
invasion of epithelial ovarian cancer cells by targeting the
Twist/Wnt/β-catenin signaling pathway. Gene. 637:25–32. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Fu X, Li Y, Alvero A, Li J, Wu Q, Xiao Q,
Peng Y, Hu Y, Li X, Yan W, et al: MicroRNA-222-3p/GNAI2/AKT axis
inhibits epithelial ovarian cancer cell growth and associates with
good overall survival. Oncotarget. 7:80633–80654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Wu X, Ruan Y, Jiang H and Xu C:
MicroRNA-424 inhibits cell migration, invasion, and epithelial
mesenchymal transition by downregulating doublecortin-like kinase 1
in ovarian clear cell carcinoma. Int J Biochem Cell Biol. 85:66–74.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zhang J, Liu L, Sun Y, Xiang J, Zhou D,
Wang L, Xu H, Yang X, Du N, Zhang M, et al: MicroRNA-520g promotes
epithelial ovarian cancer progression and chemoresistance via DAPK2
repression. Oncotarget. 7:26516–26534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Zhang L, Li Z, Gai F and Wang Y:
MicroRNA-137 suppresses tumor growth in epithelial ovarian cancer
in vitro and in vivo. Mol Med Rep. 12:3107–3114. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Liu J, Jin S and Wang R: MicroRNA-139
suppressed tumor cell proliferation, migration and invasion by
directly targeting HDGF in epithelial ovarian cancer. Mol Med Rep.
16:3379–3386. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Xu L, Li H, Su L, Lu Q and Liu Z:
MicroRNA-455 inhibits cell proliferation and invasion of epithelial
ovarian cancer by directly targeting Notch1. Mol Med Rep.
16:9777–9785. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Yan J, Jiang J, Meng XN, Xiu YL and Zong
ZH: MiR-23b targets cyclin G1 and suppresses ovarian cancer
tumorigenesis and progression. J Exp Clin Cancer Res. 35:312016.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Lin J, Zhang L, Huang H, Huang Y, Huang L,
Wang J, Huang S, He L, Zhou Y, Jia W, et al: MiR-26b/KPNA2 axis
inhibits epithelial ovarian carcinoma proliferation and metastasis
through downregulating OCT4. Oncotarget. 6:23793–23806. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Xu J, Jiang N, Shi H, Zhao S, Yao S and
Shen H: miR-28-5p promotes the development and progression of
ovarian cancer through inhibition of N4BP1. Int J Oncol.
50:1383–1391. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Wang Y, Zhang X, Tang W, Lin Z, Xu L, Dong
R, Li Y, Li J, Zhang Z, Li X, et al: miR-130a upregulates mTOR
pathway by targeting TSC1 and is transactivated by NF-κB in
high-grade serous ovarian carcinoma. Cell Death Differ.
24:2089–2100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Wang L, He J, Xu H, Xu L and Li N: MiR-143
targets CTGF and exerts tumor-suppressing functions in epithelial
ovarian cancer. Am J Transl Res. 8:2716–2726. 2016.PubMed/NCBI
|
|
140
|
Dong M, Yang P and Hua F: miR-191
modulates malignant transformation of endometriosis through
regulating TIMP3. Med Sci Monit. 21:915–920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Niu K, Shen W, Zhang Y, Zhao Y and Lu Y:
MiR-205 promotes motility of ovarian cancer cells via targeting
ZEB1. Gene. 574:330–336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Dai C, Xie Y, Zhuang X and Yuan Z: MiR-206
inhibits epithelial ovarian cancer cells growth and invasion via
blocking c-Met/AKT/mTOR signaling pathway. Biomed Pharmacother.
104:763–770. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Xia B, Yang S, Liu T and Lou G: miR-211
suppresses epithelial ovarian cancer proliferation and cell-cycle
progression by targeting Cyclin D1 and CDK6. Mol Cancer. 14:572015.
