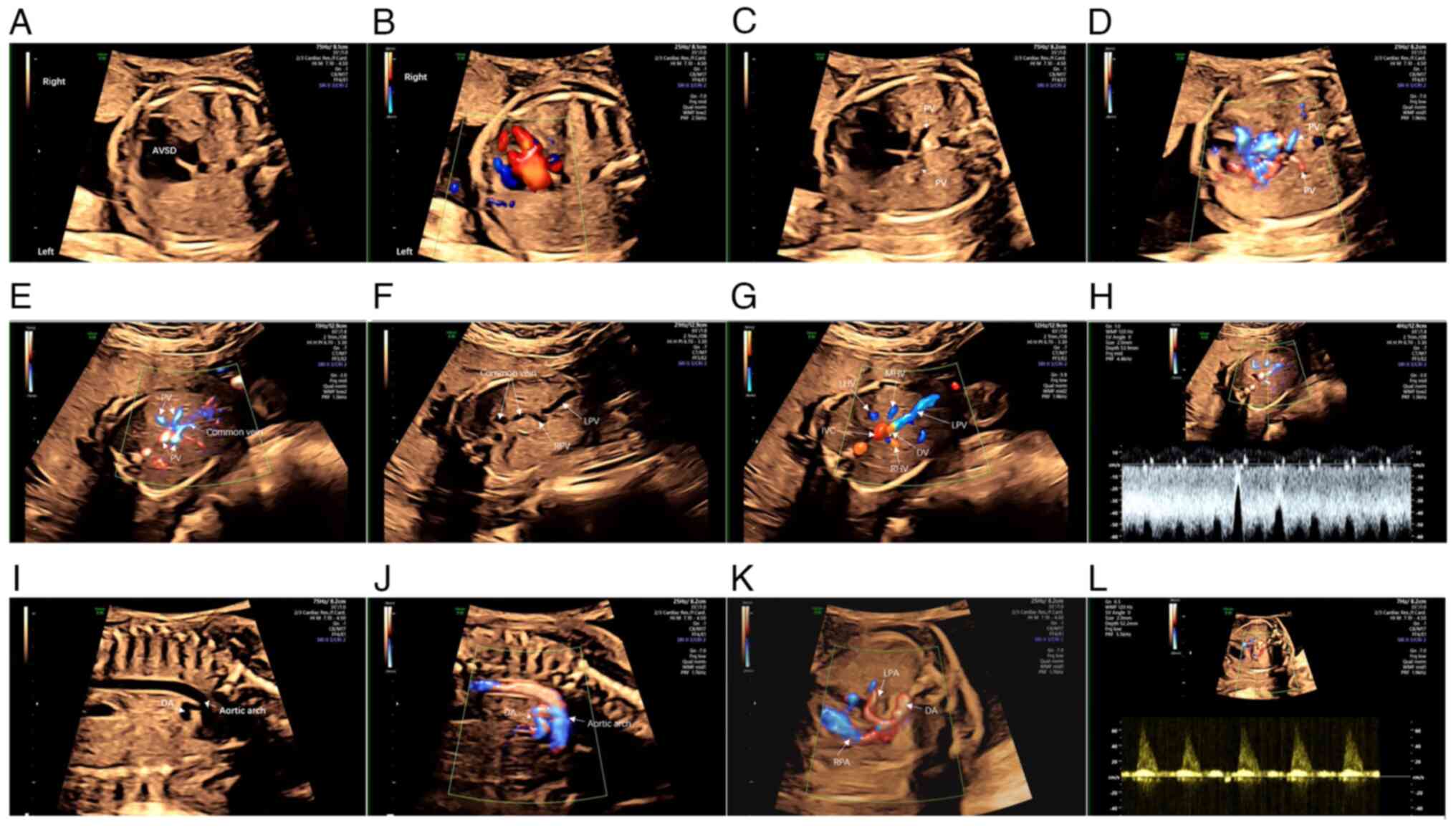
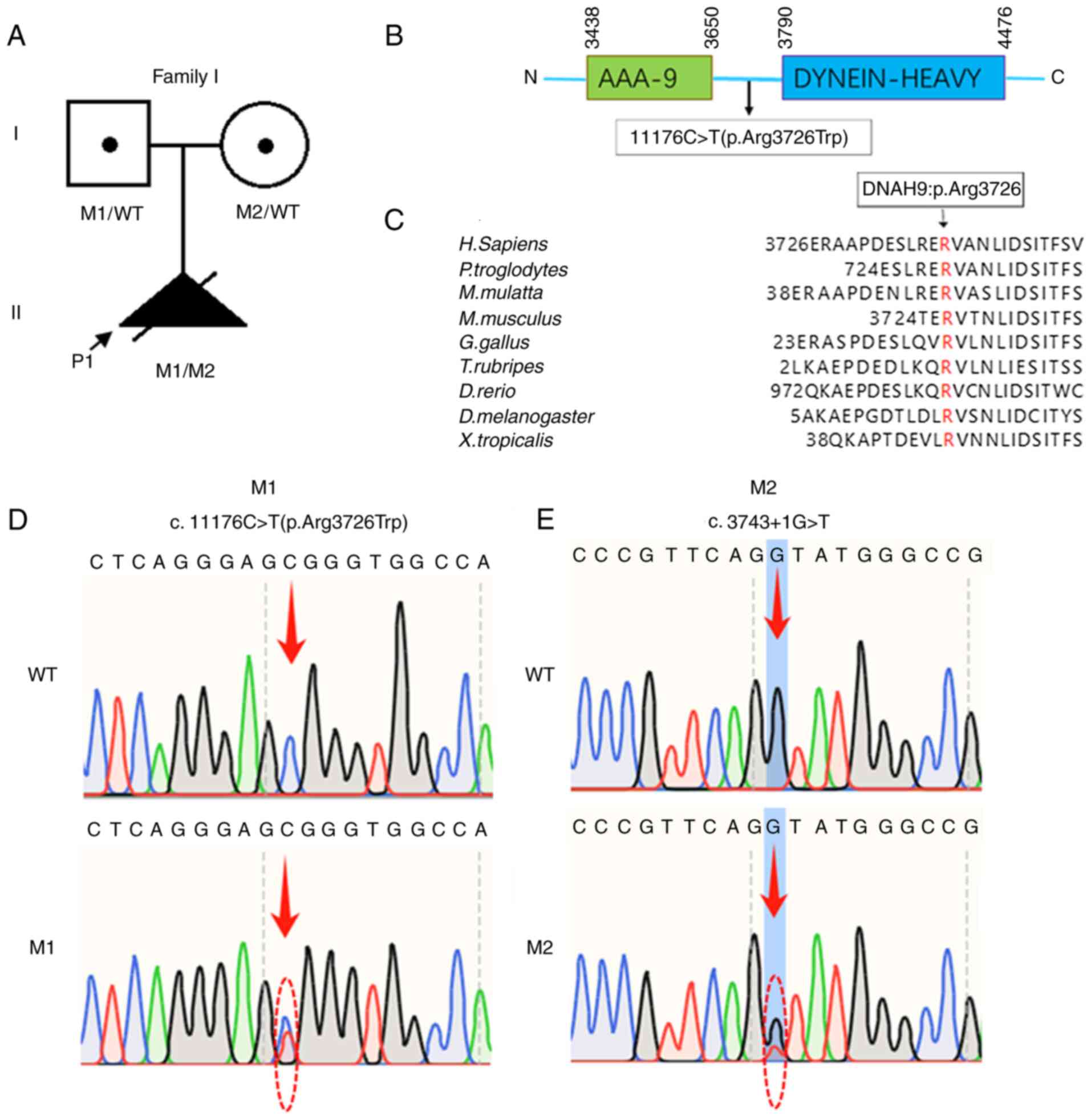
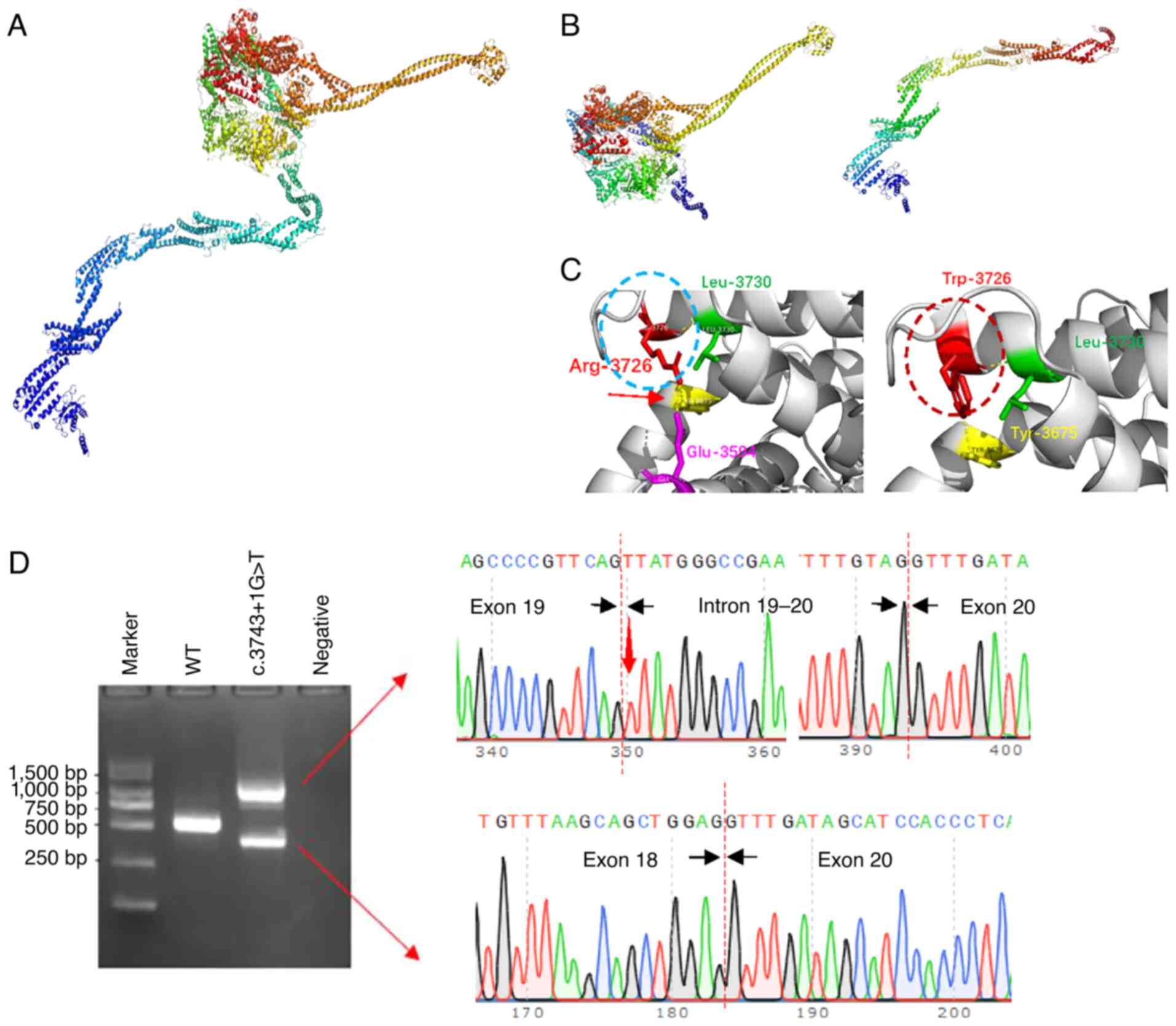
|
1
|
Hoffman JI, Kaplan S and Liberthson RR:
Prevalence of congenital heart disease. Am Heart J. 147:425–439.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lambrechts D, Devriendt K, Driscoll DA,
Goldmuntz E, Gewillig M, Vlietinck R, Collen D and Carmeliet P: Low
expression VEGF haplotype increases the risk for tetralogy of
Fallot: A family-based association study. J Med Genet. 42:519–522.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kalisch-Smith JI, Ved N and Sparrow DB:
Environmental risk factors for congenital heart disease. Cold
Spring Harb Perspect Biol. 12:a0372342020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Williams K, Carson J and Lo C: Genetics of
