Introduction
Osteoporosis is a systemic skeletal disorder closely
related to ageing, which is characterized by changes in bone
structure and the loss of bone density (1). According to the International
Osteoporosis Foundation 2024 report (2), ~500 million individuals aged 50 years
and older worldwide are affected by osteoporosis, with 37 million
fragility fractures occurring annually (equivalent to 70 fractures
per minute). This includes over 10 million hip fractures, a subset
of fragility fractures that account for significant morbidity and
mortality (3). Hence,
investigating the pathogenesis of osteoporosis in older individuals
is important in the search for novel intervention strategies and
treatment modalities.
Cellular ageing is a complex biological process. As
the normal cell passage time increases, the mitotic activity
decreases and genomic instability increases, leading to cell
degradation, fragment accumulation and gradual ageing (4). A study has shown that, as primary
cells continue to divide, telomeres shorten. When telomere DNA
shortens to a certain extent, cells reach the limit of normal cell
division and then automatically initiate an ageing process termed
cellular replicative ageing (5). A
number of classical signaling pathways regulate the ageing process
of bone mesenchymal stromal cells (BMSCs). For instance, research
findings have shown that micro RNA (miR)-34a can activate the
sirtuin 1/FOXO3a pathway and induce the ageing of BMSCs under
hypoxia and serum deficiency (6).
Xiang et al (7) found that
the AKT/FOXO3a/PTEN-induced kinase 1/Parkin axis suppresses the
mitosis-induced senescence of BMSCs. In addition, signaling
pathways such as p53/p21WAF1/CIP1 (8), p16/retinoblastoma (9), AKT/mTOR (10) and MAPK (11) are associated with cellular
senescence. These signaling pathways universally participate in the
cellular senescence process of BMSCs. Senescence-related signaling
pathways in BMSCs may be a potential target for small molecule
inhibitors, thereby preventing and treating age-related
degenerative diseases.
BMSCs have the potential to self-replicate and
differentiate into multiple cell types, including osteoblasts and
adipocytes (12). Age-related
functional changes in BMSCs play an essential role in the onset and
progression of osteoporosis (13).
It has been shown that the senescence-associated secretory
phenotype (SASP) secreted by BMSCs increases with age. This
secretion includes various biologically active substances that
modify the functionality of stem and progenitor cells (14). Tian et al (15) found that a reduction in crystallin
αB expression can facilitate the degradation of ferritin heavy
chain 1, leading to elevated iron-induced cell death in BMSCs and a
decline in osteogenic differentiation. Multinucleated osteoclasts
are formed by the fusion of monocyte and macrophage progenitor
cells and play a critical role in osteoporosis initiation. The
functional imbalance between osteoblasts and osteoclasts is a
significant contributing factor to osteoporosis (16). A notable study by Ma et al
(17) found that BMSCs can affect
the differentiation of osteoclasts, a process important in
maintaining bone metabolism equilibrium. Recent research indicates
that the high-fat diet-induced senescence of BMSCs impacts their
proliferation and osteogenic capacity, thereby contributing to the
development of osteoporosis (18).
A prior study on ageing BMSCs have mainly focused on their impact
on osteoblast and adipocyte differentiation (19). However, the precise mechanisms
through which ageing BMSCs influence osteoclast differentiation in
age-related osteoporosis remains ambiguous and necessitates
additional investigation.
The primary objective of the present study was to
identify genes with high expression levels in bone marrow
mesenchymal stem cells (BMSCs) of osteoporosis patients. Initially,
the GSE35959 transcriptome sequencing dataset was analyzed.
Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) functional enrichment analyses were performed to
annotate gene functions and signaling pathways. Principal component
analysis (PCA) was then conducted to visualize sample variations
and groupings, providing insights into inter-sample differences.
Gene set enrichment analysis (GSEA) was employed to determine the
enrichment of predefined gene sets associated with cellular
processes, exploring potential biological implications at the gene
set level. Furthermore, a replicative senescence model of mouse
BMSCs was established. Reverse transcription-quantitative (RT-q)
PCR and western blotting were utilized to detect the expression of
AKT1 at both mRNA and protein levels in senescent BMSCs.
Gain-of-function experiments were carried out by overexpressing
AKT1 in young BMSCs. To investigate the effects of AKT1 on BMSC
senescence and osteoclast activation, activated senescent BMSCs
were co-cultured with osteoclast precursors and osteoclast
formation was quantified. Through these comprehensive approaches,
the present study aimed to identify novel targets for the
prevention and treatment of age-related osteoporosis.
Materials and methods
Differential gene screening in BMSCs
from elderly individuals diagnosed with osteoporosis
The transcriptome sequencing data from the GSE35959
dataset were downloaded from the Gene Expression Omnibus database
(https://www.ncbi.nlm.nih.gov/geo/)
(20). In this analysis, the
population was divided into three groups, G1 (42–67 year healthy
control group, mean age 57.6±9.56 years), G2 (79–89 year
non-osteoporotic control group, mean age 81.75±4.86 years) and G3
(79–94 year osteoporosis patient group, mean age 86.2±5.89 years),
as shown in Table I. Subsequently,
the differential gene expression in the BMSCs from G2 and G3 in the
dataset was analyzed. Differential analysis was conducted using the
‘limma’ package (v3.40.2; http://bioconductor.org/packages/3.10/bioc/html/limma.html)
in R software (v4.0.3) (21),
selecting results with |log2foldchange (FC)|≥1 and Padj<0.05 as
differential genes. Volcano plots were drawn using the ‘ggplot2’
package (v3.3.5; http://cran.r-project.org/package=ggplot2) in R.
Additionally, the expression of differentially expressed genes was
visualized through the ‘pheatmap’ package (v1.0.12; http://cran.r-project.org/package=pheatmap) in R.
 | Table I.Population grouping in the present
study. |
Table I.
Population grouping in the present
study.
| Group | Average age
(years) | Osteoporosis
status | Dataset number | Age, years |
|---|
| G1 | 57.6±9.56 | None | GSM878095 | 42 |
|
|
|
| GSM878096 | 67 |
|
|
|
| GSM878097 | 61 |
|
|
|
| GSM878098 | 62 |
|
|
|
| GSM878099 | 56 |
| G2 | 81.75±4.86 | None | GSM878100 | 79 |
|
|
|
| GSM878101 | 79 |
|
|
|
| GSM878102 | 80 |
|
|
|
| GSM878103 | 89 |
| G3 | 86.2±5.89 | Present | GSM878104 | 79 |
|
|
|
| GSM878105 | 94 |
|
|
|
| GSM878106 | 87 |
|
|
|
| GSM878107 | 82 |
|
|
|
| GSM878108 | 89 |
GO and KEGG enrichment analyses
The R software package, ‘clusterProfiler’ (v3.18.0;
http://bioconductor.org/packages/3.12/bioc/html/clusterProfiler.html)
was used to perform GO and KEGG enrichment analyses on the
downregulated genes in the BMSCs from G2 and to visualize the
results in a bubble chart form.
PCA and GSEA
The R software package ‘ggplot2’ (v3.3.5) was used
to perform PCA visualization on the gene expression data from three
groups (G1, G2 and G3) and to present the results in both
two-dimensional (PC1 vs. PC2) and three-dimensional (PC1 vs. PC2
vs. PC3) scatter plot formats. The R software packages
‘clusterProfiler’ (v3.18.0) and ‘fgsea’ (v1.16.0; http://bioconductor.org/packages/3.12/bioc/html/fgsea.html)
were used to perform GSEA on the differentially expressed genes
between G2 and G3, calculate enrichment scores (ES), conduct
pathway-related analyses using signaling pathways in the KEGG
pathway database and visualize the results via enrichment plots and
running enrichment score plots.
Cell lines
Mouse BMSCs were acquired through primary isolation
from mouse bone marrow tissue by a subsidiary of the Eli Lilly and
Company (Sichuan Lilaisinuo Biotechnology Co., Ltd.) (cat. no.
LL-A1-80093). Cell lines were obtained as commercially prepared
primary cells or cell lines, without additional animal procurement
or primary isolation by the research team. RAW 264.7 cells (cat.
no. CL-0190) were procured from Procell Life Science &
Technology Co., Ltd.
Establishment of a replicative ageing
BMSCs model
Primary BMSCs were cultured in DMEM/F12 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.; cat. no.
16000-044) and 1% penicillin-streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). All cells were maintained at 37°C with 95%
humidity and 5% CO2 and regularly tested for mycoplasma
by PCR (Genlantis Diagnostics Inc.). Based on established
replicative ageing models, the P3 (early passage) and P10 (late
passage) timepoints were selected for analysis, as P10 BMSCs are
known to exhibit elevated senescence markers (22,23).
The cells were continuously passaged for 10 generations to generate
the replicative ageing cell model (P10-BMSCs).
Western blotting (WB)
The mouse BMSC samples were collected and then lysed
using RIPA buffer supplemented with protease inhibitor and
phosphatase inhibitor (all Biosharp Life Sciences). The protein
concentration was detected using a BCA protein detection kit
(Shanghai Biyuntian Biotechnology Co., Ltd.). Next, ~30 µg of
protein sample was separated on a Bis-Tris 10% high-resolution
precast gel (Shanghai Yeasen Biotechnology Co., Ltd.) and then
transferred onto an Immobilon PSQ PVDF membrane (MilliporeSigma).