View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wu Q, Ren X, Zhang Y, Fu X, Li Y, Peng Y,
Xiao Q, Li T, Ouyang C, Hu Y, et al: MiR-221-3p targets ARF4 and
inhibits the proliferation and migration of epithelial ovarian
cancer cells. Biochem Biophys Res Commun. 497:1162–1170. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Cao L, Wan Q, Li F and Tang C: MiR-363
inhibits cisplatin chemoresistance of epithelial ovarian cancer by
regulating snail-induced epithelial-mesenchymal transition. BMB
Rep. 51:456–461. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Xia B, Li H, Yang S, Liu T and Lou G:
MiR-381 inhibits epithelial ovarian cancer malignancy via YY1
suppression. Tumour Biol. 37:9157–9167. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Yuan J, Wang K and Xi M: MiR-494 inhibits
epithelial ovarian cancer growth by targeting c-Myc. Med Sci Monit.
22:617–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Li N, Zhao X, Wang L, Zhang S, Cui M and
He J: miR-494 suppresses tumor growth of epithelial ovarian
carcinoma by targeting IGF1R. Tumour Biol. 37:7767–7776. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhou QH, Zhao YM, JIA LL and Zhang Y:
Mir-595 is a significant indicator of poor patient prognosis in
epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 21:4278–4282.
2017.PubMed/NCBI
|
|
150
|
Zhang S, Zhang JY, Lu LJ, Wang CH and Wang
LH: MiR-630 promotes epithelial ovarian cancer proliferation and
invasion via targeting KLF6. Eur Rev Med Pharmacol Sci.
21:4542–4547. 217.PubMed/NCBI
|
|
151
|
Shi C and Zhang Z: miR-761 inhibits tumor
progression by targeting MSI1 in ovarian carcinoma. Tumour Biol.
37:5437–5443. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Xie X, Huang Y, Chen L and Wang J: miR-221
regulates proliferation and apoptosis of ovarian cancer cells by
targeting BMF. Oncol Lett. 16:6697–6704. 2018.PubMed/NCBI
|
|
153
|
Wen C, Liu X, Ma H, Zhang W and Li H:
miR-338-3p suppresses tumor growth of ovarian epithelial carcinoma
by targeting Runx2. Int J Oncol. 46:2277–2285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Salem M, O'Brien JA, Bernaudo S, Shawer H,
Ye G, Brkić J, Amleh A, Vanderhyden BC, Refky B, Yang BB, et al:
miR-590-3p promotes ovarian cancer growth and metastasis via a
Novel FOXA2-versican pathway. Cancer Res. 78:4175–4190. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Lin Z, Zhao J, Wang X, Zhu X and Gong L:
Overexpression of microRNA-497 suppresses cell proliferation and
induces apoptosis through targeting paired box 2 in human ovarian
cancer. Oncol Rep. 36:2101–2107. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Lin M, Xia B, Qin L, Chen H and Lou G:
S100A7 regulates ovarian cancer cell metastasis and chemoresistance
through MAPK signaling and is targeted by miR-330-5p. DNA Cell
Biol. 37:491–500. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Chen JL, Chen F, Zhang TT and Liu NF:
Suppression of SIK1 by miR-141 in human ovarian cancer cell lines
and tissues. Int J Mol Med. 37:1601–1610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Zuberi M, Khan I, Gandhi G, Ray PC and
Saxena A: The conglomeration of diagnostic, prognostic and
therapeutic potential of serum miR-199a and its association with
clinicopathological features in epithelial ovarian cancer. Tumour
Biol. 37:11259–11266. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Guan X, Zong ZH, Chen S, Sang XB, Wu DD,
Wang LL, Liu Y and Zhao Y: The role of miR-372 in ovarian carcinoma
cell proliferation. Gene. 624:14–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Li J, Li D and Zhang W: Tumor suppressor
role of miR-217 in human epithelial ovarian cancer by targeting
IGF1R. Oncol Rep. 35:1671–1679. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Zhang X, Liu J, Zang D, Wu S, Liu A, Zhu
J, Wu G, Li J and Jiang L: Upregulation of miR-572
transcriptionally suppresses SOCS1 and p21 and contributes to human
ovarian cancer progression. Oncotarget. 6:15180–15193. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Zhou J, Gong G, Tan H, Dai F, Zhu X, Chen