congenital heart disease. Biomolecules. 9:8792019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van der Bom T, Zomer AC, Zwinderman AH,
Meijboom FJ, Bouma BJ and Mulder BJ: The changing epidemiology of
congenital heart disease. Nat Rev Cardiol. 8:50–60. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shaikh Qureshi WM and Hentges KE:
Functions of cilia in cardiac development and disease. Ann Hum
Genet. 88:4–26. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang X, Wang Q, Li T, Zhou Y, Gao J, Ma W,
Zhao N, Liu X, Ai Z, Cheng SY, et al: A splicing variant in EFCAB7
hinders ciliary transport and disrupts cardiac development. J Biol
Chem. 301:1082492025. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saric N and Ishibashi N: The role of
primary cilia in congenital heart defect-associated neurological
impairments. Front Genet. 15:14602282024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang D, Sha Y, Gao Y, Zhang J, Cheng H,
Zhang J, Ni X, Wang C, Xu C, Geng H, et al: Novel variants in Dnah9
lead to nonsyndromic severe asthenozoospermia. Reprod Biol
Endocrinol. 19:272021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fassad MR, Shoemark A, Legendre M, Hirst
RA, Koll F, le Borgne P, Louis B, Daudvohra F, Patel MP, Thomas L,
et al: Mutations in outer dynein arm heavy chain DNAH9 cause motile
cilia defects and situs inversus. Am J Hum Genet. 103:984–994.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen W, Zhang Y, Shen L, Zhu J, Cai K, Lu
Z, Zeng W, Zhao J and Zhou X: Biallelic DNAH9 mutations are
identified in Chinese patients with defective left-right patterning
and cilia-related complex congenital heart disease. Hum Genet.