The membrane was subsequently incubated with 5% skimmed milk (Wuhan
Servicebio Technology, Co., Ltd.) for 1 h at room temperature. The
membrane was then incubated with primary antibodies against AKT1
(ABclonal Biotech Co., Ltd.; cat. no. A17909; 1:1,000), p16
(ABclonal Biotech Co., Ltd.; cat. no. A23882; 1:1,000), p21
(Proteintech Group, Inc.; cat. no. 28248-1-AP, 1:2,000) and β-actin
(ABclonal Biotech Co., Ltd.; cat. no. AC026; 1:50,000) overnight at
4°C. After washing, the membrane was incubated with a Goat Anti
Rabbit IgG (H+L) HRP-conjugated secondary antibody (Affinity
Biosciences; cat. no. S0001; 1:5,000) at room temperature for 1 h.
Finally, protein bands were visualized using the ‘Torchlight’
Hypersensitive ECL Western HRP Substrate (Chengdu Zen-Bioscience
Co., Ltd.; cat. no. 17046) and imaged using a 5200 Multi
chemiluminescence image analysis system (Tanon Science and
Technology Co., Ltd.). Densitometric analysis of the bands was
performed using Gel-Pro Analyzer software (version 4.0; Media
Cybernetics, Inc.).
RNA isolation and RT-qPCR
Total RNA was extracted from mouse sample cells
using the Molpure® Cell/Tissue Total RNA Kit (Shanghai
Yeasen Biotechnology Co., Ltd.). For RNA extraction, cells were
lysed with 350 µl lysis buffer LB (supplied in the kit) at a
density of 1–5×106 cells per sample (the lysate was
repeatedly pipetted until no cell clumps were visible). After
quantification, 1 µg RNA was reverse transcribed using the
PrimeScript RT kit (Baoriyi Biotechnology Co., Ltd.) with a
protocol of 37°C for 15 min followed by 85°C for 5 sec; this step
was performed according to the manufacturer's protocol. qPCR was
performed using TB Green® Premix Ex Taq II (Tli RNaseH
Plus; Bao Ri Yi Biotechnology Co., Ltd.), with thermocycling
conditions as follows: 95°C for 30 sec (pre-denaturation), followed
by 45 cycles of 95°C for 5 sec (denaturation), 55°C for 30 sec
(annealing) and 72°C for 30 sec (extension with fluorescence
acquisition); qPCR was also performed according to the
manufacturer's protocol. The Cq (quantification cycle) values of
the samples were detected using QuantStudio Design & Analysis
SE Software (Thermo Fisher Scientific, Inc.). GAPDH was used as the
reference gene and quantification was performed using the
2−ΔΔCq method (24).
All experiments (RNA extraction, cDNA synthesis, and qPCR) were
independently replicated three times. The primer sequences used in
this experiment are shown in Table
II.
 | Table II.Primer sequences used in the present
study. |
Table II.
Primer sequences used in the present
study.
| Primer | Forward primer,
5′-3′ | Reverse primer,
5′-3′ |
|---|
| β-actin |
CTACCTCATGAAGATCCTGACC |
CACAGCTTCTCTTTGATGTCAC |
| AKT1 |
TGCACAAACGAGGGGAATATAT |
CGTTCCTTGTAGCCAATAAAGG |
| NFATc1 |
GAGAATCGAGATCACCTCCTAC |
TTGCAGCTAGGAAGTACGTCTT |
| RANK |
CTGAAAAGCACCTGACAAAAGA |
CTGTGTAGCCATCTGTTGAGTT |
| TRAF6 |
GAAAATCAACTGTTTCCCGACA |
ACTTGATGATCCTCGAGATGTC |
Plasmid transfection
Logarithmically growing bone marrow mesenchymal stem
cells (BMSCs) were washed with PBS, trypsinized, centrifuged at 250
× g for 5 min at room temperature, resuspended in culture medium,
counted, adjusted to 1×105 cells/ml and seeded at 2
ml/well in 6-well plates. After adhesion, transfection complexes
were prepared by adding 15 µg AKT1 cDNA recombinant plasmid
[pcDNA3.1(+); plasmid (Invitrogen; Thermo Fisher Scientific, Inc.)
containing mouse AKT1 ORF sequence (cat. no. MG50254-M; Sino
Biological Inc.)] to 300 µl Opti-MEM I in one tube and 30 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) to 300 µl Opti-MEM I in another, incubating both
for 5 min, combining them, incubating for 10 min and adjusting the
volume to 6 ml with Opti-MEM I. Control cells (NC group) were
transfected with 15 µg empty cloning vector (Sino Biological Inc.).
After 6 h at 37°C, the medium was replaced with a complete medium
(cat. no. CM-M131; Procell Life Science & Technology Co.,
Ltd.), and the cells underwent further culturing such that the
total duration from transfection to subsequent experiments was 48
h. For validation, supernatants were collected, cells were washed
with PBS, trypsinized, centrifuged at 250 × g for 5 min at room
temperature, washed again with PBS and centrifuged at 250 × g for 5
min at room temperature to obtain cell pellets for qPCR and western
blot analysis.
Senescence-associated β-galactosidase
(SA-β-gal) staining
Logarithmically growing RAW264.7 cells were
dissociated into a single-cell suspension, centrifuged at 250 × g
for 5 min at room temperature and resuspended in culture medium.
Cells were counted and seeded at 0.5×105 cells/ml (2
ml/well) in 6-well plates, then incubated at 37°C with 5%
CO2. After adhesion, transfection complexes were
prepared by mixing 15 µg of AKT1 cDNA ORF Clone (plasmid backbone:
Cloning Vector; cat. no. MG50254-M; Sino Biological Inc.) in 300 µl
Opti-MEM I Reduced Serum Medium (Gibco; Thermo Fisher Scientific,
Inc.) with 30 µl Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) in 300 µl Opti-MEM I, incubating both
tubes for 5 min, combining them and further incubating for 10 min.
The mixture was adjusted to 6 ml with Opti-MEM I before adding to
cells. Control cells (NC group) were transfected with an equal mass
of empty cloning vector. After 48 h, cells were washed with PBS,
fixed with 1 ml β-galactosidase staining fixative (Beyotime
Institute of Biotechnology; C0602) for 15 min at room temperature
and washed twice with PBS (3 min each). Staining working solution
(1 ml/well) was prepared by mixing 10 µl β-galactosidase staining
solution A, 10 µl solution B, 930 µl solution C and 50 µl X-Gal
solution (Beyotime Institute of Biotechnology; cat. no. C0602) and
applied to cells overnight at 37°C. Images were captured using an
inverted biological microscope (DMI1; Leica Microsystems,
Ltd.).
Osteolysis induction and co-culturing
of RAW264.7 cells and osteoclasts
Logarithmically growing RAW264.7 cells were
dissociated into a single-cell suspension, centrifuged at 250 × g
for 5 min at room temperature, resuspended in culture medium,
counted, adjusted to 0.5×105 cells/ml and seeded at 2
ml/well (1×105 cells/well) in 6-well plates. After
adhesion, cells were induced to differentiate into osteoclasts with
α-MEM medium (cat. no. 41061029; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 50 µg/l RANK ligand (cat. no. C28A;
Novoprotein Scientific Inc.), incubated at 37°C in a 5%
CO2 atmosphere and the medium was replaced every 2 days
for 5 days. Transfected BMSCs were then counted, adjusted to
2×105 cells/ml and 2 ml/well (4×105
cells/well) were seeded into the upper chamber of a Transwell
plate, while the induced osteoclasts remained in the lower chamber
of the 6-well plate for 48 h of co-culture under the same
conditions before subsequent staining.
Tartrate-resistant acid phosphatase
(TRAP) staining
After removing the supernatant, co-cultured cells
were fixed with 4% paraformaldehyde for 15 min at room temperature
and washed three times with distilled water. Cells were
permeabilized with 0.2% Triton X-100 at room temperature for 30
min, followed by three light washes with distilled water. The TRAP
staining kit (cat. no. G1492; Beijing Solarbio Science &
Technology Co., Ltd.) was used according to the manufacturer's
instructions. Mature osteoclasts were identified as multinucleated
cells (≥3 nuclei) and images were captured using an inverted
biological microscope (DMI1; Leica Microsystems, Ltd.) (25).
Statistical analysis
GraphPad Prism 8.0 statistical software (Dotmatics)
was used to perform graphical analysis and statistical testing on
experimental data, which are expressed as the mean ± standard
deviation. For experimental datasets, comparisons between two
groups were conducted using an unpaired two-tailed Student's
t-test. For comparisons among three or more groups, one-way
analysis of variance was used for normally distributed data with
homogeneous variances, followed by Tukey's Honestly Significant
Difference test for pairwise comparisons when overall differences
were significant, and Kruskal-Wallis test was applied to
non-normally distributed or heteroscedastic data, with Dunn's
multiple comparisons test for post hoc pairwise comparisons upon
significant differences. For bioinformatics data analyses (such as
high-throughput sequencing or -omics data), R software 4.2.2 was
used to handle complex statistical modeling and computational
pipelines. P<0.05 was considered to indicate a statistically
significant difference.
