Introduction
Liver fibrosis represents a reparative response to
sustained hepatic injury caused by autoimmune hepatitis, primary
cholangitis, alcoholic liver disease and hepatocellular carcinoma,
with advanced fibrotic stages often progressing to cirrhosis
(1,2). The activation of hepatic stellate
cells (HSCs) is a central event in the onset and progression of
fibrosis, and its inhibition has been shown to attenuate disease
severity (3,4). Disruption of hepatic circadian
regulation has also been implicated in the pathogenesis of both
acute and chronic liver disorders (5,6). The
circadian clock, a ubiquitous endogenous regulatory mechanism, is
conserved across nearly all living organisms and enables
physiological adaptation to temporal environmental fluctuations
(7). Core components of the
circadian machinery include Clock, brain and muscle arnt-like
protein 1 (Bmal1), period circadian regulator (Per)1/2/3,
cryptochrome (Cry)1/2 and nuclear receptor subfamily 1 group D
member 1 (Rev-erbα), among others (6,8).
Genetic ablation of Per1/2 or Bmal1 in mouse models resulted in
impaired hepatic function, persistent inflammation and fibrotic
remodeling (9–11). In a cholestasis-induced liver
injury model, Per2 deficiency led to upregulation of liver
fibrosis-related genes and excessive extracellular matrix
deposition (11,12), indicating that circadian disruption
contributes to chronic hepatic inflammation and fibrogenesis.
Rev-erbα regulates transcriptional networks involved
in metabolism, circadian rhythm and inflammation, positioning it as
a therapeutic target for metabolic disorders, malignancies,
epilepsy, inflammatory conditions and neurodegenerative diseases
(13–19). Notably, a marked reduction in both
the baseline expression and oscillatory amplitude of Rev-erbα has
been observed in carbon tetrachloride (CCl4)-induced
mouse liver fibrosis models and in vitro activated HSCs
(10,20). Previous functional studies
indicated that Rev-erbα knockdown activated the cyclic GMP-AMP
synthase (cGAS) pathway, promoting a pro-inflammatory
microenvironment and accelerating fibrogenic progression (21), whereas its overexpression
suppressed cGAS signaling and mitigated fibrosis (21). Additionally, Rev-erbα has been
identified as a key negative regulator of NLR family domain
containing protein 3 (NLRP3) inflammasome expression, with evidence
showing its protective role against ulcerative colitis in mice
(22). Given that NLRP3
inflammasomes can directly activate HSCs and exacerbate liver
fibrosis (23,24), the mechanistic association between
Rev-erbα and NLRP3 signaling in hepatic fibrosis warrants further
elucidation.
To investigate the regulatory role of Rev-erbα in
liver fibrosis and its interaction with the NLRP3 inflammasome, in
the present study, in vivo experiments employed the Rev-erbα
agonist GSK4112 and antagonist SR8278 to pharmacologically enhance
or suppress Rev-erbα expression in a CCl4-induced mouse
model of hepatic fibrosis. In parallel, stable LX-2 cell lines with
Rev-erbα knockdown or overexpression were generated via lentiviral
transduction. Following 24-h transforming growth factor-β1 (TGF-β1)
stimulation, cellular activation and downstream signaling events
were evaluated. Based on a comprehensive analysis of the molecular
mechanisms underlying hepatic fibrosis pathogenesis, the present
study aims to elucidate the role of key regulatory factors in
disease progression, thus laying the theoretical groundwork for
developing more effective therapeutic strategies.
Materials and methods
Reagents
CCl4 (cat. no. 5623-5) for liver fibrosis
induction was obtained from National Pharmaceutical Chemical
Reagent (https://www.reagent.com.cn/). TGF-β1
(cat. no. PRT221-0020), used to stimulate LX-2 cell activation, was
sourced from Beyotime Institute of Biotechnology. The Rev-erbα
agonist GSK4112 (cat. no. 1216744-19-2) and antagonist SR8278 (cat.
no. 1254944-66-5) were purchased from MedChemExpress. Serum alanine
aminotransferase (ALT) and aspartate aminotransferase (AST) levels
were quantified using ALT (cat. no. C009-2-1) and AST (cat. no.
C010-2-1) assay kits from Nanjing Jiancheng Bioengineering
Institute. For liver tissue staining, Masson's trichrome kit (cat.
no. G1006), hematoxylin solution (cat. no. G1077) and
diaminobenzidine (DAB) chromogenic kit (cat. no. G1212) were
obtained from Wuhan Servicebio Technology Co., Ltd., while the
Sirius Red staining kit (cat. no. 2610-10-8) was sourced from
ChemicalBook Inc. Xylene (cat. no. 1330-20-7), glycerol (cat. no.
56-81-5), citrate buffer (cat. no. 6132-04-3) and glacial acetic
acid (cat. no. 64-19-7), used for immunohistochemistry, were
supplied by Sinopharm Chemical Reagent Co., Ltd. Cell culture
reagents included DMEM (cat. no. 8119054) and FBS (cat. no.
A5670701) from Gibco; Thermo Fisher Scientific, Inc. The CCK-8 cell
viability assay kit (cat. no. 40203ES80) was provided by Shanghai
Yeasen Biotechnology Co., Ltd. Custom lentiviral transfection
reagents for Rev-erbα overexpression and knockdown were obtained
from Jikai Biotechnology, and puromycin (cat. no. ST551-50mg) used
for selection was purchased from Beyotime Institute of
Biotechnology. Total RNA was extracted using a commercial RNA
extraction kit (cat. no. 19211ES60) from Shanghai Yeasen
Biotechnology Co., Ltd., followed by reverse transcription with a
kit from Tiangen Biotech Co., Ltd. (cat. no. KR118). SYBR Green
qPCR mix (cat. no. 11201E03) was purchased from Shanghai Yeasen
Biotechnology Co., Ltd. Primers targeting α-smooth muscle actin
(α-SMA), TGF-β1, Rev-erbα, NLRP3, Caspase-1, apoptosis associated
speck (ASC), interleukin (IL)-18, IL-1β and β-actin were designed
based on GenBank cDNA sequences and synthesized by Shanghai
Shanjing Molecular Biotechnology Co., Ltd. Protein extraction was
performed using RIPA lysis buffer (cat. no. WB0102) with protease
inhibitors (cat. no. WB0122), both from Shanghai Weiao
Biotechnology Co., Ltd. Protein concentration was determined using
a BCA assay kit (cat. no. MA-0082-2) from Dalian Meilun Biology
Technology Co., Ltd., and chemiluminescent detection was performed
with an ECL kit (cat. no. SW-WB012) from Shanghai Weiao
Biotechnology Co., Ltd. Primary antibodies against Rev-erbα (cat.
no. ab305753), Caspase-1 (cat. no. ab207802), IL-18 (cat. no.
ab243091), IL-1β (cat. no. ab254360), α-SMA (cat. no. ab124964),
collagen 1 (COL-1) (cat. no. ab34710) and TGF-β1 (cat. no. ab27937)
were obtained from Abcam. Antibodies against NLRP3 (cat. no. 15101)
and ASC (cat. no. 67824S) were sourced from Cell Signaling
Technology, Inc. β-actin (cat. no. GB15003) and HRP-conjugated
Affinipure Goat Anti-rabbit IgG (cat. no. GB22303) were provided by
Wuhan Servicebio Technology Co., Ltd.
Animal experiments
A total of 42 male C57BL/6 mice, SPF grade, 6 weeks
(22±2 g) were obtained from Shanghai Jiesijie Experimental Animal
Co., Ltd., and housed under standard conditions at the Animal
Facility of Shanghai Municipal Hospital of Traditional Chinese
Medicine. Mice were kept at a standard temperature of 25–27°C and a
humidity of 55–65%, in a light/dark cycle of 12/12 h, and with
ad libitum access to sterilized food and water. After a
1-week acclimatization period, animals were randomly assigned to
experimental groups as follows: i) A total of 12 mice were divided
into a control group (Con) and a CCl4-induced model
group (CCl4) (n=6 per group). Liver fibrosis was induced
by intraperitoneal injection of a 20% CCl4 solution in
olive oil (5 ml/kg), administered twice weekly for 6 weeks
(25). Control mice received an
equal volume of physiological saline. ii) The remaining 30 mice
were randomly allocated into three groups: Control (n=12), GSK4112
(n=12) and SR8278 (n=12). For pharmacological modulation of
Rev-erbα, lyophilized GSK4112 and SR8278 were reconstituted in
DMSO. Mice in the SR8278 group received daily intraperitoneal
injections of SR8278 (25 mg/kg) (26), while those in the GSK4112 group
were administered GSK4112 (20 µg/mouse) (27,28).
Control mice were treated with equivalent volumes of DMSO.
Injections were performed once daily for 14 consecutive days. After
treatment, six mice from each group were randomly selected and
reassigned to receive CCl4 treatment as described above,
forming the CCl4 group (n=6), SR8278 + CCl4
group (n=6) and GSK4112 + CCl4 group (n=6).
Upon completion of modeling, mice were anesthetized
with 1% pentobarbital sodium (50 mg/kg). Livers were harvested,
with a 1×1-cm section from the central lobe fixed in 4%
paraformaldehyde for histological analysis at ~25°C. The remaining
tissue was snap-frozen in liquid nitrogen and stored at −80°C for
subsequent experiments. Euthanasia was performed via cervical
dislocation (29). All animal
procedures were approved by the Ethics Committee of Shanghai
Municipal Hospital of Traditional Chinese Medicine (Shanghai,
China; approval no. 2022033).
