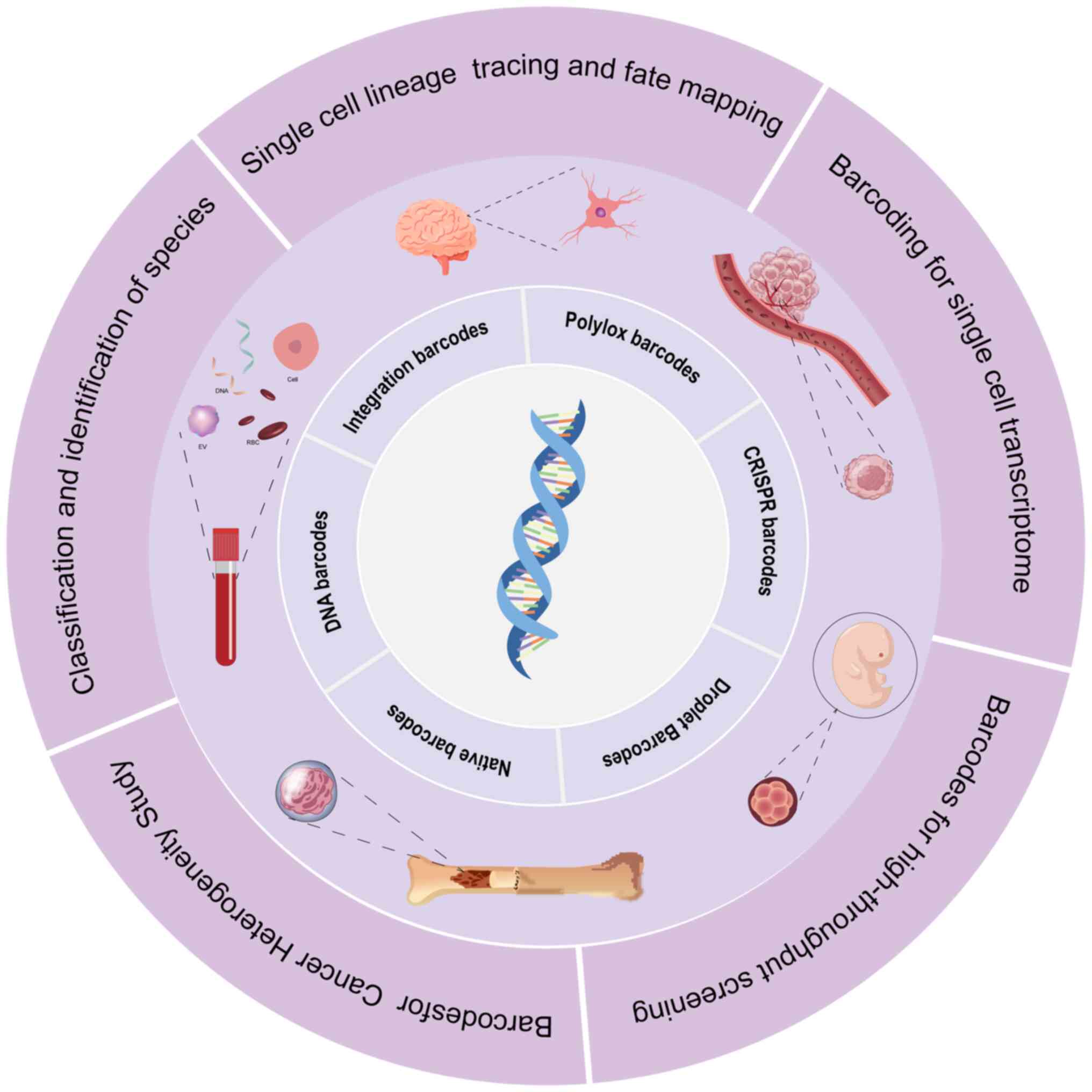
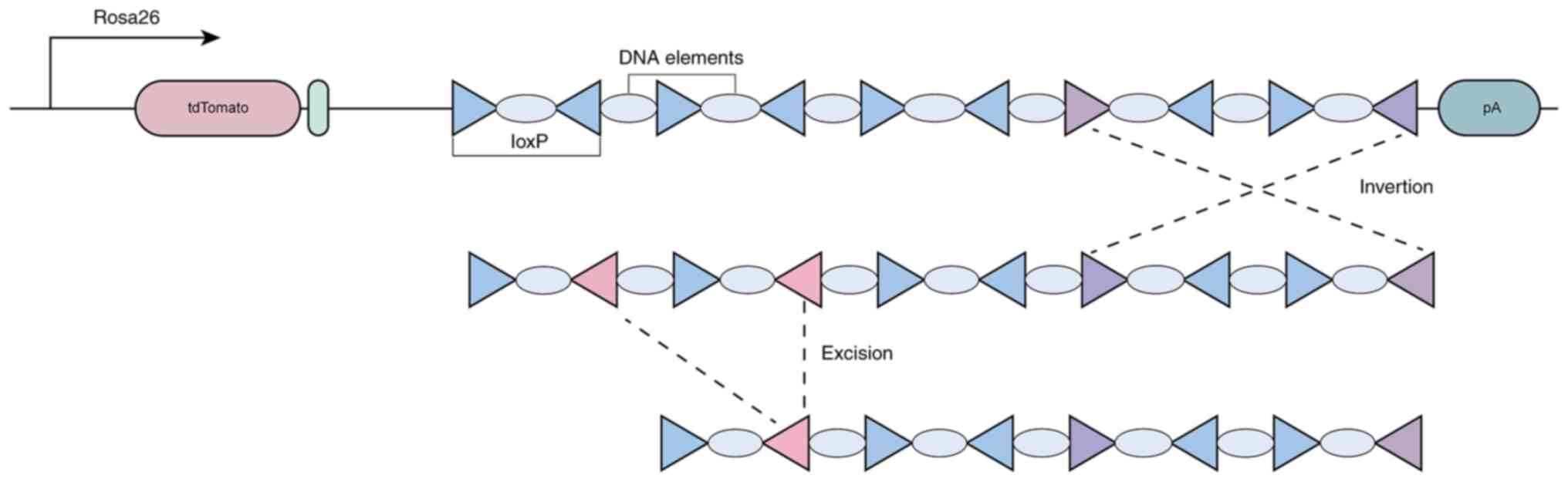
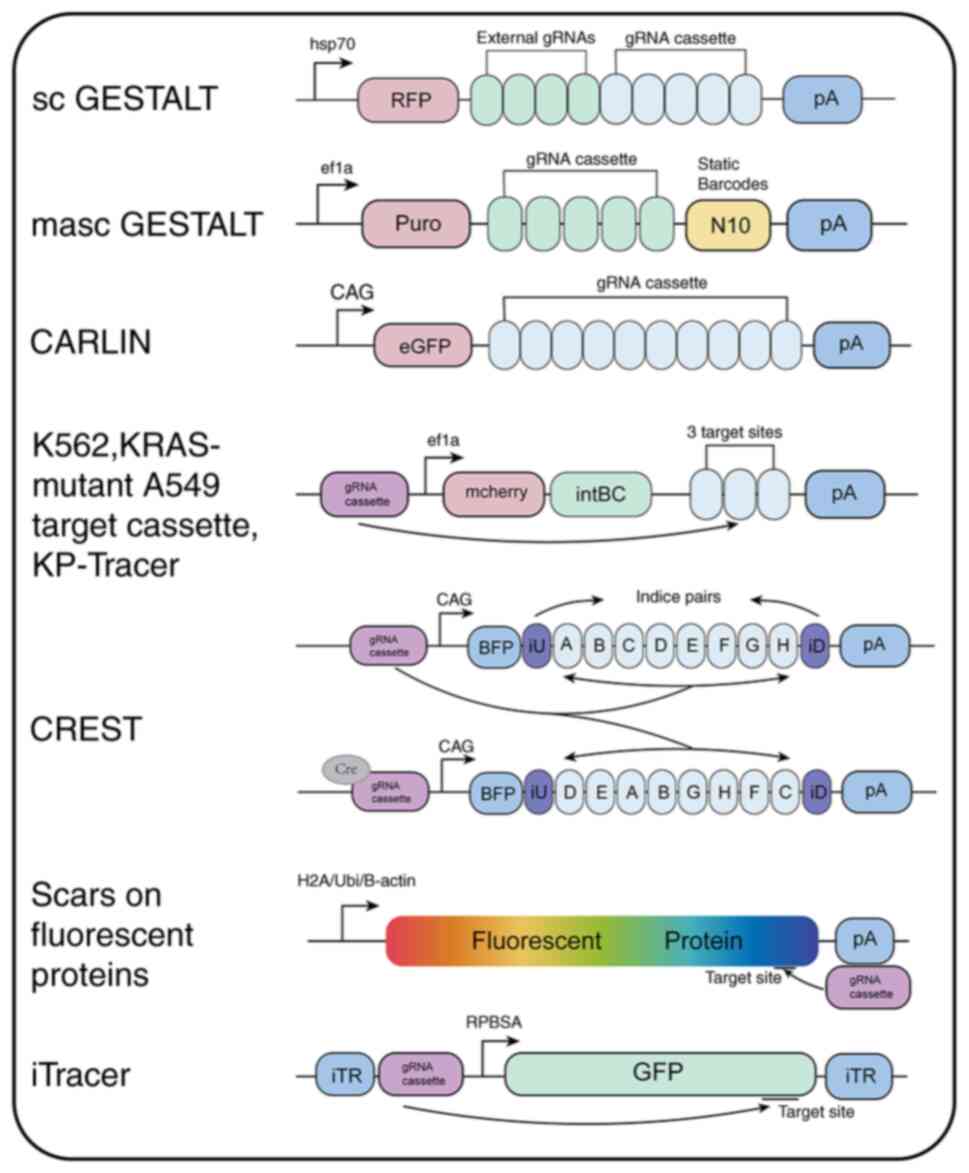
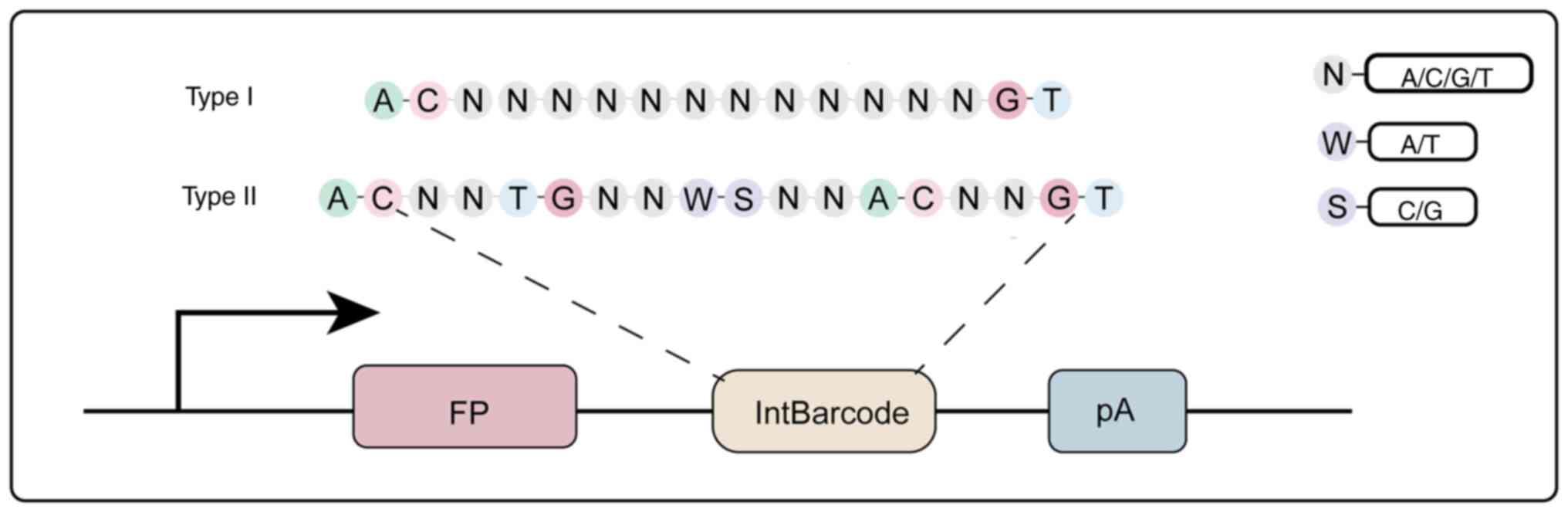
|
1
|
Loewer A and Lahav G: We are all
individuals: Causes and consequences of non-genetic heterogeneity
in mammalian cells. Curr Opin Genet Dev. 21:753–758. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang Y, Zhang X and Wang Z: Cellular
barcoding: From developmental tracing to anti-tumor drug discovery.