Results
Screening of differentially expressed
genes in BMSCs from G3
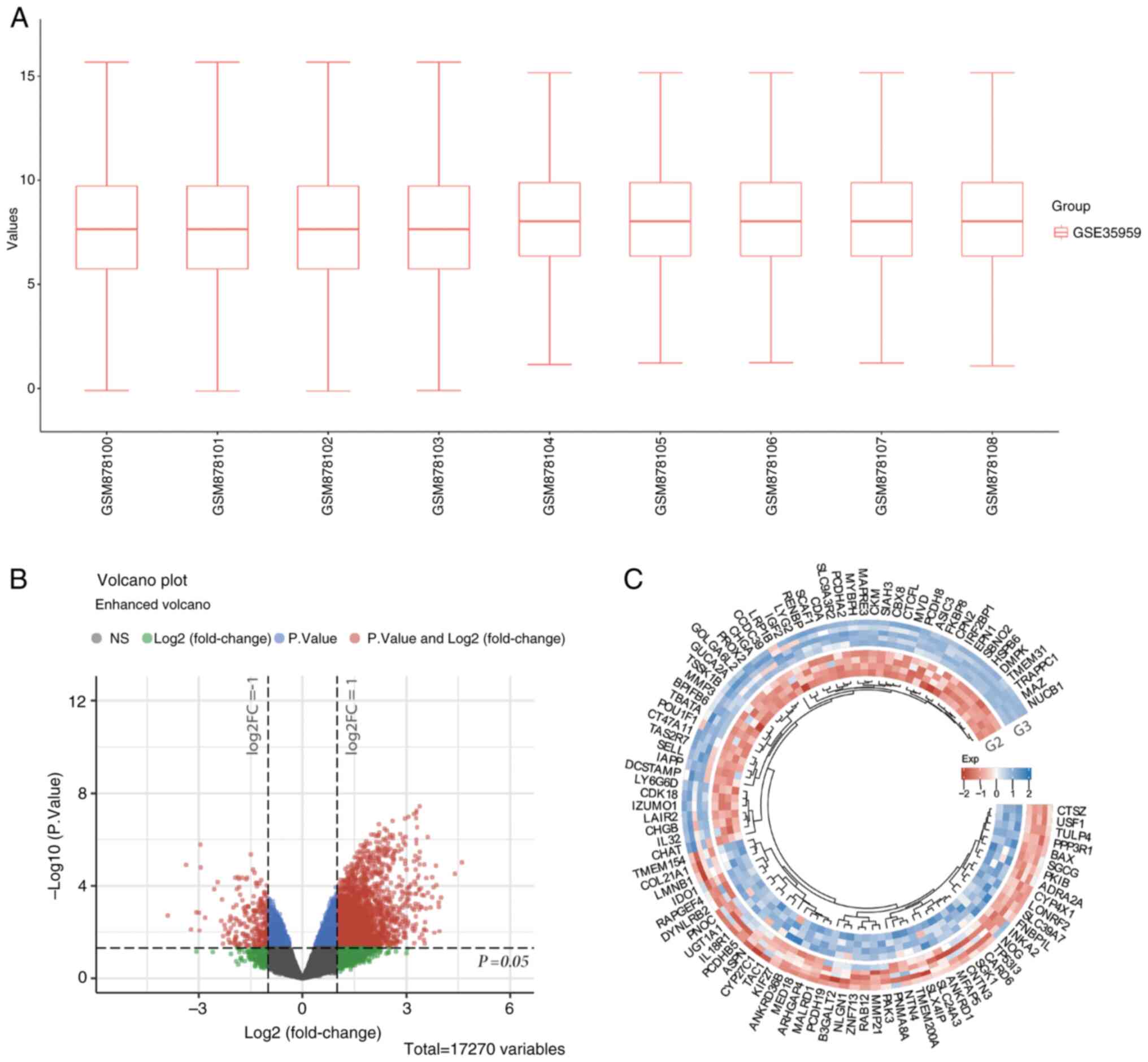
The GSE35959 dataset was selected to analyze the
effects of primary osteoporosis on BMSC gene expression. Initially,
no abnormal outliers were observed in the BMSC samples from G2 and
G3 (Fig. 1A). Subsequently, the
differential gene expression between BMSCs from G2 and G3 was
analyzed. According to the results, BMSCs obtained from G2 had 274
upregulated genes and 2,592 downregulated genes (Fig. 1B). The heatmap shown in Fig. 1C depicts the expression of the
differentially expressed genes in the two groups. Due to the large
number of differentially expressed genes, the top 50 upregulated
and the top 50 downregulated genes with the greatest differential
changes are displayed separately.
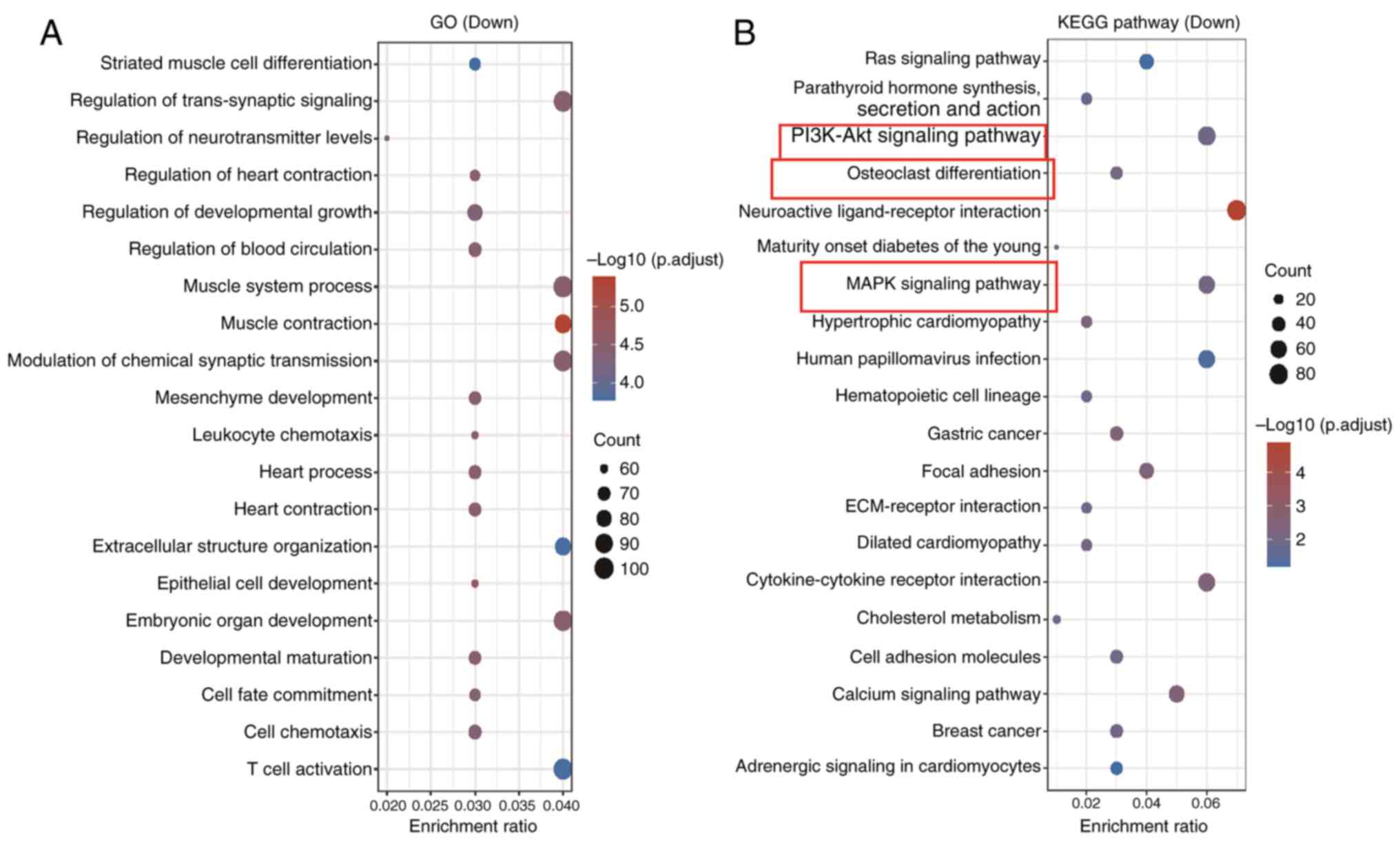
GO and KEGG enrichment analyses
GO and KEGG enrichment analyses were performed on
the 2,592 genes that were upregulated in BMSCs from G3. The GO
results showed that these genes mainly participate in processes
such as ‘striated muscle cell differentiation,’ ‘regulation of
trans-synaptic signaling’ and ‘regulation of neurotransmitter
levels’ (Fig. 2A). The KEGG
enrichment analysis revealed a significant association with the
‘PI3K/Akt signaling pathway’, ‘osteoclast differentiation’ and
‘MAPK signaling pathway’ (Fig.