Cell culturing and treatment
The human HSC LX-2 cell line was obtained from
Shanghai Kanglang Biological Technology Co., Ltd., and cultured in
DMEM supplemented with 10% FBS under standard conditions (37°C and
5% CO2).
For fibrogenic induction, LX-2 cells were divided
into a control group and a TGF-β1-treated group, with the latter
exposed to 5 ng/ml TGF-β1 for 24 h (25).
The Rev-erbα overexpression plasmid and knockdown
shRNA plasmid utilized in this study were constructed by Shanghai
Jikai Biotechnology, employing a triple-plasmid co-transfection
system in 293T cells for lentiviral production; the lentiviral
supernatant was concentrated and purified into high-titer stocks
within 48–72 h post-transfection. For Rev-erbα overexpression
lentivirus, the plasmid combination consisted of the transfer
plasmid GV208 carrying the target gene Rev-erbα (4 µg), the
packaging plasmid Helper 1.0 (3 µg), and the envelope plasmid
Helper 2.0 (1 µg). For Rev-erbα knockdown lentivirus, the plasmids
included the transfer plasmid pLKO.1-shRNA (4 µg), the packaging
plasmid psPAX2 (3 µg) and the envelope plasmid pMD2.G (1 µg). LX-2
cells were seeded in 24-well plates at 5×104 cells/ml (1
ml/well) and were assigned to the following groups: Con522
(negative control for Rev-erbα overexpression), con313 (negative
control for Rev-erbα knockdown), OvRev-erbα (Rev-erbα
overexpression; MOI=10) and Rev-erbα sh1/sh2/sh3 (shRev-erbα
knockdown groups; MOI=30/20/10, respectively). After adherence to
24-well plates and reaching 20–30% confluency, transduction was
performed with Rev-erbα lentiviruses at different multiplicities of
infection. The viral volume was calculated using the formula: Virus
volume (µl)=(MOI × cell number)/viral titer (TU/ml). After 12 h of
incubation at 37°C, the DMEM was replaced with culture medium
containing 10% FBS, and cells were cultured for an additional 72 h.
Transduction efficiency was assessed via fluorescence inverted
microscopy (Olympus IX73; Olympus Coporation) to determine optimal
conditions. Puromycin selection (2 µg/ml) was initiated at 72 h
post-transduction and maintained for 7–14 days. Successful Rev-erbα
modulation was validated by RT-qPCR for mRNA expression and western
blotting for protein expression, as performed below, with
successfully transfected cells subsequently used for further
experiments.
To assess the effect of Rev-erbα on TGF-β1-induced
activation, LX-2 cells were categorized into TGF-β1, OvRev-erbα +
TGF-β1 and shRev-erbα + TGF-β1 groups. Based on preliminary
findings, OvRev-erbα + TGF-β1 was transduced with Rev-erbα at
MOI=10 and shRev-erbα + TGF-β1 received interference at MOI=20. The
TGF-β1 group was treated with DMEM without viral particles. All
groups were subsequently treated with 5 ng/ml TGF-β1 for 24 h at
37°C and in 5% CO2.
Serum liver function analysis
For serum collection, blood samples from each mouse
group were left at room temperature (25°C) for 4 h, centrifuged at
3,000 rpm and 4°C for 15 min, and serum was harvested. AST and ALT
levels were measured using the aforementioned commercially
available detection kits, following the manufacturer's
protocols.
Masson and Sirius red staining
Fresh liver specimens (~1×1 cm) were fixed in 4%
paraformaldehyde at room temperature for 24 h. After paraffin
embedding, tissue sections were stained using Masson's trichrome
and Sirius red protocols. For liver tissues, sections of 5–7 µm in
thickness were used. Collagen fiber staining was performed at room
temperature, with Masson's trichrome staining for 10 min and Sirius
red staining for 30 min. Histopathological alterations were
assessed using a light microscope at ×200 magnification (BX41,
Beijing Ruike Zhongyi Technology Co., Ltd.).
Cellular immunofluorescence
detection
For immunofluorescence, cells from the TGF-β1,
OvRev-erbα + TGF-β1 and shRev-erbα + TGF-β1 groups were fixed with
4% paraformaldehyde at room temperature for 15 min and subjected to
10% FBS blocking at 37°C for 30 min. The TGF-β1, OvRev-erbα +
TGF-β1 and shRev-erbα + TGF-β1 groups were incubated with an α-SMA
antibody (1:200) at 4°C for 16 h, followed by secondary antibody
incubation at room temperature for 50 min. After PBS washing (pH
7.4), nuclei were counterstained with DAPI at 25°C for 10 min in
the dark. Slides were rinsed, spin-dried and mounted using an
anti-fade glycerol medium. Fluorescence imaging was performed with
a fluorescence inverted microscope (Olympus IX73 microscope).
Quantification of positive staining was carried out using ImageJ
1.8.0 software (National Institutes of Health), and statistical
analyses of protein expression levels were performed using GraphPad
Prism 8.3.0 (Dotmatics).
Immunohistochemical detection of liver
tissue
Following fixation (4% paraformaldehyde at room
temperature for 24 h), liver tissues were dehydrated at room
temperature for 30 min and embedded in paraffin (melting point
~60°C). Paraffin blocks were sectioned into 5-µm slices, rinsed and
air-dried prior to staining. The procedure included the following
steps: i) At room temperature, dewaxing with xylene for 30 min and
graded ethanol hydration (100, 95, 80 and 75%) for 20 min; ii)
rehydration through a descending ethanol series followed by
distilled water rinses; iii) nuclear staining with hematoxylin at
room temperature for 10 min; iv) rinsing in distilled water; v)
antigen retrieval by heating sections in 200 ml 0.01Μ citrate
buffer (pH6.0) at 70°C for 10 min; vi) washing in PBS three times
(5 min each), then incubating in 3% hydrogen peroxide at room
temperature for 10 min to quench endogenous peroxidase activity;
vii) blocking with 2% FBS in PBS at 37°C for 2 h; viii) incubation
with primary antibodies, α-SMA (1:500), COL-1 (1:600) and TGF-β1
(1:800), at 4°C for 16 h; ix) after PBS washes, incubation with
HRP-conjugated goat anti-mouse secondary antibody (1:200) at 37°C
for 2 h; x) DAB chromogenic development for 10 min; xi) rinsing
with distilled water; and xii) clearing with xylene for 10 min,
followed by mounting with neutral resin and drying at 70°C.
Microscopic images were captured using a Horiba BX41 light
microscope (Beijing Ruike Zhongyi Technology Co., Ltd.) from five
randomly selected fields per section at magnifications of ×100 and
×400. Quantitative analysis of antibody-positive staining areas was
performed using ImageJ 1.8.0 software (National Institutes of
Health). Statistical analysis of target protein expression levels
was conducted using GraphPad Prism 8.3.0 (Dotmatics).
Cell viability detection
Cell viability was assessed using the CCK-8 assay. A
suspension of 2×103 cells in 100 µl complete culture
medium containing 10% FBS was seeded into each well of a 96-well
plate and incubated under standard conditions (37°C and 5%
CO2) for 6 h. Subsequently, 10 µl CCK-8 solution were
added to each well. After 2 h of incubation, optical density (OD)
was measured at 450 nm using a microplate reader (BioTek Epoch2;
Agilent Technologies, Inc.). OD values were used to calculate cell
viability, and results were presented in histogram form for
comparative analysis.
Reverse transcription-quantitative PCR
(RT-qPCR)
For RT-qPCR analysis, 40–50 mg of liver tissue was
collected from each group, and total RNA was extracted using
TRIzol® (Thermo Fisher Scientific, Inc.) and
precipitated in ethanol. Reverse transcription was performed using
the FastKing gDNA Dispelling RT SuperMixJun (cat. no. KR118;
Tiangen Biotech Co., Ltd.), and the resulting cDNA was diluted
10-fold with ddH2O for amplification. qPCR was performed
using SYBR Green PCR Mastermix (cat. no. 11201E03). The results
were analyzed on an ABI StepOnePlus real-time PCR system (Applied
Biosystems) using the 2−ΔΔCq method (30). Values were normalized to β-actin.
The reaction conditions were as follows: 95°C predenaturation once
for 5 min, followed by denatured at 95°C for 10 sec, annealing at
60°C for 20 sec and extension at 72°C for 20 sec, for 40 cycles.
Primer sequences are provided in Table SI.
Western blotting analysis
The quantity of whole-protein extracts from cells
and livers of mice, extracted using RIPA lysis buffer (cat. no.
WB0102) with protease inhibitors (cat. no. WB0122), were determined
using the BCA assay. Equal amounts of total protein (20–30 µg) were
resolved by 7.5% SDS-PAGE and transferred onto PVDF membranes.
After blocking with 5% skimmed milk at room temperature for 2 h,
the membranes were incubated with specific primary antibodies for
16 h at 4°C [Rev-erbα (1:1,000), Caspase-1 (1:1,000), IL-18
(1:1,000), IL-1β (1:1,000), α-SMA (1:1,000), collagen 1 (1:1,000),
TGF-β1 (1:1,000), NLRP3 (1:1,000), ASC (1:1,000) and β-actin
(1:3,000)], followed by incubation with HRP-conjugated Affinipure
Goat Anti-rabbit IgG (1:5,000) at room temperature for 2 h. Signal
detection was performed using a chemiluminescence imaging system
(Amersham Imager 600; Cytiva), and band intensities were
semi-quantified using ImageJ 1.8.0 software.