Y, Wang J, Liu Y, Chen P, Wu X and Wen J: Urinary microRNA-30a-5p
is a potential biomarker for ovarian serous adenocarcinoma. Oncol
Rep. 33:2915–2923. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Zhang X, Li S, Dong C, Xie X and Zhang Y:
Knockdown of long noncoding RNA NR_026689 inhibits proliferation
and invasion and increases apoptosis in ovarian carcinoma HO-8910PM
cells. Oncol Res. 25:259–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Zhu L, Guo Q, Lu X, Zhao J, Shi J, Wang Z
and Zhou X: CTD-2020K17.1, a novel long non-coding RNA, promotes
migration, invasion, and proliferation of serous ovarian cancer
cells in vitro. Med Sci Monit. 24:1329–1339. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Qiu JJ, Zhang XD, Tang XY, Zheng TT, Zhang
Y and Hua KQ: ElncRNA1, a long non-coding RNA that is
transcriptionally induced by oestrogen, promotes epithelial ovarian
cancer cell proliferation. Int J Oncol. 51:507–514. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: LncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Wang Y, Wang H, Song T, Zou Y, Jiang J,
Fang L and Li P: HOTAIR is a potential target for the treatment of
cisplatin-resistant ovarian cancer. Mol Med Rep. 12:2211–2216.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Lu CW, Zhou DD, Xie T, Hao JL, Pant OP, Lu
CB and Liu XF: HOXA11 antisense long noncoding RNA (HOXA11-AS): A
promising lncRNA in human cancers. Cancer Med. 7:3792–3799. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Du W, Feng Z and Sun Q: LncRNA LINC00319
accelerates ovarian cancer progression through miR-423-5p/NACC1
pathway. Biochem Biophys Res Commun. 507:198–202. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Shu C, Yan D, Mo Y, Gu J, Shah N and He J:
Long noncoding RNA lncARSR promotes epithelial ovarian cancer cell
proliferation and invasion by association with HuR and miR-200
family. Am J Cancer Res. 8:981–992. 2018.PubMed/NCBI
|
|
171
|
Chen S, Wu DD, Sang XB, Wang LL, Zong ZH,
Sun KX, Liu BL and Zhao Y: The lncRNA HULC functions as an oncogene
by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell
Death Dis. 8:e31182017. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Qnbo L, Guan W, Ren W, Zhang L, Zhang J
and Xu G: MALAT1 affects ovarian cancer cell behavior and patient
survival. Oncol Rep. 39:2644–2652. 2018.PubMed/NCBI
|
|
173
|
Lin Q, Guan W, Ren W, Zhang L, Zhang J and
Xu G: MALAT1 affects ovarian cancer cell behavior and patient
survival. Oncol Rep. 39:2644–2652. 2018.PubMed/NCBI
|
|
174
|
Yan C, Jiang Y, Wan Y, Zhang L, Liu J,
Zhou S and Cheng W: Long noncoding RNA NBAT-1 suppresses
tumorigenesis and predicts favorable prognosis in ovarian cancer.
Onco Targets Ther. 10:1993–2002. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Liu Y, Wang Y, Fu X and Lu Z: Long
non-coding RNA NEAT1 promoted ovarian cancer cells' metastasis
through regulation of miR-382-3p/ROCK1 axial. Cancer Sci.
109:2188–2198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Chen S, Wang LL, Sun KX, Liu Y, Guan X,
Zong ZH and Zhao Y: LncRNA PCGEM1 induces ovarian carcinoma
tumorigenesis and progression through RhoA pathway. Cell Physiol
Biochem. 47:1578–1588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Huang K, Geng J and Wang J: Long
non-coding RNA RP11-552M11.4 promotes cells proliferation,
migration and invasion by targeting BRCA2 in ovarian cancer. Cancer
Sci. 109:1428–1446. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Li H, Liu C, Lu Z, Chen L, Wang J, Li Y
and Ma H: Upregulation of the long non-coding RNA SPRY4-IT1
indicates a poor prognosis and promotes tumorigenesis in ovarian
cancer. Biomed Pharmacother. 88:529–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Li TH, Zhang JJ, Liu SX and Chen Y: Long
non-coding RNA taurine-upregulated gene 1 predicts unfavorable
prognosis, promotes cells proliferation, and inhibits cells
apoptosis in epithelial ovarian cancer. Medicine (Baltimore).