2B). Several classical signaling pathways, including
PI3K/AKT/mTOR, p53/p21 and MAPK, have been demonstrated to
influence BMSC ageing (26,27).
Subsequent analysis indicated the enrichment of AKT1, MAPK3,
RELA and CSF1 in the MAPK signaling, PI3K/AKT signaling
and osteoclast differentiation pathways. Subsequently, using the
GSE35959 dataset, the levels of the aforementioned four genes in
BMSCs from G1, G2 and G3 were further analyzed (Fig. S1). According to the results, the
expression levels of the four genes were markedly lower in BMSCs
from G2 than those from G1, while the expression levels of the four
genes were markedly higher in BMSCs from G3 than those from G1.
Hence, it was hypothesized that the overactivation of AKT1,
MAPK3, RELA and CSF1 in the stem cells of G3 may
contribute to the pathogenesis of osteoporosis.
PCA and GSEA
PCA analysis revealed differences in gene expression
patterns between the three groups: G1, G2 and G3. Fig. S2A displays a two-dimensional PCA
plot, where PC1 explains 22.4% of the variation and PC2 explains
13% of the variation. The three sample groups form distinct
clusters, indicating significant differences in gene expression
between them. Fig. S2B shows the
three-dimensional PCA plot, adding PC3 (explaining 9.2% of the
variation), further reinforcing the separation pattern between the
sample groups. This clustering pattern suggests that both age (G1
vs. G2) and disease status (G2 vs. G3) have significant effects on
gene expression that can be clearly distinguished through PCA. GSEA
revealed significant positive enrichment of the
‘KEGG_PI3K-Akt_signaling_pathway’ in the comparison between the G3
and G2 groups (Fig. S2C). The
normalized enrichment score was 1.552 with an adjusted P-value of
0.004, indicating statistically significant activation of this
pathway. The running enrichment score (RES) curve displayed a
prominent positive peak (maximum RES ~0.3) and genes within the
pathway were predominantly clustered on the left side of the ranked
list (red area), reflecting their upregulated expression in the G3
group. These results suggested that the PI3K/AKT signaling pathway
is markedly activated during the progression from healthy aging
(G2) to osteoporosis (G3). This differential regulation highlights
the potential role of this pathway in the pathophysiology of
osteoporosis, offering critical insights for identifying
therapeutic targets.
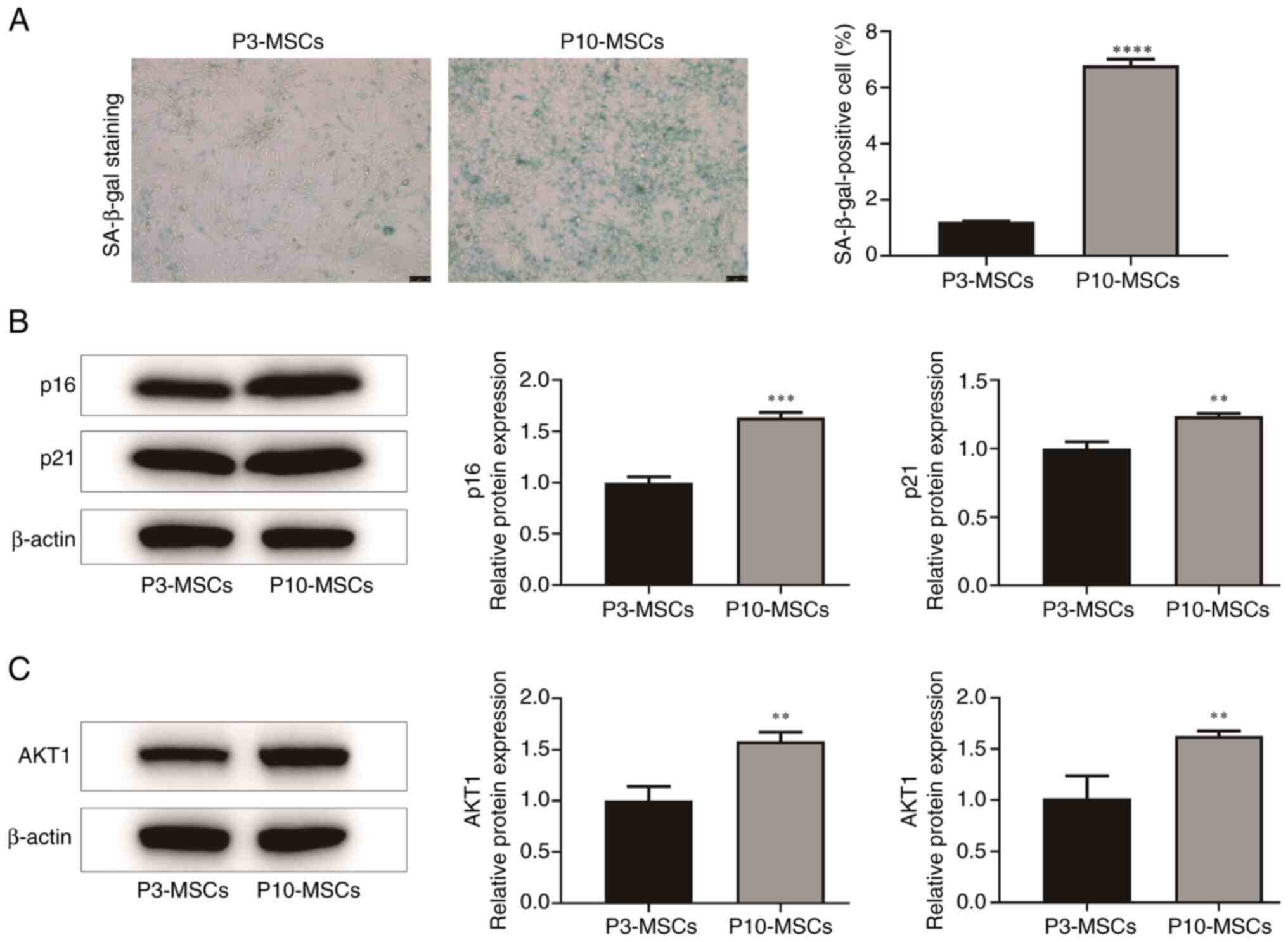
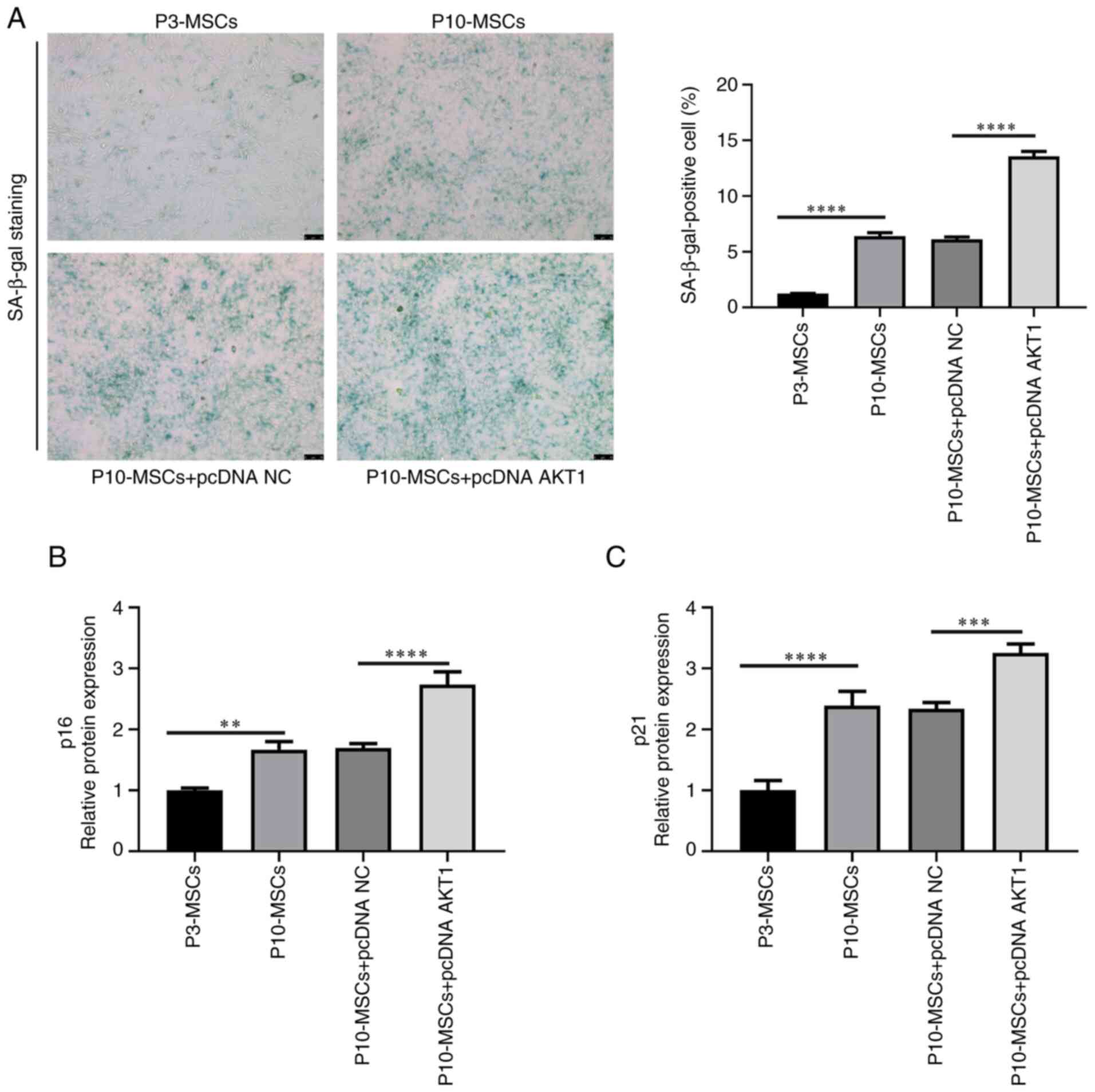
Changes in AKT1 expression levels in
BMSCs with replicative ageing
In the present study, P3 and P10 denoted early
(younger) and late (older) passage numbers of BMSCs, respectively,
with higher passage numbers reflecting increased replicative
cycles. It is well established that SA-β-gal staining, p21
expression and p16 expression serve as significant biomarkers of
the ageing process in BMSCs (28,29).