Statistical analysis
Statistical analyses were performed using GraphPad
Prism 8.3.0 (Dotmatics). Comparisons between two groups were
performed using unpaired, two-tailed t-tests, while multiple-group
comparisons were evaluated by one-way ANOVA. When comparing ≥3
groups (e.g., shRev-erbα 1, shRev-erbα 2 and shRev-erbα3) requiring
assessment of all possible pairwise comparisons (shRev-erbα 1 vs.
shRev-erbα 2, shRev-erbα 1 vs. shRev-erbα 3 and shRev-erbα 2 vs.
shRev-erbα 3), Tukey's test was employed for post-hoc evaluation.
When analyzing ≥3 groups (e.g., CCl4, GSK4112 +
CCl4 and SR8287 + CCl4) focusing on specific
pairwise comparisons between predetermined groups (CCl4
vs. GSK4112 + CCl4 and CCl4, vs. SR8287 +
CCl4), Bonferroni's correction was applied for post-hoc
statistical assessment. Data are expressed as the mean ± SEM.
P<0.05 was used to indicate a statistically significant
difference.
Results
Reduced expression of circadian clock
gene Rev-erbα in liver fibrosis
To investigate the expression dynamics of the
circadian clock gene Rev-erbα in liver fibrosis, a mouse model was
established using CCl4 administration. Compared with
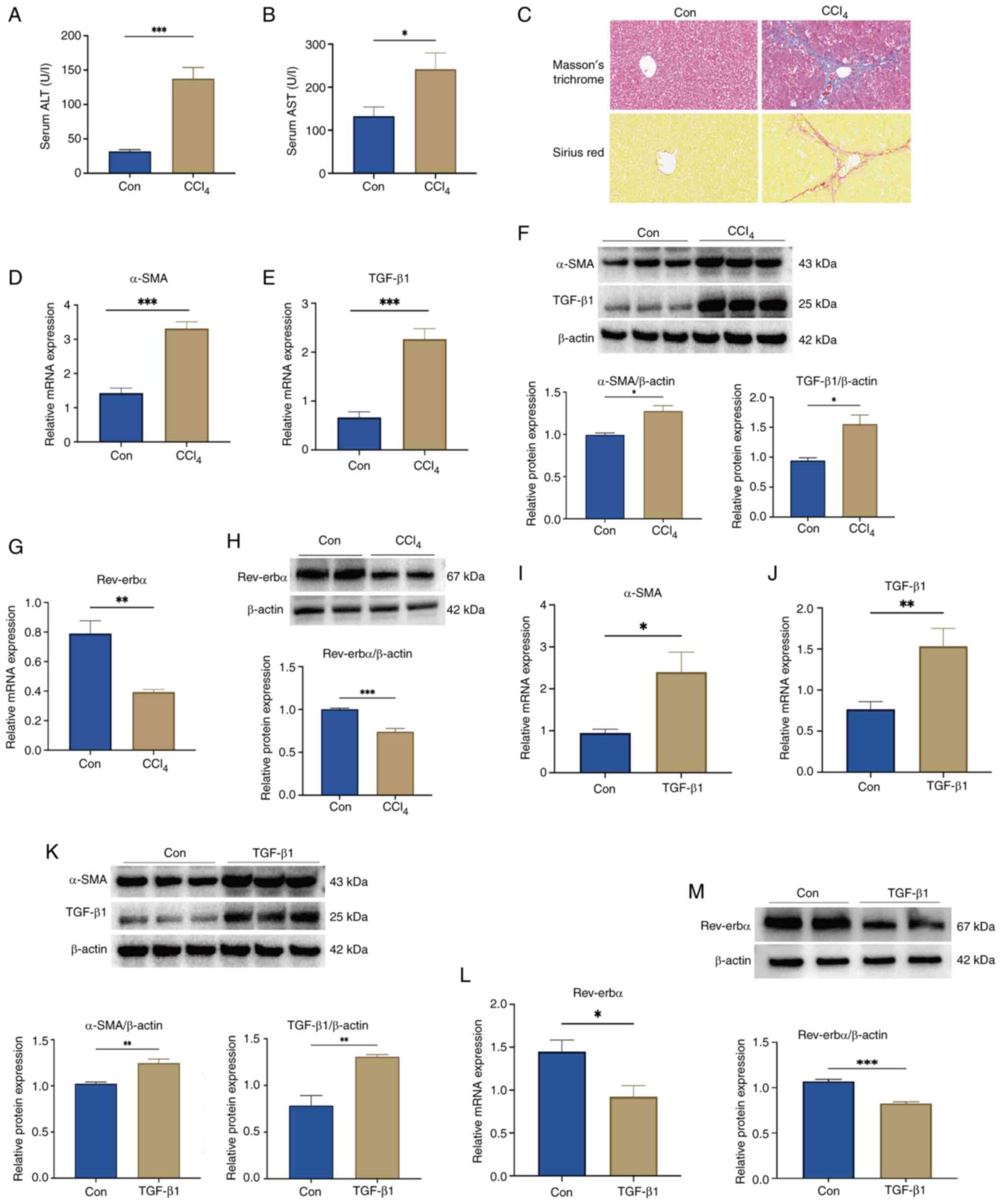
those in the Con group, serum ALT and AST levels were significantly
elevated in the CCl4-treated mice, indicating
substantial hepatic injury (Fig. 1A
and B). Histological analysis via Masson's trichrome and Sirius
red staining revealed well-preserved hepatic architecture in the
control group, whereas CCl4-treated mice exhibited
marked collagen deposition and expanded fibrotic areas, consistent
with progressive fibrosis (Fig.
1C). In parallel, mRNA and protein expression levels of TGF-β1
and α-SMA were significantly upregulated in the livers of
CCl4-treated mice compared with levels in the controls
(Fig. 1D-F), confirming successful
model induction. Notably, Rev-erbα mRNA and protein levels were
significantly reduced in the liver tissue of CCl4 group
mice relative to the controls (Fig. 1G
and H). This downregulation was further corroborated in
vitro, where TGF-β1-stimulated LX-2 cells showed significant
upregulation of fibrotic markers (α-SMA and TGF-β1) (Fig. 1I-K) and a concomitant decrease in
Rev-erbα expression at both the transcript and protein levels
(Fig. 1L and M). These results
suggest that Rev-erbα may act as a negative regulator in the
progression of liver fibrosis, with its reduced expression
potentially facilitating fibrogenic and pro-inflammatory responses
in hepatic tissue.
Rev-erbα has a significant inhibitory
impact on liver fibrosis
Rev-erbα serves as a critical transcriptional
regulator of metabolic, circadian and inflammatory pathways,
rendering it a promising therapeutic target for metabolic
disorders, malignancies, epilepsy, inflammatory conditions and
neurodegenerative diseases (22,31).
To evaluate its role in liver fibrosis progression, C57BL/6 mice
received intraperitoneal injections of the Rev-erbα agonist GSK4112
(25 mg/kg) or the antagonist SR8278 (20 µg/mouse). Western blot
analysis confirmed successful pharmacological modulation, with
Rev-erbα expression significantly reduced in the SR8278 group and
significantly elevated in the GSK4112 group (Fig. 2A).