97:e05752018. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Hong HH, Hou LK, Pan X, Wu CY, Huang H, Li
B and Nie W: Long non-coding RNA UCA1 is a predictive biomarker of
cancer. Oncotarget. 7:44442–44447. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Zhang L, Cao X, Zhang L, Zhang X, Sheng H
and Tao K: UCA1 overexpression predicts clinical outcome of
patients with ovarian cancer receiving adjuvant chemotherapy.
Cancer Chemother Pharmacol. 77:629–634. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Qiu JJ, Wang Y, Liu YL, Zhang Y, Ding JX
and Hua KQ: The long non-coding RNA ANRIL promotes proliferation
and cell cycle progression and inhibits apoptosis and senescence in
epithelial ovarian cancer. Oncotarget. 7:32478–32492. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Cao Y, Shi H, Ren F, Jia Y and Zhang R:
Long non-coding RNA CCAT1 promotes metastasis and poor prognosis in
epithelial ovarian cancer. Exp Cell Res. 359:185–194. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Hua F, Li CH, Chen XG and Liu XP: Long
Noncoding RNA CCAT2 knockdown suppresses tumorous progression by
sponging miR-424 in epithelial ovarian cancer. Oncol Res.
26:241–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Yim GW, Kim HJ, Kim LK, Kim SW, Kim S, Nam
EJ and Kim YT: Long Non-coding RNA HOXA11 antisense promotes cell
proliferation and invasion and predicts patient prognosis in serous
ovarian cancer. Cancer Res Treat. 49:656–668. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Koutsaki M, Spandidos DA and Zaravinos A:
Epithelial-mesenchymal transition-associated miRNAs in ovarian
carcinoma, with highlight on the miR-200 family: Prognostic value
and prospective role in ovarian cancer therapeutics. Cancer Lett.
351:173–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Sulaiman SA, Ab Mutalib NS and Jamal R:
miR-200c regulation of metastases in ovarian cancer: Potential role
in epithelial and mesenchymal transition. Front Pharmacol.
7:2712016. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Teng Y, Su X, Zhang X, Zhang Y, Li C, Niu
W, Liu C and Qu K: miRNA-200a/c as potential biomarker in
epithelial ovarian cancer (EOC): Evidence based on miRNA
meta-signature and clinical investigations. Oncotarget.
7:81621–81633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Muralidhar G and Barbolina M: The miR-200
Family: Versatile players in epithelial ovarian cancer. Int J Mol
Sci. 16:16833–16847. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Zuberi M, Khan I, Mir R, Gandhi G, Ray PC
and Saxena A: Utility of serum miR-125b as a diagnostic and
prognostic indicator and its alliance with a panel of tumor
suppressor genes in epithelial ovarian cancer. PLoS One.
11:e01539022016. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Faul C, Gerszten K, Edwards R, Land S,
D'Angelo G, Kelley J III and Price F: A phase I/II study of
hypofractionated whole abdominal radiation therapy in patients with
chemoresistant ovarian carcinoma: Karnofsky score determines
treatment outcome. Int J Radiat Oncol Biol Phys. 47:749–754. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Iorio GC, Martini S, Arcadipane F, Ricardi
U and Franco P: The role of radiotherapy in epithelial ovarian
cancer: A literature overview. Med Oncol. 36:642019. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Sorbe B: Consolidation treatment of
advanced ovarian carcinoma with radiotherapy after induction
chemotherapy. Int J Gynecol Cancer. 13 (Suppl 2):S192–S195. 2003.
View Article : Google Scholar
|
|
194
|
Pang L and Guo Z: Differences in
characteristics and outcomes between large-cell neuroendocrine
carcinoma of the ovary and high-grade serous ovarian cancer: A
retrospective observational cohort study. Front Oncol.