Cancer Lett. 567:2162812023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen C, Liao Y and Peng G: Connecting past
and present: Single-cell lineage tracing. Protein Cell. 13:790–807.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weinreb C and Klein AM: Lineage
reconstruction from clonal correlations. Proc Natl Acad Sci USA.
117:17041–17048. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Rop FV, Ismail JN, Bravo González-Blas
C, Hulselmans GJ, Flerin CC, Janssens J, Theunis K, Christiaens VM,
Wouters J, Marcassa G, et al: HyDrop enables droplet based
single-cell ATAC-seq and single-cell RNA-seq using dissolvable
hydrogel beads. Elife. 11:e739712022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sulston JE, Schierenberg E, White JG and
Thomson JN: The embryonic cell lineage of the nematode
Caenorhabditis elegans. Dev Biol. 100:64–119. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu R, Neff NF, Quake SR and Weissman IL:
Tracking single hematopoietic stem cells in vivo using
high-throughput sequencing in conjunction with viral genetic
barcoding. Nat Biotechnol. 29:928–933. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Weber K, Thomaschewski M, Warlich M, Volz
T, Cornils K, Niebuhr B, Täger M, Lütgehetmann M, Pollok JM,
Stocking C, et al: RGB marking facilitates multicolor clonal cell
tracking. Nat Med. 17:504–509. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weber K, Bartsch U, Stocking C and Fehse
B: A multicolor panel of novel lentiviral ‘Gene Ontology’ (LeGO)
vectors for functional gene analysis. Mol Ther. 16:698–706. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gomez-Nicola D, Riecken K, Fehse B and
Perry VH: In-vivo RGB marking and multicolour single-cell tracking
in the adult brain. Sci Rep. 4:75202014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weber K, Mock U, Petrowitz B, Bartsch U
and Fehse B: Lentiviral gene ontology (LeGO) vectors equipped with
novel drug-selectable fluorescent proteins: New building blocks for
cell marking and multi-gene analysis. Gene Ther. 17:511–520. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mohme M, Maire CL, Riecken K, Zapf S,
Aranyossy T, Westphal M, Lamszus K and Fehse B: Optical barcoding
for single-clone tracking to study tumor heterogeneity. Mol Ther.
25:621–633. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shembrey C, Smith J, Grandin M, Williams
N, Cho HJ, Mølck C, Behrenbruch C, Thomson BN, Heriot AG, Merino D
and Hollande F: Longitudinal monitoring of intra-tumoural
heterogeneity using optical barcoding of patient-derived colorectal
tumour models. Cancers (Basel). 14:5812022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schepers K, Swart E, van Heijst JW,
Gerlach C, Castrucci M, Sie D, Heimerikx M, Velds A, Kerkhoven RM,
Arens R and Schumacher TN: Dissecting T cell lineage relationships
by cellular barcoding. J Exp Med. 205:2309–2318. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van Heijst JW, Gerlach C, Swart E, Sie D,
Nunes-Alves C, Kerkhoven RM, Arens R, Correia-Neves M, Schepers K
and Schumacher TN: Recruitment of antigen-specific CD8+
T cells in response to infection is markedly efficient. Science.
325:1265–1269. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kalhor R, Kalhor K, Mejia L, Leeper K,
Graveline A, Mali P and Church GM: Developmental barcoding of whole
mouse via homing CRISPR. Science. 361:eaat98042018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kebschull JM and Zador AM: Cellular
barcoding: Lineage tracing, screening and beyond. Nat Methods.
15:871–879. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sankaran VG, Weissman JS and Zon LI:
Cellular barcoding to decipher clonal dynamics in disease. Science.
378:eabm58742022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Serrano A, Berthelet J, Naik SH and Merino
D: Mastering the use of cellular barcoding to explore cancer
heterogeneity. Nat Rev Cancer. 22:609–624. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Merino D, Weber TS, Serrano A, Vaillant F,
Liu K, Pal B, Di Stefano L, Schreuder J, Lin D, Chen Y, et al:
Barcoding reveals complex clonal behavior in patient-derived
xenografts of metastatic triple negative breast cancer. Nat Commun.
10:7662019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ko J, Wang Y, Carlson JCT, Marquard A,
Gungabeesoon J, Charest A, Weitz D, Pittet MJ and Weissleder R:
Single extracellular vesicle protein analysis using immuno-droplet
digital polymerase chain reaction amplification. Adv Biosyst.
4:e19003072020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Banijamali M, Höjer P, Nagy A, Hååg P,
Gomero EP, Stiller C, Kaminskyy VO, Ekman S, Lewensohn R, Karlström
AE, et al: Characterizing single extracellular vesicles by droplet
barcode sequencing for protein analysis. J Extracell Vesicles.
11:e122772022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yeo GHT, Lin L, Qi CY, Cha M, Gifford DK
and Sherwood RI: A multiplexed barcodelet single-cell RNA-Seq
approach elucidates combinatorial signaling pathways that drive ESC
differentiation. Cell Stem Cell. 26:938–950.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Klein AM, Mazutis L, Akartuna I,
Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA and Kirschner
MW: Droplet barcoding for single-cell transcriptomics applied to
embryonic stem cells. Cell. 161:1187–1201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Perié L, Duffy KR, Kok L, de Boer RJ and
Schumacher TN: The branching point in erythro-myeloid
differentiation. Cell. 163:1655–1662. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schueder F, Unterauer EM, Ganji M and
Jungmann R: DNA-Barcoded fluorescence microscopy for spatial omics.
Proteomics. 20:e19003682020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Howland KK and Brock A: Cellular barcoding
tracks heterogeneous clones through selective pressures and
phenotypic transitions. Trends Cancer. 9:591–601. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kretzschmar K and Watt FM: Lineage
Tracing. Cell. 148:33–45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Perié L and Duffy KR: Retracing the in
vivo haematopoietic tree using single-cell methods. FEBS Lett.
590:4068–4083. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kok L, Masopust D and Schumacher TN: The
precursors of CD8+ tissue resident memory T cells: From lymphoid
organs to infected tissues. Nat Rev Immunol. 22:283–293. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Naik SH, Perié L, Swart E, Gerlach C, van
Rooij N, de Boer RJ and Schumacher TN: Diverse and heritable
lineage imprinting of early haematopoietic progenitors. Nature.