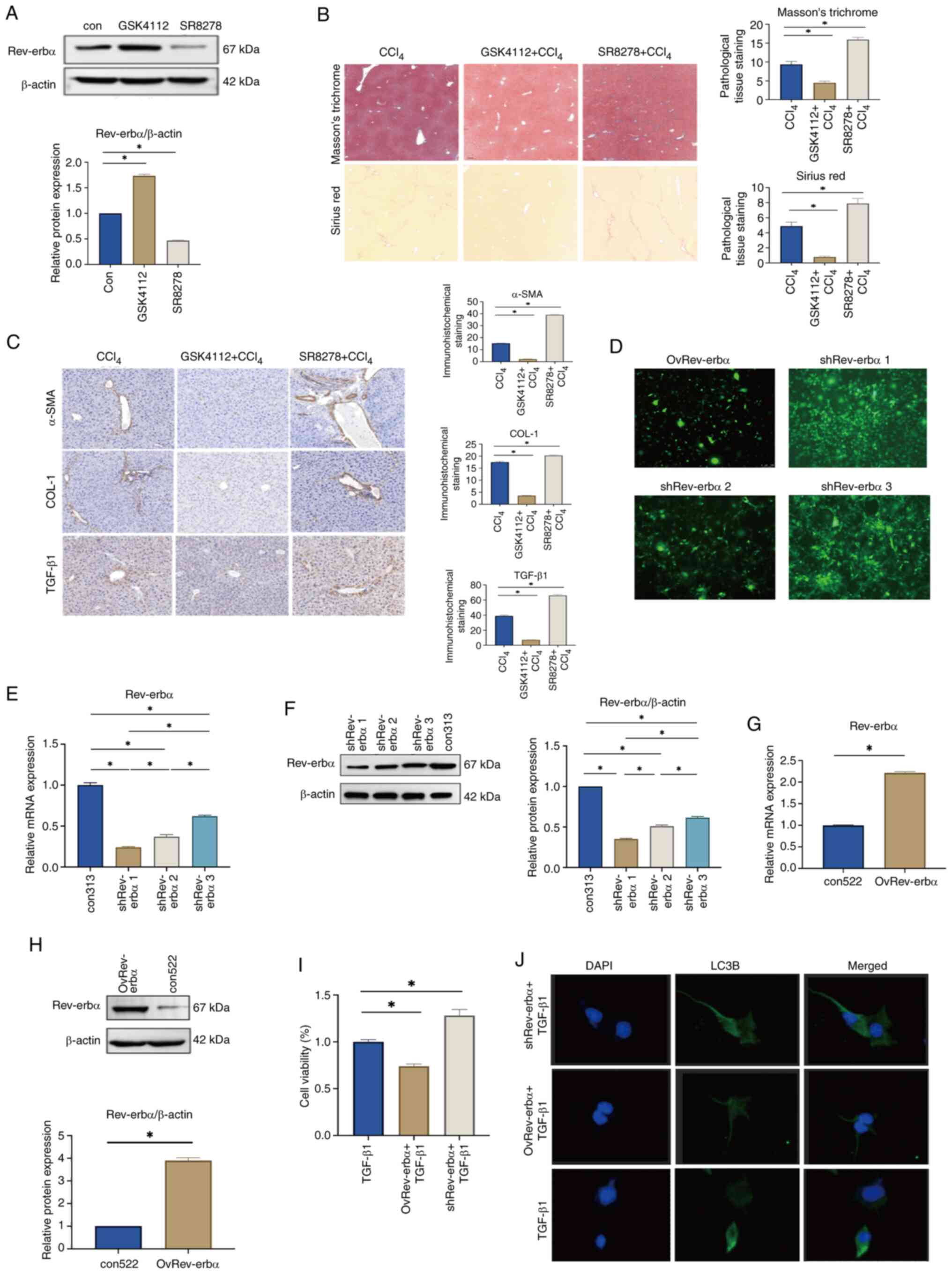 | Figure 2.Rev-erbα significantly inhibits liver
fibrosis. (A) Rev-erbα protein expression in mouse liver tissue
following administration of Rev-erbα agonist GSK4112 or inhibitor
SR8278 (n=3). (B) Liver histopathology in the CCl4,
GSK4112 + CCl4 and SR8278 + CCl4 groups
(Masson's trichrome and Sirius red staining; ×100 magnification)
(n=3). (C) Expression of α-SMA, COL-1 and TGF-β1 in liver tissue of
CCl4-induced liver fibrosis mice from in the
CCl4, GSK4112 + CCl4 and SR8278 +
CCl4 groups (×400 magnification) (n=3). (D) Lentiviral
fluorescence expression in stable Rev-erbα gene knockdown or
overexpression cell lines (×200 magnification). The greater the
extent of green fluorescent areas, the better the lentiviral
transduction efficacy. (E) The mRNA expression of Rev-erbα in
shRev-erbα cell groups (n=6). (F) Protein expression of Rev-erbα in
the shRev-erbα cell group (n=6). (G) The mRNA expression of
Rev-erbα in the OvRev-erbα cell group (n=6). (H) Protein expression
of Rev-erbα in the OvRev-erbα cell group (n=6). (I) LX-2 cell
viability in the TGF-β1, OvRev-erbα + TGF-β1 and shRev-erbα +
TGF-β1 groups (n=3). (J) Expression of α-SMA in the TGF-β1,
OvRev-erbα + TGF-β1 and shRev-erbα + TGF-β1 groups
(immunofluorescence; ×200 magnification). Data are expressed as the
mean ± SEM; *P<0.05. Con, control; Rev-erbα, nuclear receptor
subfamily 1 group D member 1; TGF-β1, transforming growth
factor-β1; α-SMA, α-smooth muscle actin; CCl4, carbon
tetrachloride; COL-1, collagen 1; DAPI,
4′,6-diamidino-2-phenylindole; LC3B, microtubule-associated protein
1A/1B light chain 3β. |
Following model establishment, liver fibrosis was
induced using CCl4. Histological staining with Masson's
trichrome and Sirius red revealed pronounced pseudolobule
formation, extensive collagen accumulation and expanded fibrotic
areas in the CCl4 + SR8278 group. By contrast, the
CCl4 + GSK4112 group exhibited more preserved hepatic
architecture, reduced fibrous proliferation and an absence of
steatosis compared with the CCl4 group (Fig. 2B). Immunohistochemical analysis
further demonstrated that expression levels of α-SMA, COL-1 and
TGF-β1 were significantly upregulated in the CCl4 +
SR8278 group, whereas these markers were significantly
downregulated in the CCl4 + GSK4112 group (Fig. 2C). These results indicate that
Rev-erbα suppression exacerbates fibrogenesis, while its activation
attenuates hepatic inflammation, collagen deposition and fibrotic
remodeling. Lentiviral transduction was employed to establish LX-2
cell lines with stable Rev-erbα knockdown or overexpression for
subsequent in vitro investigations. Fluorescence microscopy
confirmed that Rev-erbα sh1 (MOI=20) achieved the most effective
knockdown (Fig. 2D), with
significantly reduced mRNA and protein expression levels of
Rev-erbα in LX-2 cells (Fig. 2E and
F). This condition was subsequently selected for downstream
experiments. Fluorescence microscopy confirmed efficient
overexpression of Rev-erbα in the MOI=10 group (Fig. 2D), accompanied by a significant
increase in both mRNA and protein levels of Rev-erbα in LX-2 cells
(Fig. 2G and H). LX-2 cells were
then allocated into three groups: TGF-β1, OvRev-erbα + TGF-β1 and
shRev-erbα + TGF-β1. Following lentiviral transduction at the
defined MOIs, cells were stimulated with 5 ng/ml TGF-β1 for 24 h to
induce activation. CCK-8 assays revealed a significant reduction in
cell viability in the OvRev-erbα + TGF-β1 group, while the
shRev-erbα + TGF-β1 group exhibited a significant increase in
viability, compared with the TGF-β1 group (Fig. 2I). Immunofluorescence analysis
further demonstrated that α-SMA expression was substantially
downregulated in the OvRev-erbα + TGF-β1 group, whereas notable
upregulation was observed in the shRev-erbα + TGF-β1 group
(Fig. 2J). These results suggest
that decreased Rev-erbα expression promotes HSC activation and
fibrogenic responses, whereas its overexpression exerts a
suppressive, anti-fibrotic effect.
Rev-erbα inhibits NLRP3 and alleviates
liver fibrosis
The NLRP3 inflammasome consists of ASC, caspase-1
and NLRP3, with ASC serving as a key adaptor that promotes the
recruitment and activation of caspase-1 and NLRP3. Caspase-1
processes the inactive precursors of IL-1β and IL-18 into their
mature, bioactive forms, thereby initiating pyroptosis and
inflammatory responses that play central roles in the progression
of alcoholic liver disease, viral hepatitis and liver fibrosis
(32,33).
In the present study, following CCl4
induction in three groups of mice, Rev-erbα protein expression was
assessed. Compared with those in the CCl4 group, the
protein levels of Rev-erbα in the GSK4112 + CCl4 group
were significantly elevated and those in the SR8278 +
CCl4 group were significantly decreased (Fig. 3A). To examine whether the
anti-fibrotic effect of Rev-erbα is mediated through modulation of
the NLRP3 inflammasome, liver fibrosis models were generated in
C57BL/6 mice subjected to CCl4 combined with either
GSK4112 or SR8278 treatment. Expression levels of NLRP3, caspase-1,
ASC and downstream proinflammatory cytokines IL-18 and IL-1β were
analyzed in liver tissue. Relative to the CCl4 group,
mRNA expression of NLRP3, caspase-1, ASC, IL-18 and IL-1β was
significantly reduced in the GSK4112 + CCl4 group,
whereas significant increases were observed in the SR8278 +
CCl4 group (Fig. 3B),
with protein expression showing consistent trends (Fig. 3C). To further substantiate these
findings, validation was performed in three groups of LX2 cells.
Following TGF-β1 stimulation, Rev-erbα protein expression was
assessed. Compared with that in the TGF-β1 group, Rev-erbα
expression was significantly upregulated in the OvRev-erbα + TGF-β1
group and markedly downregulated in the shRev-erbα + TGF-β1 group
(Fig. 3D). Subsequent analysis
focused on the regulatory effect of Rev-erbα on the NLRP3
inflammasome in LX2 cells. RT-qPCR and western blotting results
demonstrated that, relative to the TGF-β1 group, mRNA and protein
levels of NLRP3, caspase-1, ASC and the downstream cytokines, IL-18
and IL-1β, were significantly suppressed in the OvRev-erbα + TGF-β1
group, whereas marked increases were observed in the ShRev-erbα +
TGF-β1 group (Fig. 3E and F).