The results indicated that the number of SA-β-gal positive cells
was markedly higher in the P10-MSCs group than in the P3-MSCs group
(P<0.0001). Additionally, the relative protein expression levels
of the cell ageing markers, p16 (P<0.001) and p21 (P<0.01),
were also markedly increased in the P10-MSCs group compared with
the P3-MSCs group. These results suggested the successful
establishment of a replicative ageing model of BMSCs in the
P10-MSCs group (Fig. 3A and
B).
AKT is a serine/threonine kinase that is dependent
on PI3K. Research indicates that the PI3K/AKT/mTOR pathway is
essential for regulating cell differentiation (30). As a member of the AKT family, AKT1
is integral to the PI3K/AKT signaling pathway. Consequently, a
further examination of the AKT1 expression levels in both the
P3-MSCs and the P10-MSCs groups was conducted. The results of the
PCR and WB analyses indicated that, compared with the P3-MSCs
group, P10-MSCs had markedly higher AKT1 protein and mRNA
expression levels (P<0.05; Fig.
3C). These findings suggest that replicative ageing may enhance
the expression of AKT1 protein in BMSCs.
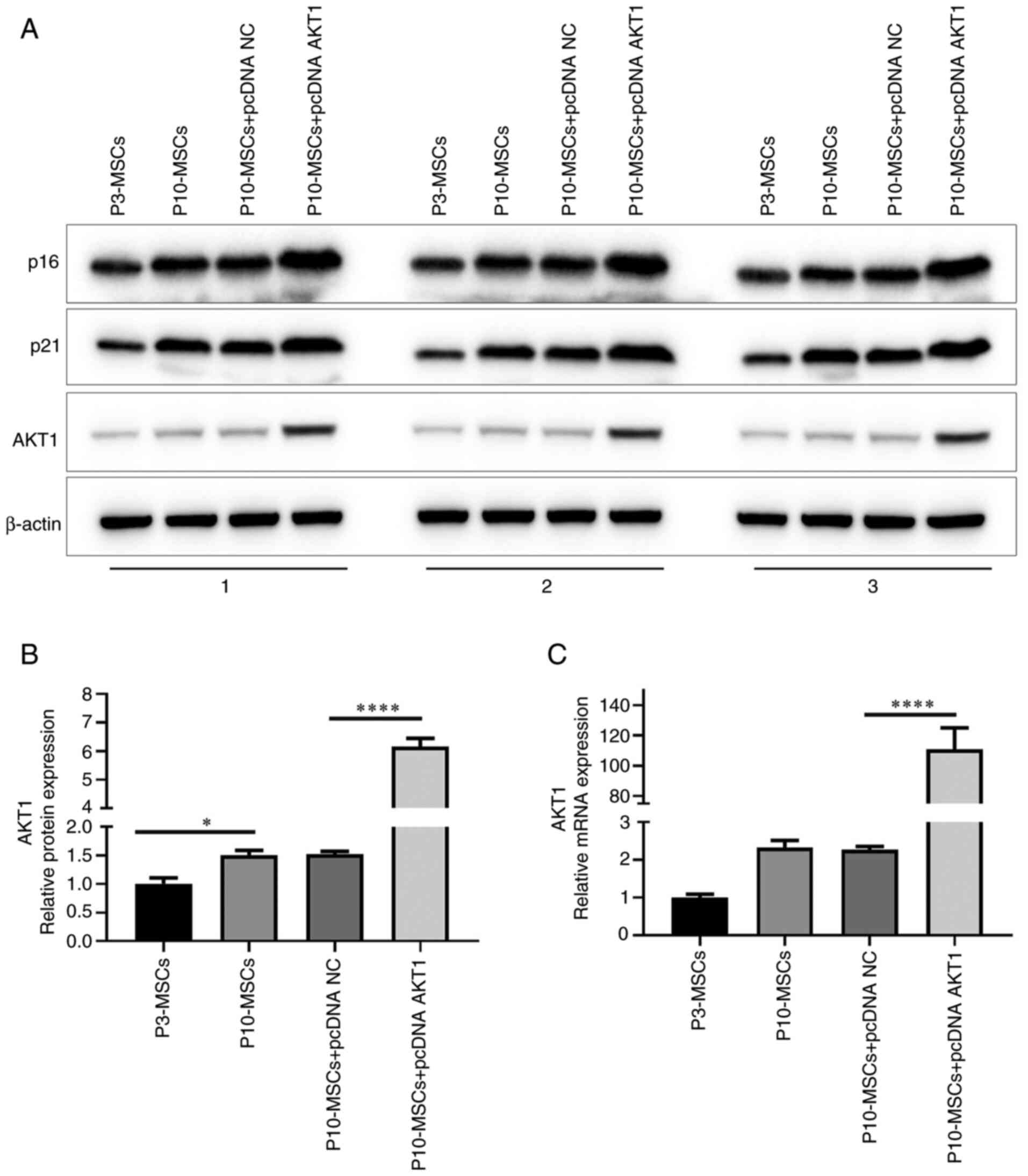
Confirmation of the transfection
efficiency of the AKT1 overexpression plasmid
To further investigate the effect of AKT1 on the
ageing of BMSCs, the gene encoding the AKT1 protein was cloned into
a pcDNA expression vector and subsequently transfected into
P10-MSCs. As shown in Fig. 4A, the
WB results showed that P10-MSCs transfected with pcDNA AKT1 had
higher levels of AKT1 protein. Following statistical analysis of
the results, it was found that in transfected P10-MSCs, AKT1
protein levels exhibited a significant increase compared with
transfected P3-MSCs (P<0.05). This finding suggests that
replicative ageing has a notable effect on elevating AKT1 protein
levels in BMSCs. In addition, compared with the P10-MSCs + pcDNA
negative control (NC) group, the P10-MSCs + pcDNA AKT1 group
exhibited a statistically significant increase in AKT1 protein
levels (P<0.0001; Fig. 4B).
Finally, qPCR demonstrated that, compared with the P10-MSCs + pcDNA
NC group, the relative mRNA expression level of AKT1 was
statistically elevated in the P10-MSCs + pcDNA AKT1 group
(P<0.0001; Fig. 4C). In
summary, these experimental findings indicated the successful
transfection of pcDNA AKT1 into P10-MSCs.
Effect of AKT1 overexpression on the
replicative ageing of BMSCs
Subsequently, the effect of AKT1
overexpression on the replicative ageing of BMSCs was verified.
Following SA-β-gal staining, the P10-MSCs + pcDNA AKT1 group had a
markedly higher proportion of SA-β-gal positive cells than the
P10-MSCs + pcDNA group (Fig. 5A).
Subsequently, a comprehensive analysis of the protein levels of the
cell ageing markers, p16 and p21, in all four experimental groups
was conducted through WB experiments (Fig. 4A). In addition, compared with the
P10 MSCs + pcDNA group, the levels of the p16 and p21 proteins were
markedly increased in the P10 MSCs + pcDNA AKT1 group (P<0.001;
Fig. 5B and C). In conclusion,
these findings indicated that the overexpression of AKT1
promotes the ageing process of BMSCs.
Effect of AKT1 overexpression on
regulating osteoclast differentiation in replicative ageing
BMSCs
In the aforementioned enrichment analysis, it was
indicated that AKT1 may be markedly associated with the
differentiation of osteoclasts in G3. Therefore, experiments using
replication-aged BMSCs were performed to further test the
regulatory role of AKT1 in osteoclast differentiation.
Initially, the supernatant derived from BMSCs was co-cultured with
osteoclasts, as shown in Fig. 6.
It was found that the quantity of TRAP+ multinucleated
cells in the P10-MSCs group was higher than that in the P3-MSCs
group, but this difference was not statistically significant
(P>0.05). Additionally, the P10-MSCs showed a tendency to
express osteoclast differentiation markers. The nuclear factor of
activated T cells, cytoplasmic 1 (NFATc1), tumor necrosis
factor receptor-associated factor 6 (TRAF6) and RANK
mRNA levels were higher in P10-MSCs than in the P3-MSCs group, but
this difference was not statistically significant (P>0.05).