496:229–232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dhimolea E, De Matos Simoes R, Kansara D,
Al'Khafaji A, Bouyssou J, Weng X, Sharma S, Raja J, Awate P,
Shirasaki R, et al: An embryonic diapause-like adaptation with
suppressed myc activity enables tumor treatment persistence. Cancer
Cell. 39:240–256.e11. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Echeverria GV, Ge Z, Seth S, Zhang X,
Jeter-Jones S, Zhou X, Cai S, Tu Y, McCoy A, Peoples M, et al:
Resistance to neoadjuvant chemotherapy in triple-negative breast
cancer mediated by a reversible drug-tolerant state. Sci Transl
Med. 11:eaav09362019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Echeverria GV, Powell E, Seth S, Ge Z,
Carugo A, Bristow C, Peoples M, Robinson F, Qiu H, Shao J, et al:
High-resolution clonal mapping of multi-organ metastasis in triple
negative breast cancer. Nat Commun. 9:50792018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blundell JR and Levy SF: Beyond genome
sequencing: Lineage tracking with barcodes to study the dynamics of
evolution, infection, and cancer. Genomics. 104((6 Pt A)): 417–430.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Macosko EZ, Basu A, Satija R, Nemesh J,
Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck
EM, et al: Highly parallel genome-wide expression profiling of
individual cells using nanoliter droplets. Cell. 161:1202–1214.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Naik SH, Schumacher TN and Perié L:
Cellular barcoding: A technical appraisal. Exp Hematol. 42:598–608.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Spanjaard B, Hu B, Mitic N,
Olivares-Chauvet P, Janjuha S, Ninov N and Junker JP: Simultaneous
lineage tracing and cell-type identification using
CRISPR-Cas9-induced genetic scars. Nat Biotechnol. 36:469–473.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wagner DE and Klein AM: Lineage tracing
meets single-cell omics: opportunities and challenges. Nat Rev
Genet. 21:410–427. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cai D, Cohen KB, Luo T, Lichtman JW and
Sanes JR: Improved tools for the Brainbow toolbox. Nat Methods.
10:540–547. 2013. View Article : Google Scholar
|
|
41
|
Pei W, Feyerabend TB, Rössler J, Wang X,
Postrach D, Busch K, Rode I, Klapproth K, Dietlein N, Quedenau C,
et al: Polylox barcoding reveals haematopoietic stem cell fates
realized in vivo. Nature. 548:456–460. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pei W, Wang X, Rössler J, Feyerabend TB,
Höfer T and Rodewald HR: Using Cre-recombinase-driven Polylox
barcoding for in vivo fate mapping in mice. Nat Protoc.
14:1820–1840. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
McLellan MA, Rosenthal NA and Pinto AR:
Cre- loxP-mediated recombination: General principles and
experimental considerations. Curr Protoc Mouse Biol. 7:1–12. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pei W, Shang F, Wang X, Fanti AK, Greco A,
Busch K, Klapproth K, Zhang Q, Quedenau C, Sauer S, et al:
Resolving fates and single-cell transcriptomes of hematopoietic
stem cell clones by polyloxexpress barcoding. Cell Stem Cell.
27:383–395.e8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Becher B, Waisman A and Lu LF: Conditional
gene-targeting in mice: Problems and solutions. Immunity.
48:835–836. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
McDavid A, Finak G, Chattopadyay PK,
Dominguez M, Lamoreaux L, Ma SS, Roederer M and Gottardo R: Data
exploration, quality control and testing in single-cell qPCR-based
gene expression experiments. Bioinformatics. 29:461–467. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kharchenko PV, Silberstein L and Scadden
DT: Bayesian approach to single-cell differential expression
analysis. Nat Methods. 11:740–742. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan J, Slowikowski K and Zhang F:
Single-cell transcriptomics in cancer: Computational challenges and
opportunities. Exp Mol Med. 52:1452–1465. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu T, Park S, Zhu J, Wang Y, Zhan X, Wang
X, Wang L, Zhu H and Wang T: Overcoming expressional drop-outs in
lineage reconstruction from single-cell RNA-sequencing data. Cell
Rep. 34:1085892021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang JY and Doudna JA: CRISPR technology:
A decade of genome editing is only the beginning. Science.
379:eadd86432023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Raj B, Wagner DE, McKenna A, Pandey S,
Klein AM, Shendure J, Gagnon JA and Schier AF: Simultaneous
single-cell profiling of lineages and cell types in the vertebrate
brain. Nat Biotechnol. 36:442–450. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
McKenna A, Findlay GM, Gagnon JA, Horwitz
MS, Schier AF and Shendure J: Whole-organism lineage tracing by
combinatorial and cumulative genome editing. Science.
353:aaf79072016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Raj B, Gagnon JA and Schier AF:
Large-scale reconstruction of cell lineages using single-cell
readout of transcriptomes and CRISPR-Cas9 barcodes by scGESTALT.
Nat Protoc. 13:2685–2713. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Simeonov KP, Byrns CN, Clark ML, Norgard
RJ, Martin B, Stanger BZ, Shendure J, McKenna A and Lengner CJ:
Single-cell lineage tracing of metastatic cancer reveals selection
of hybrid EMT states. Cancer Cell. 39:1150–1162.e9. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bowling S, Sritharan D, Osorio FG, Nguyen
M, Cheung P, Rodriguez-Fraticelli A, Patel S, Yuan WC, Fujiwara Y,
Li BE, et al: An engineered CRISPR-Cas9 mouse line for simultaneous
readout of lineage histories and gene expression profiles in single
cells. Cell. 181:1410–1422.e27. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chan MM, Smith ZD, Grosswendt S, Kretzmer
H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, et al:
Molecular recording of mammalian embryogenesis. Nature. 570:77–82.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Quinn JJ, Jones MG, Okimoto RA, Nanjo S,
Chan MM, Yosef N, Bivona TG and Weissman JS: Single-cell lineages
reveal the rates, routes, and drivers of metastasis in cancer
xenografts. Science (New York, NY). 371:eabc19442021. View Article : Google Scholar
|
|
58
|
Yang D, Jones MG, Naranjo S, Rideout WM
III, Min KHJ, Ho R, Wu W, Replogle JM, Page JL, Quinn JJ, et al:
Lineage tracing reveals the phylodynamics, plasticity, and paths of
tumor evolution. Cell. 185:1905–1923.e25. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Xie L, Liu H, You Z, Wang L, Li Y, Zhang
X, Ji X, He H, Yuan T, Zheng W, et al: Comprehensive spatiotemporal
mapping of single-cell lineages in developing mouse brain by
CRISPR-based barcoding. Nat Methods. 20:1244–1255. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Alemany A, Florescu M, Baron CS,
Peterson-Maduro J and Van Oudenaarden A: Whole-organism clone
tracing using single-cell sequencing. Nature. 556:108–112. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Allen F, Crepaldi L, Alsinet C, Strong AJ,
Kleshchevnikov V, De Angeli P, Páleníková P, Khodak A, Kiselev V,
Kosicki M, et al: Predicting the mutations generated by repair of
Cas9-induced double-strand breaks. Nat Biotechnol. Nov
27–2018.(Epub ahead of print).