141:1339–1353. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Loges NT, Antony D, Maver A, Deardorff MA,
Güleç EY, Gezdirici A, Nöthe-Menchen T, Höben IM, Jelten L, Frank
D, et al: Recessive DNAH9 loss-of-function mutations cause
laterality defects and subtle respiratory ciliary-beating defects.
Am J Hum Genet. 103:995–1008. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang T, Yuan H, Zhu H, Ying Y, Ding J,
Ding H, Shi X, He Y, Pan H and Zhong Y: Fetal congenital heart
disease caused by compound heterozygous mutations in the DNAH9
gene: A case report. Front Genet. 12:7717562021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Feng J, Li J, Du Y, Shi T, Sharma L and
Jie Z: Case report: Rare dynein axonemal heavy chain 9 mutations in
a han-Chinese patient with kartagener syndrome. Front Med
(Lausanne). 9:8939682022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Takeuchi K, Xu Y, Ogawa S, Ikejiri M,
Nakatani K, Gotoh S, Usui S, Masuda S, Nagao M and Fujisawa T: A
pediatric case of productive cough caused by novel variants in
DNAH9. Hum Genome Var. 8:32021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Isa HM, Alkharsi FA, Busehail MY and
Haider F: A Novel DNAH9 gene mutation causing primary ciliary
dyskinesia with an unusual association of jejunal atresia in a
bahraini child. Cureus. 14:e329642022.PubMed/NCBI
|
|
17
|
Tate G: Whole-exome sequencing reveals a
combination of extremely rare single-nucleotide polymorphism of
DNAH9 and RSPH1 genes in a Japanese fetus with situs viscerum
inversus. Med Mol Morphol. 54:275–280. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Y, Chen Y, Shi C, Huang Z, Zhang Y,
Li S, Li Y, Ye J, Yu C, Li Z, et al: SOAPnuke: A MapReduce
acceleration-supported software for integrated quality control and
preprocessing of high-throughput sequencing data. Gigascience.
7:1–6. 2018. View Article : Google Scholar
|
|
19
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
He WB, Tu CF, Liu Q, Meng LL, Yuan SM, Luo
AX, He FS, Shen J, Li W, Du J, et al: DMC1 mutation that causes
human non-obstructive azoospermia and premature ovarian
insufficiency identified by whole-exome sequencing. J Med Genet.
55:198–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qi H, Pan D, Zhang Y, Zhu Y, Zhang X and
Fu T: NEXMIF combined with KIDINS220 gene mutation caused
neurodevelopmental disorder and epilepsy: One case report. Actas
Esp Psiquiatr. 52:588–594. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li G, Chen Y, Han X, Li N and Li S:
Concurrent of compound heterozygous variant of a novel in-frame
deletion and the common hypomorphic haplotype in TBX6 and inherited
17q12 microdeletion in a fetus. BMC Pregnancy Childbirth.
24:4562024. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A MapReduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hamada H, Meno C, Watanabe D and Saijoh Y:
Establishment of vertebrate left-right asymmetry. Nat Rev Genet.
3:103–113. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Komatsu Y and Mishina Y: Establishment of
left-right asymmetry in vertebrate development: The node in mouse
embryos. Cell Mol Life Sci. 70:4659–4666. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nonaka S, Shiratori H, Saijoh Y and Hamada
H: Determination of left-right patterning of the mouse embryo by
artificial nodal flow. Nature. 418:96–99. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Essner JJ, Vogan KJ, Wagner MK, Tabin CJ,
Yost HJ and Brueckner M: Conserved function for embryonic nodal
cilia. Nature. 418:37–38. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Djenoune L, Mahamdeh M, Truong TV, Nguyen
CT, Fraser SE, Brueckner M, Howard J and Yuan S: Cilia function as
calcium-mediated mechanosensors that instruct left-right asymmetry.
Science. 379:71–78. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katoh TA, Omori T, Mizuno K, Sai X,
Minegishi K, Ikawa Y, Nishimura H, Itabashi T, Kajikawa E, Hiver S,
et al: Immotile cilia mechanically sense the direction of fluid
flow for left-right determination. Science. 379:66–71. 2023.
View Article : Google Scholar : PubMed/NCBI
|