Subsequently, a comparative analysis of the number of
TRAP+ multinucleated cells and the relative mRNA
expression levels of osteoclast differentiation markers, including
NFATc1, TRAF6 and RANK, between the P10-MSCs group
and the P10-MSCs + pcDNA NC group was conducted. There were no
significant differences (P>0.05) in these markers between these
groups. However, the results of the PCR experiment indicated that,
compared with the P10-MSCs + pcDNA NC group, the relative mRNA
expression levels of the osteoclast differentiation markers,
NFATc1 (P<0.01), TRAF6 (P<0.0001) and
RANK (P<0.001), were markedly increased in the P10-MSCs +
pcDNA AKT1 group. In summary, these findings suggested that BMSCs
experiencing replicative ageing may facilitate the differentiation
of osteoclasts. Furthermore, an elevated AKT1 expression
level in ageing BMSCs may be associated with the promotion of
osteoclast differentiation.
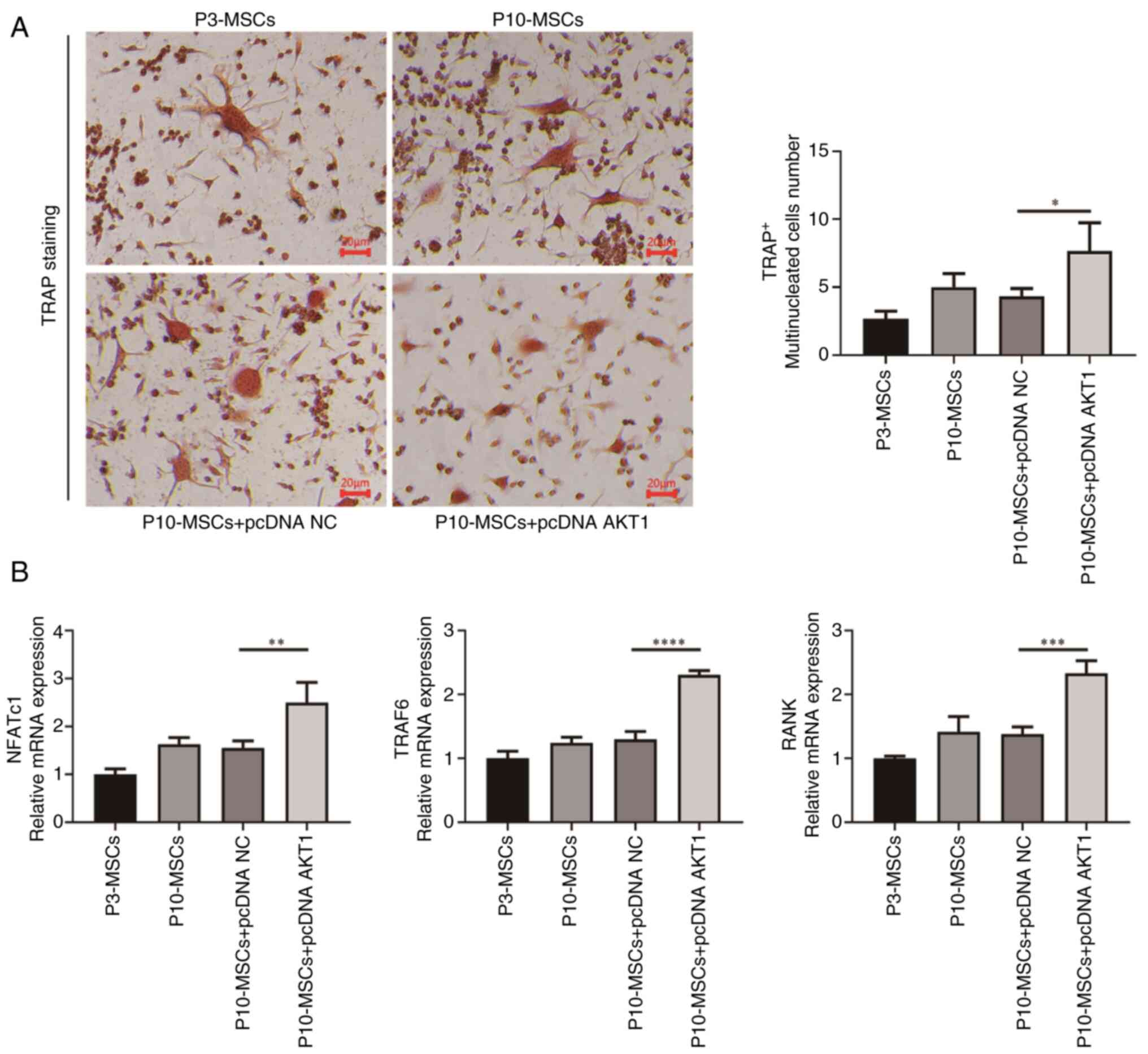 | Figure 6.Replicative-aged BMSCs regulate
osteoclast differentiation. (A) TRAP staining to detect osteoclast
differentiation (×200 magnification). (B) PCR to detect the mRNA
expression of the osteoblast markers, NFATc1, TRAF6 and
RANK, in each group. *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001. BMSCs, bone mesenchymal stromal cells; TRAP,
tartrate-resistant acid phosphatase; NFATc1, nuclear factor of
activated T cells, cytoplasmic 1; TRAF6, tumor necrosis factor
receptor-associated factor 6; RANK, receptor activator of nuclear
factor κB. |
Discussion
Research has demonstrated that the ageing of BMSCs
influences the equilibrium between bone formation and resorption,
serving as a significant contributor to age-related bone diseases
(31). Jin et al (32) found that optineurin facilitates the
osteogenic differentiation of BMSCs and inhibits adipogenic
differentiation by delaying cellular ageing and enhancing
autophagy. According to Hu et al (22), nucleosome assembly protein 1 like 2
is responsible for driving the ageing of mesenchymal stem cells and
preventing their differentiation into osteoblasts. However, current
research on the effects of ageing BMSCs in age-related bone
diseases predominantly emphasizes the differentiation processes
between osteoblasts and adipocytes. By contrast, there is a paucity
of studies investigating the influence of ageing BMSCs on
osteoclast differentiation. Ageing BMSCs can promote the
differentiation of bone marrow monocytes into osteoclasts,
ultimately leading to the occurrence of osteoporosis (33). Research on the novel mechanisms
that ageing BMSCs use to influence osteoclast activation is
essential for age-related osteoporosis discovery, prevention and
treatment. In the present study, it was observed that replicative
ageing enhanced the expression of AKT1 protein in BMSCs and this
increase in AKT1 expression further exacerbated the ageing process
of BMSCs. Furthermore, the overexpression of AKT1 was shown to
promote osteoclast activation in ageing BMSCs, which may contribute
to the development of osteoporosis.
Previous research has confirmed that cellular ageing
is influenced by the regulation of various signaling pathways. For
instance, Prince et al (34) identified a correlation between the
AKT/mTOR pathway and the progression of ageing in ACP epithelial
cells. In addition, Li et al (35) demonstrated that deoxynivalenol
induces cellular senescence in RAW264.7 macrophages through
modulation of the hypoxia-inducible factor-1α/p53/p21 signaling
pathway. The present study initially conducted a screening of
highly expressed genes in BMSCs from G3, using bioinformatics
methodologies. Then, an enrichment analysis of the highly expressed
genes was conducted. Bubble plots revealed a significant
association between these genes and the PI3K/AKT and MAPK signaling
pathways, as well as osteoclast differentiation processes. It is
noteworthy that the four genes, AKT1, MAPK3, RELA and
CSF1, exhibited significant enrichment in the aforementioned
three pathways. PCA confirmed transcriptional divergence with aging
and disease and GSEA revealed significant enrichment of the
PI3K/AKT pathway in G3 vs. G2. Previous research has indicated that
AKT is crucial in the ageing process across various cell types
(36,37). Nevertheless, there has been a lack
of research to ascertain whether the expression of AKT1
influences the replicative ageing of BMSCs.
Ageing-related biomarkers such as SA-β-gal staining,
p21 expression and p16 expression are used to evaluate the process
of cellular ageing (38,39). The present study initially observed
a significant increase in ageing-related markers, specifically in
the p21 and p16 protein levels as well as the number of SA-β-gal
positive cells, in the P10-MSCs group (cultured continuously for 10
generations) compared with the P3-MSCs group (cultured for three
generations). This indicated that a replicative ageing model was
successfully established using P10-MSCs. In addition, it was found
that P10-MSCs expressed markedly higher levels of AKT1 mRNA and
protein, which indicated that senescent BMSCs have higher AKT1
protein levels. Additionally, the overexpression of AKT1 in
the P10-MSCs group resulted in an increased proportion of SA-β-gal
positive cells, as well as higher p21 and p16 levels. This finding
suggested that AKT1 may facilitate the promotion of
senescence in BMSCs. Consistent with these findings, the study
conducted by Xiang et al (7) demonstrated that CSE can induce ageing
in BMSCs through the upregulation of phosphorylated AKT. Overall,
in the present study, the expression of AKT1 was found to be
elevated in senescent BMSCs and the overexpression of AKT1 was
shown to promote senescence in these cells. Accordingly, it is
hypothesized that AKT1 represents a novel therapeutic target
for treating and preventing replicative senescence in BMSCs. To the
best of our knowledge, the present study is the first to indicate
that AKT1 upregulation enhances the regulation of osteoclast
differentiation in BMSCs undergoing replicative ageing.