12:8916992022. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Patel SC, Frandsen J, Bhatia S and Gaffney
D: Impact on survival with adjuvant radiotherapy for clear cell,
mucinous, and endometriod ovarian cancer: The SEER experience from
2004 to 2011. J Gynecol Oncol. 27:e452016. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Pestell KE, Medlow CJ, Titley JC, Kelland
LR and Walton MI: Characterisation Of The P53 Status, Bcl-2
expression and radiation and platinum drug sensitivity of a panel
of human ovarian cancer cell lines. Int J Cancer. 77:913–918. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Zielske SP: Epigenetic DNA methylation in
radiation biology: On the field or on the sidelines? J Cell
Biochem. 116:212–217. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Kurrey NK, Jalgaonkar SP, Joglekar AV,
Ghanate AD, Chaskar PD, Doiphode RY and Bapat SA: Snail and slug
mediate radioresistance and chemoresistance by antagonizing
p53-Mediated apoptosis and acquiring a stem-like phenotype in
ovarian cancer cells. Stem Cells. 27:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Liu WJ, Huang YX, Wang W, Zhang Y, Liu BJ,
Qiu JG, Jiang BH and Liu LZ: NOX4 signaling mediates cancer
development and therapeutic resistance through HER3 in ovarian
cancer cells. Cells. 10:16472021. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Chen J, Jia Y, Jia ZH, Zhu Y and Jin YM:
Silencing the expression of MTDH increases the radiation
sensitivity of SKOV3 ovarian cancer cells and reduces their
proliferation and metastasis. Int J Oncol. 53:2180–2190.
2018.PubMed/NCBI
|
|
201
|
Zhao Y, Liu S, Wen Y and Zhong L: Effect
of MicroRNA-210 on the growth of ovarian cancer cells and the
efficacy of radiotherapy. Gynecol Obstet Invest. 86:71–80. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Xing Y, Cui D, Wang S, Wang P, Xing X and
Li H: Oleuropein represses the radiation resistance of ovarian
cancer by inhibiting hypoxia and microRNA-299-targetted heparanase
expression. Food Funct. 8:2857–2864. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Marques C, Ferreira da Silva F, Sousa I
and Nave M: Chemotherapy-free treatment of recurrent advanced
ovarian cancer: Myth or reality? Int J Gynecol Cancer. 33:607–618.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Marchetti C, De Felice F, Romito A,
Iacobelli V, Sassu CM, Corrado G, Ricci C, Scambia G and Fagotti A:
Chemotherapy resistance in epithelial ovarian cancer: Mechanisms
and emerging treatments. Semin Cancer Biol. 77:144–166. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Falzone L, Bordonaro R and Libra M:
SnapShot: Cancer chemotherapy. Cell. 186:1816–1816.e1. 2023.
View Article : Google Scholar
|
|
206
|
Raab M, Sanhaji M, Zhou S, Rödel F,
El-Balat A, Becker S and Strebhardt K: Blocking mitotic exit of
ovarian cancer cells by pharmaceutical inhibition of the
anaphase-promoting complex reduces chromosomal instability.
Neoplasia. 21:363–375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Swanton C, Nicke B, Schuett M, Eklund AC,
Ng C, Li Q, Hardcastle T, Lee A, Roy R, East P and Kschischo M:
Chromosomal instability determines taxane response. Proc Natl Acad
Sci USA. 106:8671–8676. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Pradhan M, Risberg BÅ, Tropé CG, van de
Rijn M, Gilks CB and Lee CH: Gross genomic alterations and gene
expression profiles of high-grade serous carcinoma of the ovary
with and without BRCA1 inactivation. BMC Cancer. 10:4932010.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Tang Z, Yang J, Wang X, Zeng M, Wang J,
Wang A, Zhao M, Guo L, Liu C, Li D and Chen J: Active DNA end
processing in micronuclei of ovarian cancer cells. BMC Cancer.