|
|
62
|
Schmidt ST, Zimmerman SM, Wang J, Kim SK
and Quake SR: Quantitative analysis of synthetic cell lineage
tracing using nuclease barcoding. ACS Synth Biol. 6:936–942. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Varanasi A and Wilson E: The Applications
of the CRISPR/Cas9 Gene-Editing System in Treating Human Diseases.
J Stud Res. 11:1–17. 2022. View Article : Google Scholar
|
|
64
|
Choubisa V and Sharma V: Unveiling neural
network potential in forecasting CRISPR effects and off-target
prophecies for gene editing. Int J Sci Res Arch. 10:252–259. 2023.
View Article : Google Scholar
|
|
65
|
Moreno-Ayala R and Junker JP: Single-cell
genomics to study developmental cell fate decisions in zebrafish.
Brief Funct Genomics. Mar 30–2021.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yabe IM, Truitt LL, Espinoza DA, Wu C,
Koelle S, Panch S, Corat MAF, Winkler T, Yu KR, Hong SG, et al:
Barcoding of macaque hematopoietic stem and progenitor cells: A
robust platform to assess vector genotoxicity. Mol Ther Methods
Clin Dev. 11:143–154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Amrani N, Gao XD, Liu P, Edraki A, Mir A,
Ibraheim R, Gupta A, Sasaki KE, Wu T, Donohoue PD, et al: NmeCas9
is an intrinsically high-fidelity genome-editing platform. Genome
Biol. 19:2142018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhao L, Liu Z, Levy SF and Wu S:
Bartender: A fast and accurate clustering algorithm to count
barcode reads. Bioinformatics. 34:739–747. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Johnson MS, Venkataram S and Kryazhimskiy
S: Best practices in designing, sequencing, and identifying random
DNA barcodes. J Mol Evol. 91:263–280. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Störtz F and Minary P: crisprSQL: A novel
database platform for CRISPR/Cas off-target cleavage assays.
Nucleic Acids Res. 49((D1)): D855–D861. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Cradick TJ, Qiu P, Lee CM, Fine EJ and Bao
G: COSMID: A web-based tool for identifying and validating
CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 3:e2142014.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Bystrykh LV and Belderbos ME: Clonal
analysis of cells with cellular barcoding: When numbers and sizes
matter. Methods Mol Biol. 1516:57–89. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Rodriguez-Fraticelli AE, Wolock SL,
Weinreb CS, Panero R, Patel SH, Jankovic M, Sun J, Calogero RA,
Klein AM and Camargo FD: Clonal analysis of lineage fate in native
haematopoiesis. Nature. 553:212–216. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Bramlett C, Jiang D, Nogalska A, Eerdeng
J, Contreras J and Lu R: Clonal tracking using embedded viral
barcoding and high-throughput sequencing. Nat Protoc. 15:1436–1458.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ordon J, Thouin J, Nakano RT, et al:
Simultaneous tracking of near-isogenic bacterial strains in
synthetic Arabidopsis microbiota by chromosomally-integrated
barcodes. 2023.
|
|
76
|
Leibovich N and Goyal S: Limitations and
Optimizations of Cellular Lineages Tracking. biorxiv. doi:.
https://doi.org/10.1101/2023.03.15.532767
|
|
77
|
Frenkel M, Hujoel MLA, Morris Z and Raman
S: Discovering chromatin dysregulation induced by protein-coding
perturbations at scale. bioRxiv. doi:. https://doi.org/10.1101/2023.09.20.555752
|
|
78
|
Blois S, Goetz BM, Bull JJ and Sullivan
CS: Interpreting and de-noising genetically engineered barcodes in
a DNA virus. PLoS Comput Biol. 18:e10101312022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Anwar SL, Wulaningsih W and Lehmann U:
Transposable elements in human cancer: Causes and consequences of
deregulation. Int J Mol Sci. 18:9742017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Ohigashi I, Yamasaki Y, Hirashima T and
Takahama Y: Identification of the transgenic integration site in
immunodeficient tgε26 human CD3ε transgenic mice. PLoS One.
5:e143912010. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wolff JH and Mikkelsen JG: Delivering
genes with human immunodeficiency virus-derived vehicles: Still
state-of-the-art after 25 years. J Biomed Sci. 29:792022.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Porter SN, Baker LC, Mittelman D and
Porteus MH: Lentiviral and targeted cellular barcoding reveals
ongoing clonal dynamics of cell lines in vitro and in vivo. Genome
Biol. 15:R752014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kivioja T, Vähärautio A, Karlsson K, Bonke
M, Enge M, Linnarsson S and Taipale J: Counting absolute numbers of
molecules using unique molecular identifiers. Nat Methods. 9:72–74.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Fan HC, Fu GK and Fodor SP: Combinatorial
labeling of single cells for gene expression cytometry. Science.
347:12583672015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hashimshony T, Wagner F, Sher N and Yanai
I: CEL-Seq: Single-cell RNA-Seq by multiplexed linear
amplification. Cell Rep. 2:666–673. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Picelli S, Björklund ÅK, Faridani OR,
Sagasser S, Winberg G and Sandberg R: Smart-seq2 for sensitive
full-length transcriptome profiling in single cells. Nat Methods.
10:1096–1098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z,
Huang Y and Wang J: Comparative analysis of droplet-based
ultra-high-throughput single-cell RNA-Seq systems. Mol Cell.