The overactivation of osteoclasts is a significant
contributor to the pathogenesis of osteoporosis. A previous study
showed that the pro-inflammatory SASP released by senescent cells
activates osteoclast precursors to promote osteoclast
differentiation, ultimately leading to a decrease in bone density
(40). In addition, a study has
shown that CSF1 regulated by nuclear paraspeckle assembly
transcript 1 can be delivered extracellularly through paracrine
pathways in ageing BMSCs, thereby promoting osteoclast
differentiation (41). In the
present study, a greater percentage of TRAP+
multinucleated cells was observed in the P10-MSCs group than in the
P3-MSCs group, it has been documented that RANK, NFATc1 and
TRAF6 serve as significant biomarkers in the differentiation
process of osteoclasts (42,43).
The present study found that the RANK, NFATc1 and
TRAF6 mRNA expression levels were increased in P10-MSCs.
These findings suggested that ageing BMSCs may facilitate the
activation of osteoclasts. Xie et al (44) found that the SHIP1
activator, AQX-1125, modulates the differentiation of osteoblasts
and osteoclasts via the PI3K/AKT and NF-κB signaling pathways.
However, there are some shortcomings in the present
research. First, further investigation is needed on how the
upregulation of AKT1 promotes the replicative ageing of
BMSCs. A previous report has indicated that epigenetics can
regulate gene transcription and translation processes by modifying
gene expression through various pathways and cellular ageing is
regulated by DNA methylation, histone modification, chromatin
remodeling and non-coding RNA (45). While the present study links AKT1
upregulation to senescence, the upstream triggers (such as DNA
damage and telomere attrition) remain unexplored. Future research
should assess AKT1 phosphorylation and its interaction with the
PI3K/AKT pathway components to elucidate mechanistic drivers. The
present study has certain room for improvement in the construction
of the senescence model and mechanistic analysis. The current
research only selected two timepoints, passage 3 (early passage)
and passage 10 (late passage), to establish the senescence model.
While this selection was based on the classical replicative
senescence model (where passage 10 BMSCs are known to highly
express senescence markers), it indeed failed to fully characterize
the continuous trajectory of BMSCs transitioning from a highly
proliferative state to degenerative senescence. Such a jump-type
analysis between two passages may have obscured key
intermediate-stage changes during the senescence process. The
present study started from molecular characteristics of clinical
senescent samples to validate pathological key pathways, selecting
senescent cells as subjects. The role of AKT1 in proliferative
cells needs upstream mechanism exploration via independent
experiments. AKT1 overexpression may have ‘enhancing/inducing
senescence’ dual effects, with mechanisms needing further study.
Expanding this area will aid revealing the role of AKT1 in the
senescence continuum and provide early osteoporosis intervention
targets. In addition, a recent study found that aldehyde
dehydrogenase 2 regulates mesenchymal stem cell ageing by
maintaining mitochondrial homeostasis (46), which provides direction for future
research. Second, regarding how ageing BMSCs regulate osteoclast
differentiation, the study by Yin et al (33) demonstrated that the extracellular
vesicles containing miR-144-5p secreted by ageing BMSCs play a
significant role in osteoclast differentiation but the specific
mechanism is unclear. Third, while rigorous thresholds (|log2FC|≥1,
Padj<0.05) support the findings of the present study, the
limited sample size of the GSE35959 dataset may affect
generalizability. Larger-scale studies incorporating broader age
ranges are warranted to validate the current observations. Future
studies should incorporate functional assays (such as bone
resorption pits) to validate the osteoclast-promoting effects of
AKT1-overexpressing BMSCs.
In conclusion, the findings of the present study
indicated that ageing increased the expression of AKT1 in
BMSCs and this elevated expression of AKT1 further
exacerbates the ageing process. In addition, ageing BMSCs that
upregulate AKT1 can promote osteoclast differentiation. The
present study provided new insights into the mechanisms of
age-related osteoporosis, as well as potential prevention and
treatment targets.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the funding of the ‘Kunlun
Talent Plateau Famous Doctor’ Talent Project of Qinghai Province in
2022.
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
CL was responsible for writing the original draft,
reviewing and editing, methodology (including design of
experimental protocols and analytical approaches) and investigation
(acquisition of data). XP contributed to core study conception and
design, managed data acquisition, organization, and statistical
analysis/interpretation of key results, revised core content
(conclusion rigor, discussion depth), reviewed and approved the
final publication version, and took responsibility for the work's
accuracy and integrity. BZ performed experimental data validation
for reliability in addition to data archiving and structured
storage. QY focused on experimental quality control (instrument
calibration and reagent validation). BD was responsible for
validation of the data analysis methods and results, as well as
analysis and interpretation of key data. GK was responsible for
validation, reviewing, writing and conception of the study design.
WG was responsible for validation of the data analysis methods and
results, as well as analysis and interpretation of key data. PK was
responsible for validation, reviewing, writing and interpretation
of experimental data. KL was responsible for resources,
supervision, project administration, funding acquisition, and
conception and design of the overall study. BD and WG confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All procedures, including the use of primary BMSCs,
were approved by the Research Ethics Committee of Qinghai
University Affiliated Hospital (approval no. P-SL-202481). The
extraction of primary BMSCs from mice used in the experiment was
approved by the Experimental Animal Ethics Committee of Sichuan
Lilaisinuo Biotechnology Co., Ltd. (approval no. LLSN-2023170).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BMSCs
|
bone mesenchymal stromal cells
|
|
TRAP
|
tartrate-resistant acid
phosphatase
|
|
SASP
|
senescence-associated secretory
phenotype
|
|
MSCs
|
mouse BMSCs
|
|
SA-β-gal
|
senescence-associated
β-galactosidase
|
|
RANK
|
receptor activator of nuclear factor κ
B
|
|
NFATc1
|
nuclear factor of activated T cells,
cytoplasmic 1
|
|
CSE
|
cystathionine γ-lyase
|
|
TRAF6
|
tumor necrosis factor
receptor-associated factor 6
|
|
SHIP1
|
src homology 2 domain-containing
inositol 5-phosphatase 1
|
References
|
1
|
Roux C and Briot K: The crisis of
inadequate treatment in osteoporosis. Lancet Rheumatol.
2:e110–e119. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
International Osteoporosis Foundation
(IOF), . Epidemiology of osteoporosis and fragility fractures. IOF
Official Report. 2024.https://www.osteoporosis.foundation/facts-statistics/epidemiology-of-osteoporosis-and-fragility-fracturesMay
21–2025
|
|
3
|
Salari N, Darvishi N, Bartina Y, Larti M,
Kiaei A, Hemmati M, Shohaimi S and Mohammadi M: Global prevalence
of osteoporosis among the world older adults: A comprehensive
systematic review and meta-analysis. J Orthop Surg Res. 16:6692021.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ogrodnik M: Cellular aging beyond cellular
senescence: Markers of senescence prior to cell cycle arrest in
vitro and in vivo. Aging Cell. 20:e133382021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu Y, Liu X, Ding X, Wang F and Geng X:
Telomere and its role in the aging pathways: Telomere shortening,
cell senescence and mitochondria dysfunction. Biogerontology.
20:1–16. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang F, Cui J, Liu X, Lv B, Liu X, Xie Z
and Yu B: Roles of microRNA-34a targeting SIRT1 in mesenchymal stem
cells. Stem Cell Res Ther. 6:1952015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiang K, Ren M, Liu F, Li Y, He P, Gong X,
Chen T, Wu T, Huang Z, She H, et al: Tobacco toxins trigger bone
marrow mesenchymal stem cells aging by inhibiting mitophagy.
Ecotoxicol Environ Saf. 277:1163922024. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng Y, Wang S, Zhang H, Lee JS, Ni C,
Guo J, Chen E, Wang S, Acharya A, Chang TC, et al: A non-canonical
role for a small nucleolar RNA in ribosome biogenesis and
senescence. Cell. 187:4770–4789.e23. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chicas A, Wang X, Zhang C, McCurrach M,
Zhao Z, Mert O, Dickins RA, Narita M, Zhang M and Lowe SW:
Dissecting the unique role of the retinoblastoma tumor suppressor
during cellular senescence. Cancer Cell. 17:376–387. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu G, Li X, Yang F, Qi J, Shang L, Zhang
H, Li S, Xu F, Li L, Yu H, et al: C-phycocyanin ameliorates the
senescence of mesenchymal stem cells through ZDHHC5-mediated
autophagy via PI3K/AKT/mTOR pathway. Aging Dis. 14:1425–1440.
2023.PubMed/NCBI
|
|
11
|
Wan D, Ai S, Ouyang H and Cheng L:
Activation of 4-1BB signaling in bone marrow stromal cells triggers
bone loss via the p-38 MAPK-DKK1 axis in aged mice. Exp Mol Med.