These results indicate that Rev-erbα knockdown promotes
inflammatory responses during liver fibrosis by enhancing NLRP3
inflammasome activation in HSCs, thereby elevating the expression
of IL-18 and IL-1β. By contrast, overexpression of Rev-erbα
reverses these effects.
 | Figure 3.Rev-erbα inhibits NLRP3 and reduces
liver fibrosis. (A) Protein expression of Rev-erbα in the
CCl4, GSK4112 + CCl4 and SR8278 +
CCl4 groups (n=3). (B) The mRNA expression of NLRP3,
caspase-1, ASC, IL-18 and IL-1β in the CCl4, GSK4112 +
CCl4 and SR8278 + CCl4 groups (n=3). (C)
Protein expression of NLRP3, caspase-1, ASC, IL-18 and IL-1β in the
CCl4, GSK4112 + CCl4 and SR8278 +
CCl4 groups (n=3). (D) Protein expression of Rev-erbα in
the TGF-β1, OvRev-erbα + TGF-β1 and shRev-erbα + TGF-β1 groups
(n=3). (E) The mRNA expression of NLRP3, caspase-1, ASC, IL-18 and
IL-1β in the TGF-β1, OvRev-erbα + TGF-β1 and shRev-erbα + TGF-β1
groups (n=3). (F) Protein expression of NLRP3, caspase-1, ASC,
IL-18 and IL-1β in the TGF-β1, OvRev-erbα + TGF-β1 and shRev-erbα +
TGF-β1 groups (n=3). Data are expressed as the mean ± SEM; ns, not
significant; *P<0.05. Rev-erbα, nuclear receptor
subfamily 1 group D member 1; TGF-β1, transforming growth
factor-β1; CCl4, carbon tetrachloride; NLRP3, NLR family
domain containing protein 3; ASC, apoptosis associated speck; IL,
interleukin. |
Collectively, the data support a role for Rev-erbα
as an upstream negative regulator of the NLRP3 inflammasome,
contributing to its anti-fibrotic function by attenuating
inflammasome-mediated inflammation in liver fibrosis and activated
HSCs.
Discussion
Liver fibrosis represents a key pathological process
in the development of cirrhosis. Hepatic injury and inflammation,
triggered by factors such as drug toxicity, excessive alcohol
intake, viral infections and autoimmune responses, drive this
progression (34). Upon
stimulation, HSCs undergo transdifferentiation into
myofibroblast-like cells, which secrete extracellular matrix
components that accumulate as scar tissue, thereby advancing
fibrogenesis (1,3). Accumulating evidence indicates that
disruptions in circadian rhythms significantly impact the
pathogenesis of various liver disorders, including fibrosis
(10,35,36).
While the master circadian clock resides in the suprachiasmatic
nucleus of the hypothalamus, peripheral tissues and organs,
including the liver, kidney, myocardium, pancreas and skeletal
muscle, possess autonomous clocks that regulate local rhythmic gene
expression to maintain physiological homeostasis (37).
Alterations in hepatic circadian regulation
profoundly affect the initiation and progression of liver disease.
Bmal1 is a core component of the molecular clock and plays a
pivotal role in regulating lipid, bile acid and glucose metabolism;
its suppression leads to metabolic dysregulation and hepatic
dysfunction (38). Bmal1-deficient
mice, especially under chronic alcohol exposure, exhibit
exacerbated hepatic steatosis and injury (39). Clock gene dysfunction promotes
lipid accumulation in the liver, contributing to the development of
non-alcoholic steatohepatitis (NASH) (40). In addition, NAD-dependent protein
deacetylase sirtuin-1 modulates hepatic insulin sensitivity through
the Clock/Bmal1 axis (41).
Genetic ablation of Clock and Bmal1 results in hypoglycemia,
whereas loss of Per and Cry leads to hyperinsulinemia (42). Notably, Rev-erbα knockdown
activates the cGAS pathway, induces a localized proinflammatory
microenvironment and accelerates liver fibrosis progression
(21).
The liver clock genes Rev-erbα and Rev-erbβ, both
classified as orphan nuclear receptors, cooperatively regulate
circadian rhythms and metabolic homeostasis (14,43).
Rev-erbα has been shown to modulate bile acid and cholesterol
biosynthesis by regulating SREBP regulating gene protein expression
in mice, and its genetic deletion leads to impaired bile acid
metabolism (44). Simultaneous
ablation of both Rev-erbα and Rev-erbβ in double-knockout models
severely disrupts the rhythmic expression of lipid
metabolism-related genes (14,45).
Beyond hepatic functions, Rev-erbα also plays a critical role in
maintaining intestinal barrier permeability and treating NASH
(46). In NASH mouse models
induced by a high-cholesterol, high-fat diet, intestinal expression
of Rev-erbα and tight junction-associated genes is downregulated,
resulting in increased intestinal permeability. Pharmacological
activation of Rev-erbα with SR9009 ameliorates hepatic lipid
accumulation, insulin resistance, inflammation and fibrosis, in
part by enhancing intestinal barrier function (46). Additionally, Rev-erbα has been
implicated in the regulation of the long non-coding RNA Platr4,
which attenuates NASH progression by suppressing NLRP3 inflammasome
activation (47). Another study
demonstrated that intestinal Rev-erbα deficiency exacerbates
high-fat diet (HFD)-induced obesity. Pharmacological modulation of
the Rev-erbα/Bmal1 axis using small-molecule compounds alleviated
HFD-induced metabolic dysfunction (48). Collectively, these findings
highlight the critical role of the gut circadian clock,
particularly Rev-erbα, in regulating lipid absorption and energy
balance, offering potential therapeutic targets for obesity and
related metabolic disorders.
Rev-erbα also plays a pivotal role in liver
ischemia-reperfusion (I/R) injury (49). Additionally, Rev-erbα deficiency
exacerbates hepatic I/R injury in mice, as evidenced by elevated
plasma levels of ALT and AST, higher histopathological injury
scores and increased hepatic myeloperoxidase activity. Moreover,
the absence of Rev-erbα leads to heightened expression of
pro-inflammatory cytokines, enhanced NLRP3 inflammasome activation
and greater infiltration of inflammatory cells. Pharmacological
activation of Rev-erbα significantly mitigates hepatic injury and
the associated inflammatory response, indicating its protective
role through suppression of inflammation (49). Rev-erbα is also critically involved
in the pathogenesis of alcoholic fatty liver (AFL) (50). In ethanol-fed mice and
ethanol-treated L02 hepatocytes, Rev-erbα expression is markedly
upregulated, and its activation promotes hepatic steatosis.
Inhibition or downregulation of Rev-erbα alleviates lipid
accumulation, primarily by restoring autophagic activity.
Mechanistically, Rev-erbα impairs autophagy via Bmal1-dependent
pathways (50). Treatment with the
Rev-erbα antagonist SR8278 reduces hepatic lipid deposition and
enhances autophagic flux in ethanol-fed mice (50), suggesting that Rev-erbα acts as a
key modulator in AFL progression and represents a potential
therapeutic target. Furthermore, Rev-erbα deficiency in ALD mice
increases CYP4A expression, lipid accumulation and oxidative
stress. Intervention with either the Rev-erbα agonist SR9009 or the
CYP4A inhibitor HET0016 attenuates alcohol-induced hepatic
steatosis and injury, highlighting both Rev-erbα and CYP4A as
viable therapeutic targets in ALD management (51). In addition, the Rev-erbα agonist
GSK4112 has demonstrated protective effects in Fas-induced acute
liver injury (26). In a mouse
model established by administration of the anti-Fas antibody Jo2,
GSK4112 treatment (25 mg/kg, intraperitoneally) significantly
reduced plasma ALT and AST levels, ameliorated liver histological
damage and improved survival. Mechanistically, GSK4112 suppressed
caspase-3 and caspase-8 activity, decreased hepatocyte apoptosis,
downregulated Fas expression and enhanced Akt phosphorylation
(26).
In the context of liver fibrosis research, Wang
et al (52) reported that
Rev-erbα binds directly to the promoter region of the pro-fibrotic
coagulation regulator PAI-1, thereby suppressing its expression and
exerting anti-fibrotic effects. A recent study further revealed
rhythmic oscillations in the expression of TGF-β signaling
components and fibrosis-associated genes in liver tissue from
hepatocyte-specific Rev-erbα/β knockout mice (53). In LX2 cells subjected to circadian
rhythm synchronization, silencing of Rev-erbα led to upregulation
of pro-fibrotic gene expression. Activation of Rev-erbα by SR9009
was shown to reduce SMAD2/3 phosphorylation in TGF-β-stimulated
human lung myofibroblasts, indicating that Rev-erbα activation
impairs TGF-β signaling in both HSCs and myofibroblasts (53). To evaluate the therapeutic
potential of targeting Rev-erbα in MASH-induced fibrosis, the
pharmacological agonist SR9009 was administered to mice fed a
choline-deficient, amino acid-defined, HFD. SR9009 treatment
significantly attenuated hepatic fibrosis, as evidenced by reduced
collagen deposition (Sirius red staining), diminished HSC
activation (α-SMA expression) and downregulation of
fibrosis-related genes (53).
These findings support the therapeutic efficacy of Rev-erbα
agonists in mitigating fibrosis progression. Consistent with these
observations, the present study identified a significant decrease
in Rev-erbα expression in both CCl4-induced liver
fibrosis mouse models and TGF-β-stimulated LX2 cells. These results
suggest that reduced Rev-erbα expression may contribute to the
progression of liver fibrosis.
Studies have identified porphyrin heme as the
endogenous ligand of Rev-erbα. Binding of heme is essential for
Rev-erbα to recruit corepressor complexes, such as nuclear receptor
co-repressor (NCoR), enabling it to actively repress transcription
of its target genes, including Bmal1, glucose-6-phosphatase
(G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK) (26,54).