73:130–142.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zheng GX, Terry JM, Belgrader P, Ryvkin P,
Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et
al: Massively parallel digital transcriptional profiling of single
cells. Nat Commun. 8:140492017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Rotem A, Ram O, Shoresh N, Sperling RA,
Schnall-Levin M, Zhang H, Basu A, Bernstein BE and Weitz DA:
High-throughput single-cell labeling (Hi-SCL) for RNA-Seq using
drop-based microfluidics. PLoS One. 10:e01163282015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Pinglay S, Lalanne JB, Daza RM, Koeppel J,
Li X, Lee DS and Shendure J: Multiplex generation and single-cell
analysis of structural variants in mammalian genomes. Science.
387:eado59782025. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lan F, Demaree B, Ahmed N and Abate AR:
Single-cell genome sequencing at ultra-high-throughput with
microfluidic droplet barcoding. Nat Biotechnol. 35:640–646. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Li J, Zhang R, Li T, et al:
Ultra-high-throughput microbial single-cell whole genome sequencing
for genome-resolved metagenomics. 2022. View Article : Google Scholar
|
|
93
|
Shembekar N, Chaipan C, Utharala R and
Merten CA: Droplet-based microfluidics in drug discovery,
transcriptomics and high-throughput molecular genetics. Lab Chip.
16:1314–1331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Suea-Ngam A, Howes PD, Srisa-Art M and
deMello AJ: Droplet microfluidics: From proof-of-concept to
real-world utility? Chem Commun (Camb). 55:9895–9903. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Sheng C, Lopes R, Li G, et al:
Probabilistic machine learning ensures accurate ambient denoising
in droplet-based single-cell omics. 2022. View Article : Google Scholar
|
|
96
|
Imoto Y: Comprehensive Noise Reduction in
Single-Cell Data with the RECODE Platform. biorxiv. doi:.
https://doi.org/10.1101/2024.04.18.590054
|
|
97
|
Sun C, Kathuria K, Emery SB, Kim B,
Burbulis IE, Shin JH; Brain Somatic Mosaicism Network, ; Weinberger
DR, Moran JV, Kidd JM, et al: Mapping recurrent mosaic copy number
variation in human neurons. Nat Commun. 15:42202024. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Bo L, Liang Y and Lin G: Cancer
classification based on multiple dimensions: SNV patterns. Comput
Biol Med. 151((Pt A)): 1062702022.PubMed/NCBI
|
|
99
|
McConnell MJ, Lindberg MR, Brennand KJ,
Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS,
Vermeesch JR, Hall IM and Gage FH: Mosaic copy number variation in
human neurons. Science. 342:632–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Addis L, Ahn JW, Dobson R, Dixit A,
Ogilvie CM, Pinto D, Vaags AK, Coon H, Chaste P, Wilson S, et al:
Microdeletions of ELP4 are associated with language impairment,
autism spectrum disorder, and mental retardation. Hum Mutat.
36:842–850. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Xiong E, Liu P, Deng R, Zhang K, Yang R
and Li J: Recent advances in enzyme-free and enzyme-mediated
single-nucleotide variation assay in vitro. Natl Sci Rev.
11:nwae1182024. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Kadam PS, Yang Z, Lu Y, Zhu H, Atiyas Y,
Shah N, Fisher S, Nordgren E, Kim J, Issadore D and Eberwine J:
Single-mitochondrion sequencing uncovers distinct mutational
patterns and heteroplasmy landscape in mouse astrocytes and
neurons. BMC Biol. 22:1622024. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Dondi A, Borgsmüller N, Ferreira PF, Haas
BJ, Jacob F and Heinzelmann-Schwarz V; Tumor Profiler Consortium;
Beerenwinkel N, : De novo detection of somatic variants in
high-quality long-read single-cell RNA sequencing data. bioRxiv
[Preprint]. 2024.03.06.583775. 2024.
|
|
104
|
Ganz J, Luquette LJ, Bizzotto S, Miller
MB, Zhou Z, Bohrson CL, Jin H, Tran AV, Viswanadham VV, McDonough
G, et al: Contrasting somatic mutation patterns in aging human
neurons and oligodendrocytes. Cell. 187:1955–1970.e23. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Csordas A, Sipos B, Kurucova T, Volfova A,
Zamola F, Tichy B and Hicks DG: Cell Tree Rings: The structure of
somatic evolution as a human aging timer. Geroscience.
46:3005–3019. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
You X, Cao Y, Suzuki T, Shao J, Zhu B,
Masumura K, Xi J, Liu W, Zhang X and Luan Y: Genome-wide direct
quantification of in vivo mutagenesis using high-accuracy
paired-end and complementary consensus sequencing. Nucleic Acids
Res. 51:e1092023. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Weng C, Weissman JS and Sankaran VG:
Robustness and reliability of single-cell regulatory multi-omics
with deep mitochondrial mutation profiling. bioRxiv [Preprint].
2024.08.23.609473. 2024.
|
|
108
|
Evrony GD, Lee E, Mehta BK, Benjamini Y,
Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ and Walsh
CA: Cell lineage analysis in human brain using endogenous
retroelements. Neuron. 85:49–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Redon R, Ishikawa S, Fitch KR, Feuk L,
Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et
al: Global variation in copy number in the human genome. Nature.
444:444–454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wei CJ and Zhang K: RETrace: Simultaneous
retrospective lineage tracing and methylation profiling of single
cells. Genome Res. 30:602–610. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Fink J, Andersson-Rolf A and Koo BK: Adult
stem cell lineage tracing and deep tissue imaging. BMB Rep.
48:655–667. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Woodworth MB, Girskis KM and Walsh CA:
Building a lineage from single cells: Genetic techniques for cell
lineage tracking. Nat Rev Genet. 18:230–244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Li L, Bowling S, McGeary SE, Yu Q, Lemke
B, Alcedo K, Jia Y, Liu X, Ferreira M, Klein AM, et al: A mouse
model with high clonal barcode diversity for joint lineage,
transcriptomic, and epigenomic profiling in single cells. Cell.
186:5183–5199.e22. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Walsh C and Cepko CL: Widespread
dispersion of neuronal clones across functional regions of the
cerebral cortex. Science. 255:434–440. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Gerrits A, Dykstra B, Kalmykowa OJ, Klauke
K, Verovskaya E, Broekhuis MJ, de Haan G and Bystrykh LV: Cellular
barcoding tool for clonal analysis in the hematopoietic system.