18:4262018. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Morden CR, Farrell AC, Sliwowski M,
Lichtensztejn Z, Altman AD, Nachtigal MW and McManus KJ: Chromosome
instability is prevalent and dynamic in high-grade serous ovarian
cancer patient samples. Gynecol Oncol. 161:769–778. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Gorringe KL, Chin SF, Pharoah P, Staines
JM, Oliveira C, Edwards PA and Caldas C: Evidence that both genetic
instability and selection contribute to the accumulation of
chromosome alterations in cancer. Carcinogenesis. 26:923–930. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Bayani J, Paderova J, Murphy J, Rosen B,
Zielenska M and Squire JA: Distinct patterns of structural and
numerical chromosomal instability characterize sporadic ovarian
cancer. Neoplasia. 10:1057–1065. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Birkbak NJ, Eklund AC, Li Q, McClelland
SE, Endesfelder D, Tan P, Tan IB, Richardson AL, Szallasi Z and
Swanton C: Paradoxical relationship between chromosomal instability
and survival outcome in cancer. Cancer Res. 71:3447–3452. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Hille S, Rein DT, Riffelmann M, Neumann R,
Sartorius J, Pfützner A, Kurbacher CM, Schöndorf T and Breidenbach
M: Anticancer drugs induce mdr1 gene expression in recurrent
ovarian cancer. Anticancer Drugs. 17:1041–1044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Zhang C, Wang M, Shi C, Shi F and Pei C:
Long non-coding RNA Linc00312 modulates the sensitivity of ovarian
cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway.
Biosci Trends. 12:309–316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Cui Y, Qin L, Tian D, Wang T, Fan L, Zhang
P and Wang Z: ZEB1 Promotes chemoresistance to cisplatin in ovarian
cancer cells by suppressing SLC3A2. Chemotherapy. 63:262–271. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Sen T, Sen N, Brait M, Begum S, Chatterjee
A, Hoque MO, Ratovitski E and Sidransky D: Np63 confers tumor cell
resistance to cisplatin through the AKT1 transcriptional
regulation. Cancer Res. 71:1167–1176. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Kumar S, Kumar A, Shah PP, Rai SN,
Panguluri SK and Kakar SS: MicroRNA signature of cis-platin
resistant vs. cis-platin sensitive ovarian cancer cell lines. J
Ovarian Res. 4:172011. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Leung AWY, Veinotte CJ, Melong N, Melong
N, Oh MH, Chen K, Enfield KSS, Backstrom I, Warburton C, Yapp D, et
al: In vivo validation of PAPSS1 (3′-phosphoadenosine
5′-phosphosulfate synthase 1) as a cisplatin-sensitizing
therapeutic target. Clin Cancer Res. 23:6555–6566. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
220
|
Kritsch D, Hoffmann F, Steinbach D, Jansen
L, Mary Photini S, Gajda M, Mosig AS, Sonnemann J, Peters S,
Melnikova M, et al: Tribbles 2 mediates cisplatin sensitivity and
DNA damage response in epithelial ovarian cancer. Int J Cancer.
141:1600–1614. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Nam EJ, Kim S, Lee TS, Kim HJ, Lee JY, Kim
SW, Kim JH and Kim YT: Primary and recurrent ovarian high-grade
serous carcinomas display similar microRNA expression patterns
relative to those of normal ovarian tissue. Oncotarget.
7:70524–70534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
222
|
Chong GO, Jeon HS, Han HS, Son JW, Lee YH,
Hong DG, Lee YS and Cho Y: Differential MicroRNA expression
profiles in primary and recurrent epithelial ovarian cancer.
Anticancer Res. 7:2611–2617. 2015.
|
|
223
|
Chong GO, Jeon HS, Han HS, Son JW, Lee YH,
Hong DG, Park HJ, Lee YS and Cho YL: Overexpression of
microRNA-196b accelerates invasiveness of cancer cells in recurrent
epithelial ovarian cancer through regulation of homeobox A9. Cancer
Genomics Proteomics. 14:137–142. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Zhou Y, Wang M, Wu J, Jie Z, Chang S and
Shuang T: The clinicopathological significance of miR-1307 in
chemotherapy resistant epithelial ovarian cancer. J Ovarian Res.
8:232015. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Chen C, Hu Y and Li L: NRP1 is targeted by
miR-130a and miR-130b, and is associated with multidrug resistance
in epithelial ovarian cancer based on integrated gene network
analysis. Mol Med Rep. 13:188–196. 2016. View Article : Google Scholar : PubMed/NCBI
|