GSK4112 (also known as SR6452), a synthetic non-porphyrin small
molecule, functions as a Rev-erbα agonist by mimicking the action
of heme (54). Conversely, SR8278
acts as an antagonist, inhibiting the transcriptional repression
activity of Rev-erbα. Structural analyses show that SR8278 closely
resembles agonist molecules, and treatment of HepG2 cells with
SR8278 results in increased expression of Rev-erbα target genes
such as Bmal1, G6Pase and PEPCK, consistent with its ability to
block the effects of endogenous heme (28,55).
Both GSK4112 and SR8278 serve as valuable chemical tools for
probing Rev-erbα functions in transcriptional repression, circadian
regulation and metabolic pathways (26,27).
The repressive function of Rev-erbα is mediated through recruitment
of the NCoR/HDAC3 corepressor complex (56,57).
In addition to transcriptional regulation, protein stability is a
critical mechanism controlling Rev-erbα activity. Rev-erbα is
intrinsically unstable, with a half-life of less than 1 h,
indicating continuous proteolytic turnover (58). Phosphorylation by glycogen synthase
kinase 3β (GSK3β) at serine residues 55 and 59 has been implicated
in modulating Rev-erbα stability (59). Furthermore, a ubiquitin E3 ligase
complex composed of ADP-ribosylation factor binding protein 1 and
MYC binding protein 2 has been shown to mediate lithium-induced
degradation of Rev-erbα (58).
SR8278 (administered via micro slow injection at 20 µg/mouse) has
also been reported to restore the DNA-binding activity of Rev-erbα
and nuclear receptor subfamily 4 group A member 2 at the tyrosine
hydroxylase promoter, promoting enrichment at R/N motifs [The ‘R/N
motif’ specifically refers to a DNA sequence pattern recognized by
transcription factors, wherein R denotes a purine base (adenine (A)
or guanine (G)), and N represents any nucleotide (A, T, C or G)]
recognized by both transcription factors (28). Additionally, SR8278 exerts
antidepressant and anxiolytic effects in 6-OHDA-induced mouse
models in a circadian rhythm-dependent manner, restoring rhythmic
patterns of emotion-related behaviors (27).
To investigate the role of Rev-erbα in liver
fibrosis, in vivo experiments were conducted using the
Rev-erbα agonist GSK4112 (25 mg/kg) and the antagonist SR8278 (20
µg/mouse) to pharmacologically activate or inhibit Rev-erbα
signaling in mice. Liver fibrosis was induced through
intraperitoneal administration of a CCl4 mixture. In
parallel, in vitro experiments involved stable lentiviral
transfection to generate LX-2 cell lines with either Rev-erbα
overexpression or knockdown, followed by TGF-β1 stimulation for 24
h to induce cellular activation. Inhibition of Rev-erbα expression
in mice resulted in increased collagen deposition and elevated
hepatic levels of α-SMA, COL-1 and TGF-β1, while activation of
Rev-erbα reversed these pathological features. Comparable effects
were observed in LX-2 cells, consistent with the established role
of activated HSCs in upregulating α-SMA and COL-1 expression
(60), with α-SMA serving as a
well-recognized marker of HSC activation and fibrosis severity
(61,62). TGF-β1, a key pro-fibrotic cytokine,
is known to drive HSC activation and sustain fibrogenesis (63,64).
Taken together, these findings indicate that downregulation of
Rev-erbα promotes liver fibrosis, whereas its upregulation exerts a
protective, anti-fibrotic effect.
The NLRP3 inflammasome has been implicated in
various inflammatory conditions, including colitis, type 2 diabetes
and atherosclerosis. Loss of Rev-erbα has been shown to exacerbate
atherosclerotic plaque vulnerability and rupture by activating the
NF-κB/NLRP3 axis, thereby increasing macrophage infiltration,
oxidative stress and inflammatory cytokine production (65). In breast cancer cells, Rev-erbα
inhibition activates the cGAS-stimulator of interferon genes
pathway, leading to elevated levels of type I interferons and
downstream chemokines, chemokine (C-C motif) ligand 5 and CXC motif
chemokine ligand 10, enhancing antitumor immune responses (66). In HSCs, Rev-erbα degradation
impairs mitochondrial fission and promotes mitochondrial DNA
(mtDNA) release, triggering cGAS activation and contributing to a
pro-inflammatory microenvironment that accelerates liver fibrosis
(21). Additionally, Rev-erbα
regulates the circadian rhythm of the NLRP3 inflammasome activity,
and its activation has been shown to alleviate hepatitis in mouse
models (31). The Rev-erbα agonist
SR9009 exerts potent anti-inflammatory effects by suppressing
pro-inflammatory cytokine expression and preventing monocyte
infiltration during inflammation. Notably, SR9009 alleviated
colitis in wild-type mice but failed to confer protection in NLRP3-
or Rev-erbα-deficient models (67). In models of neuroinflammation,
SR9009 inhibited NLRP3 activation, and reduced IL-6, IL-1β and
IL-18 production, attenuating microglial and astrocyte activation,
and ameliorating status epilepticus (31). Similarly, in degenerative disc
disease, Rev-erbα activation suppressed NLRP3 inflammasome assembly
and IL-1β production, reducing local inflammation (68). These findings underscore the
central role of the Rev-erbα/NLRP3 axis in inflammatory disease
progression. Consistent with this mechanistic framework, the
present study demonstrated that Rev-erbα inhibition in both in
vivo and in vitro liver fibrosis models led to
upregulation of NLRP3, caspase-1, ASC, IL-18 and IL-1β. Conversely,
pharmacological or genetic activation of Rev-erbα suppressed the
expression of these inflammasome components. These results support
the hypothesis that Rev-erbα functions as a negative upstream
regulator of the NLRP3 inflammasome in activated HSCs and fibrotic
liver tissue, thereby mediating its anti-fibrotic effects through
suppression of inflammasome-driven inflammation (Fig. 4).
 | Figure 4.Mechanism of Rev-erbα promoting the
progression of liver fibrosis. The decrease in the expression of
the biological clock gene Rev-erbα promotes the activation of NLRP3
inflammasomes (ASC, caspase-1 and NLRP3) and downstream
inflammatory pathways (IL-18 and IL-1β), and promotes the
progression of liver fibrosis. ASC, apoptosis-associated speck;
α-SMA, α-smooth muscle actin; CCl4, carbon
tetrachloride; COL-1, collagen 1; IL, interleukin; NLRP3, NLR
family domain containing protein 3; Rev-erbα, nuclear receptor
subfamily 1 group D member 1; TGF-β1, transforming growth
factor-β1. |
Although significant progress has been made in
elucidating the role of Rev-erbα in liver fibrosis and its
association with the NLRP3 inflammasome, several limitations remain
that require further investigation. First, the molecular mechanisms
underlying these observations have not been fully explored. While
the present study demonstrates that Rev-erbα negatively regulates
the NLRP3 inflammasome, the specific molecular interactions remain
unclear. Future research should focus on understanding how Rev-erbα
interacts with the various components of the NLRP3 inflammasome and
how this interaction influences the onset and progression of liver
fibrosis. Second, the animal model used in this study,
CCl4-induced liver fibrosis, only partially mimics the
pathological processes of human liver fibrosis and does not fully
reflect clinical conditions. Third, the study's focus on HSCs
limits its scope. Liver fibrosis is a complex, multifactorial
condition involving interactions between various cell types,
including hepatocytes, fibroblasts and immune cells. A broader
examination of the role of these other cell types would provide a
more comprehensive understanding of the disease process. Finally,
while Rev-erbα shows promise as a therapeutic target in liver
fibrosis, translating this potential into clinical applications
faces significant challenges. These include the optimization of
drug delivery systems, addressing inter-individual variability, and
ensuring safety and efficacy in clinical settings.
In light of these limitations, several future
research directions are proposed. Regarding the molecular
mechanisms, building upon existing findings and recent
international advancements, a preliminary hypothesis can be formed
concerning how Rev-erbα regulates the NLRP3 inflammasome in liver
fibrosis. The first potential mechanism involves direct
transcriptional regulation. As a nuclear receptor, Rev-erbα may
bind directly to the promoter regions of NLRP3 inflammasome-related
genes and suppress their transcription. For instance, Rev-erbα
could bind to the promoters of NLRP3, caspase-1 or ASC, inhibiting
their expression and thus reducing NLRP3 inflammasome activity. The
second mechanism involves indirect signaling pathways. Rev-erbα may
modulate NLRP3 inflammasome activity by influencing other cellular
signaling pathways, such as inhibiting the NF-κB pathway, thereby
reducing NLRP3 activation. Additionally, Rev-erbα could regulate
oxidative stress and mitochondrial function, indirectly affecting
inflammasome activity. The third mechanism entails protein-protein
interactions, where Rev-erbα may physically interact with one or
more components of the NLRP3 inflammasome, interfering with its
assembly and function. For example, Rev-erbα might bind to NLRP3,
caspase-1 or ASC, preventing the formation of active inflammasome
complexes. To validate these hypotheses and elucidate the precise
molecular mechanisms by which Rev-erbα regulates the NLRP3
inflammasome in liver fibrosis, advanced molecular biology
techniques such as chromatin immunoprecipitation and RNA sequencing
should be employed. These tools will enable the exploration of how
Rev-erbα interacts with each component of the inflammasome and how
these interactions influence the progression of liver fibrosis.
Furthermore, the development of more clinically relevant animal
models is needed. Models that better replicate the etiology of
human liver fibrosis, such as cholestatic liver disease or viral
hepatitis models, would provide more accurate insights into the
role of Rev-erbα in different types of liver fibrosis. The
interaction between various cell types should also be explored.