Blood. 115:2610–2618. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Golden JA, Fields-Berry SC and Cepko CL:
Construction and characterization of a highly complex retroviral
library for lineage analysis. Proc Natl Acad Sci USA. 92:5704–5708.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Khozyainova AA, Valyaeva AA, Arbatsky MS,
Isaev SV, Iamshchikov PS, Volchkov EV, Sabirov MS, Zainullina VR,
Chechekhin VI, Vorobev RS, et al: Complex analysis of single-cell
RNA sequencing data. Biochemistry (Mosc). 88:231–252. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Weinreb C, Rodriguez-Fraticelli A, Camargo
FD and Klein AM: Lineage tracing on transcriptional landscapes
links state to fate during differentiation. Science.
367:eaaw33812020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Bizzotto S, Dou Y, Ganz J, Doan RN, Kwon
M, Bohrson CL, Kim SN, Bae T, Abyzov A; NIMH Brain Somatic
Mosaicism Network, ; et al: Landmarks of human embryonic
development inscribed in somatic mutations. Science. 371:1249–1253.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Sun J, Ramos A, Chapman B, Johnnidis JB,
Le L, Ho YJ, Klein A, Hofmann O and Camargo FD: Clonal dynamics of
native haematopoiesis. Nature. 514:322–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Kim IS, Wu J, Rahme GJ, Battaglia S, Dixit
A, Gaskell E, Chen H, Pinello L and Bernstein BE: Parallel
single-cell RNA-Seq and genetic recording reveals lineage decisions
in developing embryoid bodies. Cell Rep. 33:1082222020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Wagenblast E, Soto M, Gutiérrez-Ángel S,
Hartl CA, Gable AL, Maceli AR, Erard N, Williams AM, Kim SY,
Dickopf S, et al: A model of breast cancer heterogeneity reveals
vascular mimicry as a driver of metastasis. Nature. 520:358–362.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Akimov Y, Bulanova D, Timonen S,
Wennerberg K and Aittokallio T: Improved detection of
differentially represented DNA barcodes for high-throughput clonal
phenomics. Mol Syst Biol. 16:e91952020. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
McKenna A and Gagnon JA: Recording
development with single cell dynamic lineage tracing. Development.
146:dev1697302019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Wang MY, Zhou Y, Lai GS, Huang Q, Cai WQ,
Han ZW, Wang Y, Ma Z, Wang XW, Xiang Y, et al: DNA barcode to trace
the development and differentiation of cord blood stem cells
(Review). Mol Med Rep. 24:8492021. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Petropoulos S, Edsgärd D, Reinius B, Deng
Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R
and Lanner F: Single-cell RNA-Seq reveals lineage and X chromosome
dynamics in human preimplantation embryos. Cell. 165:1012–1026.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Farrell JA, Wang Y, Riesenfeld SJ, Shekhar
K, Regev A and Schier AF: Single-cell reconstruction of
developmental trajectories during zebrafish embryogenesis. Science.
360:eaar31312018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
McFarland JM, Paolella BR, Warren A,
Geiger-Schuller K, Shibue T, Rothberg M, Kuksenko O, Colgan WN,
Jones A, Chambers E, et al: Multiplexed single-cell transcriptional
response profiling to define cancer vulnerabilities and therapeutic
mechanism of action. Nat Commun. 11:42962020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Birnbaum KD: Power in numbers: Single-cell
RNA-Seq strategies to dissect complex tissues. Annu Rev Genet.
52:203–221. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Lan F, Saba J, Ross TD, Zhou Z, Krauska K,
Anantharaman K, Landick R and Venturelli OS: Massively parallel
single-cell sequencing of diverse microbial populations. Nat
Methods. 21:228–235. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Cusanovich DA, Daza R, Adey A, Pliner HA,
Christiansen L, Gunderson KL, Steemers FJ, Trapnell C and Shendure
J: Multiplex single-cell profiling of chromatin accessibility by
combinatorial cellular indexing. Science. 348:910–914. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Zhu C, Yu M, Huang H, Juric I, Abnousi A,
Hu R, Lucero J, Behrens MM, Hu M and Ren B: An ultra
high-throughput method for single-cell joint analysis of open
chromatin and transcriptome. Nat Struct Mol Biol. 26:1063–1070.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Wu B, Bennett HM, Ye X, Sridhar A,
Eidenschenk C, Everett C, Nazarova EV, Chen HH, Kim IK, Deangelis
M, et al: Overloading And unpacKing (OAK)-droplet-based
combinatorial indexing for ultra-high throughput single-cell
multiomic profiling. Nat Commun. 15:91462024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Mathur L, Szalai B, Du NH, Utharala R,
Ballinger M, Landry JJM, Ryckelynck M, Benes V, Saez-Rodriguez J
and Merten CA: Combi-seq for multiplexed transcriptome-based
profiling of drug combinations using deterministic barcoding in
single-cell droplets. Nat Commun. 13:44502022. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Paddison PJ, Silva JM, Conklin DS,
Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L,
Scobie K, et al: A resource for large-scale RNA-interference-based
screens in mammals. Nature. 428:427–431. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Silva JM, Marran K, Parker JS, Silva J,
Golding M, Schlabach MR, Elledge SJ, Hannon GJ and Chang K:
Profiling essential genes in human mammary cells by multiplex RNAi
screening. Science. 319:617–620. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Datlinger P, Rendeiro AF, Schmidl C,
Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A,
Alpar D and Bock C: Pooled CRISPR screening with single-cell
transcriptome readout. Nat Methods. 14:297–301. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Duncombe TA and Dittrich PS: Droplet
barcoding: Tracking mobile micro-reactors for high-throughput
biology. Curr Opin Biotechnol. 60:205–212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Adamson B, Norman TM, Jost M, Cho MY,
Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et
al: A multiplexed single-cell CRISPR screening platform enables
systematic dissection of the unfolded protein response. Cell.
167:1867–1882.e21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Jaitin DA, Weiner A, Yofe I, Lara-Astiaso
D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A
and Amit I: Dissecting immune circuits by linking CRISPR-pooled
screens with single-cell RNA-Seq. Cell. 167:1883–1896.e15. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Emanuel G, Moffitt JR and Zhuang X:
High-throughput, image-based screening of pooled genetic-variant
libraries. Nat Methods. 14:1159–1162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Camp JG, Platt R and Treutlein B: Mapping
human cell phenotypes to genotypes with single-cell genomics.
Science. 365:1401–1405. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Xie S, Duan J, Li B, Zhou P and Hon GC:
Multiplexed engineering and analysis of combinatorial enhancer
activity in single cells. Mol Cell. 66:285–299.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Lawson MJ, Camsund D, Larsson J, Baltekin
Ö, Fange D and Elf J: In situ genotyping of a pooled strain library
after characterizing complex phenotypes. Mol Syst Biol. 13:9472017.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Marusyk A, Janiszewska M and Polyak K:
Intratumor heterogeneity: The rosetta stone of therapy resistance.