53:654–666. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao Q, Wang L, Wang S, Huang B, Jing Y and
Su J: Bone marrow mesenchymal stromal cells: Identification,
classification and differentiation. Front Cell Dev Biol.
9:7871182022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Infante A and Rodríguez CI: Osteogenesis
and aging: Lessons from mesenchymal stem cells. Stem Cell Res Ther.
9:2442018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yin Y, Chen H, Wang Y, Zhang L and Wang X:
Roles of extracellular vesicles in the aging microenvironment and
age-related diseases. J Extracell Vesicles. 10:e121542021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tian B, Li X, Li W, Shi Z, He X, Wang S,
Zhu X, Shi N, Li Y, Wan P and Zhu C: CRYAB suppresses ferroptosis
and promotes osteogenic differentiation of human bone marrow stem
cells via binding and stabilizing FTH1. Aging (Albany NY).
16:8965–8979. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gambari L, Grassi F, Roseti L, Grigolo B
and Desando G: Learning from monocyte-macrophage fusion and
multinucleation: Potential therapeutic targets for osteoporosis and
rheumatoid arthritis. Int J Mol Sci. 21:60012020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma QL, Fang L, Jiang N, Zhang L, Wang Y,
Zhang YM and Chen LH: Bone mesenchymal stem cell secretion of
sRANKL/OPG/M-CSF in response to macrophage-mediated inflammatory
response influences osteogenesis on nanostructured Ti surfaces.
Biomaterials. 154:234–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen J, Kuang S, Cen J, Zhang Y, Shen Z,
Qin W, Huang Q, Wang Z, Gao X, Huang F and Lin Z: Multiomics
profiling reveals VDR as a central regulator of mesenchymal stem
cell senescence with a known association with osteoporosis after
high-fat diet exposure. Int J Oral Sci. 16:412024. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li H, Qu J, Zhu H, Wang J, He H, Xie X, Wu
R and Lu Q: CGRP regulates the age-related switch between
osteoblast and adipocyte differentiation. Front Cell Dev Biol.
9:6755032021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benisch P, Jakob F and Ebert R: Effects of
aging, primary osteoporosis and cellular senescence on human
mesenchymal stem cells. PLoS One. 7:e514522012.PubMed/NCBI
|
|
21
|
R Core Team, . R: A language and
environment for statistical computing (version 4.0.3). R Foundation
for Statistical Computing; Vienna, Austria: 2020, https://www.R-project.org/October 16–2023
|
|
22
|
Hu M, Xing L, Zhang L, Liu F, Wang S, Xie
Y, Wang J, Jiang H, Guo J, Li X, et al: NAP1L2 drives mesenchymal
stem cell senescence and suppresses osteogenic differentiation.
Aging Cell. 21:e135512022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang Y, Zhang W, Wang X, Yang J, Cui Y,
Song H, Li W, Li W, Wu L, Du Y, et al: A passage-dependent network
for estimating the in vitro senescence of mesenchymal stromal/stem
cells using microarray, bulk and single cell RNA sequencing. Front
Cell Dev Biol. 11:9986662023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pánczél Á, Nagy SP, Farkas J, Jakus Z,
Győri DS and Mócsai A: Fluorescence-based real-time analysis of
osteoclast development. Front Cell Dev Biol. 9:6579352021.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huo S, Tang X, Chen W, Gan D, Guo H, Yao
Q, Liao R, Huang T, Wu J, Yang J, et al: Epigenetic regulations of
cellular senescence in osteoporosis. Ageing Res Rev. 99:1022352024.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qi L, Fang X, Yan J, Pan C, Ge W, Wang J,
Shen SG, Lin K and Zhang L: Magnesium-containing bioceramics
stimulate exosomal miR-196a-5p secretion to promote senescent
osteogenesis through targeting Hoxa7/MAPK signaling axis. Bioact
Mater. 33:14–29. 2023.PubMed/NCBI
|
|
28
|
Zheng Y, Wu S, Ke H, Peng S and Hu C:
Secretion of IL-6 and IL-8 in the senescence of bone marrow
mesenchymal stem cells is regulated by autophagy via FoxO3a. Exp
Gerontol. 172:1120622023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang W, Huang C, Sun A, Qiao L, Zhang X,
Huang J, Sun X, Yang X and Sun S: Hydrogen alleviates cellular
senescence via regulation of ROS/p53/p21 pathway in bone
marrow-derived mesenchymal stem cells in vivo. Biomed Pharmacother.
106:1126–1134. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tanaka Y, Sonoda S, Yamaza H, Murata S,
Nishida K, Hama S, Kyumoto-Nakamura Y, Uehara N, Nonaka K, Kukita T
and Yamaza T: Suppression of AKT-mTOR signal pathway enhances
osteogenic/dentinogenic capacity of stem cells from apical papilla.
Stem Cell Res Ther. 9:3342018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen G, Wang S, Wei R, Liu Y, Xu T, Liu Z,
Tan Z, Xie Y, Yang D, Liang Z, et al: Circular RNA circ-3626
promotes bone formation by modulating the miR-338-3p/Runx2 axis.
Joint Bone Spine. 91:1056692024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jin J, Huang R, Chang Y and Yi X: Roles
and mechanisms of optineurin in bone metabolism. Biomed
Pharmacother. 172:1162582024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yin S, Lin S, Xu J, Yang G, Chen H and
Jiang X: Dominoes with interlocking consequences triggered by zinc:
Involvement of microelement-stimulated MSC-derived exosomes in
senile osteogenesis and osteoclast dialogue. J Nanobiotechnol.
21:3462023. View Article : Google Scholar
|
|
34
|
Prince EW, Apps JR, Jeang J, Chee K,
Medlin S, Jackson EM, Dudley R, Limbrick D, Naftel R, Johnston J,
et al: Unraveling the complexity of the senescence-associated
secretory phenotype in adamantinomatous craniopharyngioma using
multimodal machine learning analysis. Neuro Oncol. 26:1109–1123.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li J, Wang X, Nepovimova E, Wu Q and Kuca
K: Deoxynivalenol induces cell senescence in RAW264.7 macrophages
via HIF-1α-mediated activation of the p53/p21 pathway. Toxicology.
506:1538682024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tragoonlugkana P, Pruksapong C, Ontong P,
Kamprom W and Supokawej A: Fibronectin and vitronectin alleviate
adipose-derived stem cells senescence during long-term culture
through the AKT/MDM2/P53 pathway. Sci Rep. 14:142422024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bai L and Wang Y: Mesenchymal stem
cells-derived exosomes alleviate senescence of retinal pigment
epithelial cells by activating PI3K/AKT-Nrf2 signaling pathway in
early diabetic retinopathy. Exp Cell Res. 441:1141702024.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Deng Z, Li Y, Wu Y, Yao R, Cao Y,
Wang M, Zhou F, Zhu H and Kang H: Ameliorative effects of
mesenchymal stromal cells on senescence associated phenotypes in
naturally aged rats. J Transl Med. 22:7222024. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Geng N, Xian M, Deng L, Kuang B, Pan Y,
Liu K, Ye Y, Fan M, Bai Z and Guo F: Targeting the
senescence-related genes MAPK12 and FOS to alleviate
osteoarthritis. J Orthop Translat. 47:50–62. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kohli J, Veenstra I and Demaria M: The
struggle of a good friend getting old: Cellular senescence in viral
responses and therapy. EMBO Rep. 22:e522432021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang H, Xu R, Li B, Xin Z, Ling Z, Zhu W,
Li X, Zhang P, Fu Y, Chen J, et al: LncRNA NEAT1 controls the
lineage fates of BMSCs during skeletal aging by impairing
mitochondrial function and pluripotency maintenance. Cell Death
Differ. 29:351–365. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang CY, Le HHT, Tsai HC, Tang CH and Yu
JH: The effect of low-level laser therapy on osteoclast
differentiation: Clinical implications for tooth movement and bone
density. J Dent Sci. 19:1452–1460. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao Y, Wang C, Qiu F, Liu J, Xie Y, Lin
Z, He J and Chen J: Trimethylamine-N-oxide promotes osteoclast
differentiation and oxidative stress by activating NF-κB pathway.
Aging (Albany NY). 16:9251–9263. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xie X, Hu L, Mi B, Panayi AC, Xue H, Hu Y,
Liu G, Chen L, Yan C, Zha K, et al: SHIP1 activator AQX-1125
regulates osteogenesis and osteoclastogenesis through PI3K/Akt and
NF-κb signaling. Front Cell Dev Biol. 10:8260232022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Hu M, Xie J, Li S and Dai L:
Dysregulation of histone modifications in bone marrow mesenchymal
stem cells during skeletal ageing: Roles and therapeutic prospects.
Stem Cell Res Ther. 14:1662023. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen Y, Hong Y, Huang X, Chen J, Li Z, Qiu
J, Liang X, Mai C, Li W, Li X and Zhang Y: ALDH2 regulates
mesenchymal stem cell senescence via modulation of mitochondrial
homeostasis. Free Radic Biol Med. 223:172–183. 2024. View Article : Google Scholar : PubMed/NCBI
|