Specifically, studying Rev-erbα expression and function in
hepatocytes, immune cells and other cell types will help clarify
how these interactions contribute to the progression of liver
fibrosis. Finally, efforts to promote clinical translation are
essential. Optimizing drug delivery systems and improving the
stability and bioavailability of Rev-erbα modulators will be key to
advancing their therapeutic potential in vivo. Clinical
trials should also be conducted to assess the efficacy and safety
of Rev-erbα regulators in patients with liver fibrosis.
In summary, by deepening our understanding of the
function and regulatory network of Rev-erbα, novel drug targets can
be identified, leading to the development of more effective
anti-fibrotic therapies. This progress not only promises to advance
medical research but also offers new hope for the treatment of
fibrotic diseases, ultimately contributing to a transformative
shift in clinical practice.
In conclusion, expression of the circadian clock
gene Rev-erbα is significantly suppressed in liver fibrosis, which
contributes to fibrogenesis by driving the upregulation of the
downstream NLRP3 inflammasome.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Shanghai Natural Science
Foundation (grant no. 22ZR1459400), the Shanghai Science and
Technology Innovation Project (grant no. 22S21901100), the National
Natural Science Foundation of China (grant no. 82304932), and the
Traditional Chinese Medicine Research Project of Shanghai Municipal
Health Commission (grant no. 2022QN052).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
JW was responsible for conceptualization,
validation, writing the original draft of the manuscript, using
software, visualization, and reviewing and editing the manuscript.
YW and LL contributed to investigation and the methodology. WP and
YL were responsible for the acquisition of funding, project
administration and formal analysis. All authors have read and
approved the final version of the manuscript. WP and YL confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
All animal experiment protocols were approved by the
Ethics Committee of Shanghai Municipal Hospital of Traditional
Chinese Medicine (Shanghai, China; approval no. 2022033).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ASC
|
apoptosis associated speck
|
|
α-SMA
|
α-smooth muscle actin
|
|
Bmal1
|
brain and muscle arnt-like protein
1
|
|
CCl4
|
carbon tetrachloride
|
|
Cry
|
cryptochrome
|
|
cGAS
|
cGMP-AMP synthase
|
|
COL-1
|
collagen 1
|
|
HSC
|
hepatic stellate cell
|
|
IL-1β
|
interleukin-1β
|
|
IL-18
|
interleukin-18
|
|
NLRP3
|
NLR family domain containing protein
3
|
|
Per
|
period circadian regulator
|
|
Rev-erbα
|
nuclear receptor subfamily 1 group D
member 1
|
|
TGF-β1
|
transforming growth factor-β1
|
References
|
1
|
Hernandez-Gea V and Friedman SL:
Pathogenesis of liver fibrosis. Annu Rev Pathol. 6:425–456. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parola M and Pinzani M: Liver fibrosis:
Pathophysiology, pathogenetic targets and clinical issues. Mol
Aspects Med. 65:37–55. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Higashi T, Friedman SL and Hoshida Y:
Hepatic stellate cells as key target in liver fibrosis. Adv Drug
Deliv Rev. 121:27–42. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tsuchida T and Friedman SL: Mechanisms of
hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol.
14:397–411. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Frazier K, Manzoor S, Carroll K, DeLeon O,
Miyoshi S, Miyoshi J, St George M, Tan A, Chrisler EA, Izumo M, et
al: Gut microbes and the liver circadian clock partition glucose
and lipid metabolism. J Clin Invest. 133:e1625152023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mukherji A, Bailey SM, Staels B and
Baumert TF: The circadian clock and liver function in health and
disease. J Hepatol. 71:200–211. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Piggins HD: Human clock genes. Ann Med.
34:394–400. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cox KH and Takahashi JS: Circadian clock
genes and the transcriptional architecture of the clock mechanism.
J Mol Endocrinol. 63:R93–R102. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsang AH, Sánchez-Moreno C, Bode B,
Rossner MJ, Garaulet M and Oster H: Tissue-specific interaction of
Per1/2 and Dec2 in the regulation of fibroblast circadian rhythms.
J Biol Rhythms. 27:478–489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
González-Fernández B, Sánchez DI, Crespo
I, San-Miguel B, de Urbina JO, González-Gallego J and Tuñón MJ:
Melatonin attenuates dysregulation of the circadian clock pathway
in mice with CCl4-Induced fibrosis and human hepatic stellate
cells. Front Pharmacol. 9:5562018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen P, Kakan X, Wang S, Dong W, Jia A,
Cai C and Zhang J: Deletion of clock gene Per2 exacerbates
cholestatic liver injury and fibrosis in mice. Exp Toxicol Pathol.
65:427–432. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen P, Han Z, Yang P, Zhu L, Hua Z and
Zhang J: Loss of clock gene mPer2 promotes liver fibrosis induced
by carbon tetrachloride. Hepatol Res. 40:1117–1127. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Laitinen S, Fontaine C, Fruchart JC and
Staels B: The role of the orphan nuclear receptor Rev-Erb alpha in
adipocyte differentiation and function. Biochimie. 87:21–25. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cho H, Zhao X, Hatori M, Yu RT, Barish GD,
Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al:
Regulation of circadian behaviour and metabolism by REV-ERB-α and
REV-ERB-β. Nature. 485:123–127. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stujanna EN, Murakoshi N, Tajiri K, Xu D,
Kimura T, Qin R, Feng D, Yonebayashi S, Ogura Y, Yamagami F, et al:
Rev-erb agonist improves adverse cardiac remodeling and survival in
myocardial infarction through an anti-inflammatory mechanism. PLoS
One. 12:e01893302017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding G, Li X, Hou X, Zhou W, Gong Y, Liu
F, He Y, Song J, Wang J, Basil P, et al: REV-ERB in GABAergic
neurons controls diurnal hepatic insulin sensitivity. Nature.
592:763–767. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
da Rocha AL, Pinto AP, Bedo BLS, Morais
GP, Oliveira LC, Carolino ROG, Pauli JR, Simabuco FM, de Moura LP,
Ropelle ER, et al: Exercise alters the circadian rhythm of
REV-ERB-α and downregulates autophagy-related genes in peripheral
and central tissues. Sci Rep. 12:200062022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo X, Song S, Qi L, Tien CL, Li H, Xu W,
Mathuram TL, Burris T, Zhao Y, Sun Z and Zhang L: REV-ERB is
essential in cardiac fibroblasts homeostasis. Front Pharmacol.
13:8996282022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Griffin P, Dimitry JM and Musiek ES:
Rev-erbs and Glia-implications for neurodegenerative diseases. J
Exp Neurosci. 13:11790695198532332019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu C, Zhao L, Tao J and Li L: Protective
role of melatonin in Early-stage and end-stage liver cirrhosis. J
Cell Mol Med. 23:7151–7162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen L, Xia S, Wang F, Zhou Y, Wang S,
Yang T, Li Y, Xu M, Zhou Y, Kong D, et al: m6A methylation-induced
NR1D1 ablation disrupts the HSC circadian clock and promotes
hepatic fibrosis hepatic fibrosis. Pharmacol Res. 189:1067042023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang S, Lin Y, Yuan X, Li F, Guo L and Wu
B: REV-ERBα integrates colon clock with experimental colitis
through regulation of NF-κB/NLRP3 axis. Nat Commun. 9:42462018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tao Y, Wang N, Qiu T and Sun X: The Role
of Autophagy and NLRP3 inflammasome in liver fibrosis. Biomed Res
Int. 2020:72691502020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wree A, McGeough MD, Inzaugarat ME, Eguchi
A, Schuster S, Johnson CD, Peña CA, Geisler LJ, Papouchado BG,
Hoffman HM, et al: NLRP3 inflammasome driven liver injury and
fibrosis: Roles of IL-17 and TNF in mice. Hepatology. 67:736–749.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang K, Lin L, Zhu Y, Zhang N, Zhou M and
Li Y: Saikosaponin d alleviates liver fibrosis by negatively
regulating the ROS/NLRP3 inflammasome through activating the ERβ
pathway. Front Pharmacol. 13:8949812022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shao R, Yang Y, Fan K, Wu X, Jiang R, Tang
L, Li L, Shen Y, Liu G and Zhang L: REV-ERBα Agonist GSK4112
attenuates Fas-induced Acute Hepatic Damage in Mice. Int J Med Sci.
18:3831–3838. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim J, Park I, Jang S, Choi M, Kim D, Sun
W, Choe Y, Choi JW, Moon C, Park SH, et al: Pharmacological rescue
with SR8278, a circadian nuclear receptor REV-ERBα antagonist as a
therapy for mood disorders in Parkinson's disease.
Neurotherapeutics. 19:592–607. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kojetin D, Wang Y, Kamenecka TM and Burris
TP: Identification of SR8278, a synthetic antagonist of the nuclear
heme receptor REV-ERB. ACS Chem Biol. 6:131–134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin L, Zhou M, Que R, Chen Y, Liu X, Zhang
K, Shi Z and Li Y: Saikosaponin-d protects against liver fibrosis
by regulating the estrogen receptor-β/NLRP3 inflammasome pathway.