Cancer Cell. 37:471–484. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Shembrey C, Huntington ND and Hollande F:
Impact of tumor and immunological heterogeneity on the anti-cancer
immune response. Cancers (Basel). 11:12172019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Lewis SM, Asselin-Labat ML, Nguyen Q,
Berthelet J, Tan X, Wimmer VC, Merino D, Rogers KL and Naik SH:
Spatial omics and multiplexed imaging to explore cancer biology.
Nat Methods. 18:997–1012. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Fennell KA, Vassiliadis D, Lam EYN,
Martelotto LG, Balic JJ, Hollizeck S, Weber TS, Semple T, Wang Q,
Miles DC, et al: Non-genetic determinants of malignant clonal
fitness at single-cell resolution. Nature. 601:125–131. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Jin X, Demere Z, Nair K, Ali A, Ferraro
GB, Natoli T, Deik A, Petronio L, Tang AA, Zhu C, et al: A
metastasis map of human cancer cell lines. Nature. 588:331–336.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Holcar M, Ferdin J, Sitar S,
Tušek-Žnidarič M, Dolžan V, Plemenitaš A, Žagar E and Lenassi M:
Enrichment of plasma extracellular vesicles for reliable
quantification of their size and concentration for biomarker
discovery. Sci Rep. 10:213462020. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Welsh JA, Goberdhan DCI, O'Driscoll L,
Buzas EI, Blenkiron C, Bussolati B, Cai H, Di Vizio D, Driedonks
TAP, Erdbrügger U, et al: Minimal information for studies of
extracellular vesicles (MISEV2023): From basic to advanced
approaches. J Extracell Vesicles. 13:e124042024. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Lozano-Andrés E, Enciso-Martinez A,
Gijsbers A, Ridolfi A, Van Niel G, Libregts SFWM, Pinheiro C, van
Herwijnen MJC, Hendrix A, Brucale M, et al: Physical association of
low density lipoprotein particles and extracellular vesicles
unveiled by single particle analysis. J Extracell Vesicles.
12:e123762023. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Chang W-H, Cerione RA and Antonyak MA:
Extracellular Vesicles and Their Roles in Cancer Progression.
Cancer Cell Signaling. vol. 2174. Robles-Flores M: Springer US; New
York, NY: pp. 143–170. 2021, View Article : Google Scholar
|
|
154
|
Ferguson S, Yang KS and Weissleder R:
Single extracellular vesicle analysis for early cancer detection.
Trends Mol Med. 28:681–692. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Yu W, Hurley J, Roberts D, Chakrabortty
SK, Enderle D, Noerholm M, Breakefield XO and Skog JK:
Exosome-based liquid biopsies in cancer: opportunities and
challenges. Ann Oncol. 32:466–477. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Verweij FJ, Balaj L, Boulanger CM, Carter
DRF, Compeer EB, D'Angelo G, El Andaloussi S, Goetz JG, Gross JC,
Hyenne V, et al: The power of imaging to understand extracellular
vesicle biology in vivo. Nat Methods. 18:1013–1026. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Crescitelli R, Lässer C and Lötvall J:
Isolation and characterization of extracellular vesicle
subpopulations from tissues. Nat Protoc. 16:1548–1580. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Ko J, Wang Y, Sheng K, Weitz DA and
Weissleder R: Sequencing-based protein analysis of single
extracellular vesicles. ACS Nano. 15:5631–5638. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Ivanova A, Chalupska R, Louro AF, Firth M,
González-King Garibotti H, Hultin L, Kohl F, Lázaro-Ibáñez E,
Lindgren J, Musa G, et al: Barcoded hybrids of extracellular
vesicles and lipid nanoparticles for multiplexed analysis of tissue
distribution. Adv Sci (Weinh). 12:e24078502025. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Kunitake K, Mizuno T, Hattori K, Oneyama
C, Kamiya M, Ota S, Urano Y and Kojima R: Barcoding of small
extracellular vesicles with CRISPR-gRNA enables comprehensive,
subpopulation-specific analysis of their biogenesis and release
regulators. Nat Commun. 15:97772024. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Farzad N, Enninful A, Bao S, Zhang D, Deng
Y and Fan R: Spatially resolved epigenome sequencing via Tn5
transposition and deterministic DNA barcoding in tissue. Nat
Protoc. 19:3389–3425. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Pandit S, Duchow M, Chao W, Capasso A and
Samanta D: DNA-barcoded plasmonic nanostructures for activity-based
protease sensing. Angew Chem Int Ed Engl. 63:e2023109642024.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Lareau CA, Ma S, Duarte FM and Buenrostro
JD: Inference and effects of barcode multiplets in droplet-based
single-cell assays. Nat Commun. 11:8662020. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Hotinger JA, Campbell IW, Hullahalli K,
Osaki A and Waldor MK: Quantification of Salmonella enterica
serovar typhimurium population dynamics in murine infection using a
highly diverse barcoded library. Elife. 13:RP1013882024. View Article : Google Scholar
|
|
165
|
Karlsson F, Kallas T, Thiagarajan D,
Karlsson M, Schweitzer M, Navarro JF, Leijonancker L, Geny S,
Pettersson E, Rhomberg-Kauert J, et al: Molecular pixelation:
spatial proteomics of single cells by sequencing. Nat Methods.
21:1044–1052. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Eyler CE, Matsunaga H, Hovestadt V,
Vantine SJ, van Galen P and Bernstein BE: Single-cell lineage
analysis reveals genetic and epigenetic interplay in glioblastoma
drug resistance. Genome Biol. 21:1742020. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Maetzig T, Morgan M and Schambach A:
Fluorescent genetic barcoding for cellular multiplex analyses. Exp
Hematol. 67:10–17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Maetzig T, Ruschmann J, Sanchez Milde L,
Lai CK, Von Krosigk N and Humphries RK: Lentiviral Fluorescent
Genetic Barcoding for Multiplex Fate Tracking of Leukemic Cells.
Mol Ther Methods Clin Dev. 6:54–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Yoon B, Kim H, Jung SW and Park J:
Single-cell lineage tracing approaches to track kidney cell
development and maintenance. Kidney Int. 105:1186–1199. 2024.
View Article : Google Scholar : PubMed/NCBI
|