Biochem Cell Biol. 99:666–674. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yue J, He J, Wei Y, Shen K, Wu K, Yang X,
Liu S, Zhang C and Yang H: Decreased expression of Rev-Erbα in the
epileptic foci of temporal lobe epilepsy and activation of Rev-Erbα
have anti-inflammatory and neuroprotective effects in the
pilocarpine model. J Neuroinflammation. 17:432020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Niu WX, Bao YY, Zhang N, Lu ZN, Ge MX, Li
YM, Li Y, Chen MH and He HW: Dehydromevalonolactone ameliorates
liver fibrosis and inflammation by repressing activation of NLRP3
inflammasome. Bioorg Chem. 127:1059712022. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang X, Kuang G, Wan J, Jiang R, Ma L,
Gong X and Liu X: Salidroside protects mice against CCl4-induced
acute liver injury via down-regulating CYP2E1 expression and
inhibiting NLRP3 inflammasome activation. Int Immunopharmacol.
85:1066622020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kisseleva T and Brenner D: Molecular and
cellular mechanisms of liver fibrosis and its regression. Nat Rev
Gastroenterol Hepatol. 18:151–166. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pan X, Mota S and Zhang B: Circadian clock
regulation on lipid metabolism and metabolic diseases. Adv Exp Med
Biol. 1276:53–66. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bailey SM: Emerging role of circadian
clock disruption in Alcohol-induced liver disease. Am J Physiol
Gastrointest Liver Physiol. 315:G364–G373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Richards J and Gumz ML: Advances in
understanding the peripheral circadian clocks. FASEB J.
26:3602–3613. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Horii R, Honda M, Shirasaki T, Shimakami
T, Shimizu R, Yamanaka S, Murai K, Kawaguchi K, Arai K, Yamashita
T, et al: MicroRNA-10a impairs liver metabolism in Hepatitis C
Virus-related cirrhosis through deregulation of the circadian clock
gene brain and Muscle Aryl hydrocarbon receptor nuclear
Translocator-Like 1. Hepatol Commun. 3:1687–1703. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang D, Tong X, Nelson BB, Jin E, Sit J,
Charney N, Yang M, Omary MB and Yin L: The hepatic
BMAL1/AKT/lipogenesis axis protects against alcoholic liver disease
in mice via promoting PPARα pathway. Hepatology. 68:883–896. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pan X, Queiroz J and Hussain MM:
Nonalcoholic fatty liver disease in CLOCK mutant mice. J Clin
Invest. 130:4282–4300. 2020.PubMed/NCBI
|
|
41
|
Zhou B, Zhang Y, Zhang F, Xia Y, Liu J,
Huang R, Wang Y, Hu Y, Wu J, Dai C, et al: CLOCK/BMAL1 regulates
circadian change of mouse hepatic insulin sensitivity by SIRT1.
Hepatology. 59:2196–2206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gnocchi D, Custodero C, Sabbà C and
Mazzocca A: Circadian rhythms: A possible new player in
Non-alcoholic fatty liver disease pathophysiology. J Mol Med
(Berl). 97:741–759. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Forman BM, Chen J, Blumberg B, Kliewer SA,
Henshaw R, Ong ES and Evans RM: Cross-talk among ROR alpha 1 and
the Rev-erb family of orphan nuclear receptors. Mol Endocrinol.
8:1253–1261. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Le Martelot G, Claudel T, Gatfield D,
Schaad O, Kornmann B, Lo Sasso G, Moschetta A and Schibler U:
REV-ERBalpha participates in circadian SREBP signaling and bile
acid homeostasis. PLoS Biol. 7:e10001812009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhou D, Wang Y, Chen L, Jia L, Yuan J, Sun
M, Zhang W, Wang P, Zuo J, Xu Z and Luan J: Evolving roles of
circadian rhythms in liver homeostasis and pathology. Oncotarget.
7:8625–8639. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ni Y, Zhao Y, Ma L, Wang Z, Ni L, Hu L and
Fu Z: Pharmacological activation of REV-ERBα improves nonalcoholic
steatohepatitis by regulating intestinal permeability. Metabolism.
114:1544092021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lin Y, Wang S, Gao L, Zhou Z, Yang Z, Lin
J, Ren S, Xing H and Wu B: Oscillating lncRNA Platr4 regulates
NLRP3 inflammasome to ameliorate nonalcoholic steatohepatitis in
mice. Theranostics. 11:426–444. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yu F, Wang Z, Zhang T, Chen X, Xu H, Wang
F, Guo L, Chen M, Liu K and Wu B: Deficiency of intestinal Bmal1
prevents obesity induced by high-fat feeding. Nat Commun.
12:53232021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lin Y, Lin L, Gao L, Wang S and Wu B:
Rev-erbα regulates hepatic ischemia-reperfusion injury in mice.
Biochem Biophys Res Commun. 529:916–921. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu Q, Xu L, Wu M, Zhou Y, Yang J, Huang
C, Xu T, Li J and Zhang L: Rev-erbα exacerbates hepatic steatosis
in alcoholic liver diseases through regulating autophagy. Cell
Biosci. 11:1292021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang Z, Smalling RV, Huang Y, Jiang Y,
Kusumanchi P, Bogaert W, Wang L, Delker DA, Skill NJ, Han S, et al:
The role of SHP/REV-ERBα/CYP4A axis in the pathogenesis of
Alcohol-associated liver disease. JCI Insight. 6:e1406872021.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang J, Yin L and Lazar MA: The orphan
nuclear receptor Rev-erb alpha regulates circadian expression of
plasminogen activator inhibitor type 1. J Biol Chem.
281:33842–33848. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Crouchet E, Dachraoui M, Jühling F,
Roehlen N, Oudot MA, Durand SC, Ponsolles C, Gadenne C,
Meiss-Heydmann L, Moehlin J, et al: Targeting the liver clock
improves fibrosis by restoring TGF-β signaling. J Hepatol.
82:120–133. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Grant D, Yin L, Collins JL, Parks DJ,
Orband-Miller LA, Wisely GB, Joshi S, Lazar MA, Willson TM and
Zuercher WJ: GSK4112, a small molecule chemical probe for the cell
biology of the nuclear heme receptor Rev-erbα. ACS Chem Biol.
5:925–932. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dong D, Sun H, Wu Z, Wu B, Xue Y and Li Z:
A validated ultra-performance liquid chromatography-tandem mass
spectrometry method to identify the pharmacokinetics of SR8278 in
normal and streptozotocin-induced diabetic rats. J Chromatogr B
Analyt Technol Biomed Life Sci. 1020:142–147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ishizuka T and Lazar MA: The N-CoR/histone
deacetylase 3 complex is required for repression by thyroid hormone
receptor. Mol Cell Biol. 23:5122–5131. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yin L and Lazar MA: The orphan nuclear
receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3
corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol.
19:1452–1459. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yin L, Joshi S, Wu N, Tong X and Lazar MA:
E3 ligases Arf-bp1 and Pam mediate lithium-stimulated degradation
of the circadian heme receptor Rev-erb alpha. Proc Natl Acad Sci
USA. 107:11614–11619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yin L, Wang J, Klein PS and Lazar MA:
Nuclear receptor Rev-erbalpha is a critical lithium-sensitive
component of the circadian clock. Science. 311:1002–1005. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kisseleva T and Brenner DA: Hepatic
stellate cells and the reversal of fibrosis. J Gastroenterol
Hepatol. 21 (Suppl 3):S84–S87. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Liu XY, Liu RX, Hou F, Cui LJ, Li CY, Chi
C, Yi E, Wen Y and Yin CH: Fibronectin expression is critical for
liver fibrogenesis in vivo and in vitro. Mol Med Rep.
14:3669–3675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Friedman SL: Hepatic stellate cells:
Protean, multifunctional, and enigmatic cells of the liver. Physiol
Rev. 88:125–172. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Xiang D, Zou J, Zhu X, Chen X, Luo J, Kong
L and Zhang H: Physalin D attenuates hepatic stellate cell
activation and liver fibrosis by blocking TGF-β/Smad and YAP
signaling. Phytomedicine. 78:1532942020. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhang J, Jiang N, Ping J and Xu L:
TGF-β1-induced autophagy activates hepatic stellate cells via the
ERK and JNK signaling pathways. Int J Mol Med. 47:256–266. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wu Z, Liao F, Luo G, Qian Y, He X, Xu W,
Ding S and Pu J: NR1D1 Deletion induces Rupture-prone vulnerable
plaques by regulating macrophage pyroptosis via the NF-κB/NLRP3
inflammasome pathway. Oxid Med Cell Longev. 2021:52175722021.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ka NL, Park MK, Kim SS, Jeon Y, Hwang S,
Kim SM, Lim GY, Lee H and Lee MO: NR1D1 Stimulates antitumor immune
responses in breast cancer by activating cGAS-STING signaling.
Cancer Res. 83:3045–3058. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kou L, Chi X, Sun Y, Han C, Wan F, Hu J,
Yin S, Wu J, Li Y, Zhou Q, et al: The circadian clock protein
Rev-erbα provides neuroprotection and attenuates neuroinflammation
against Parkinson's disease via the microglial NLRP3 inflammasome.
J Neuroinflammation. 19:1332022. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang ZN, Wang J, Wang ZY, Min LY, Ni HL,
Han YL, Tian YY, Cui YZ, Han JX and Cheng XF: SR9009 attenuates
inflammation-related NPMSC pyroptosis and IVDD through
NR1D1/NLRP3/IL-1β pathway. iScience. 27:1097332024. View Article : Google Scholar : PubMed/NCBI
|