Introduction
Atrial fibrillation (AF) is the most common
arrhythmia in clinical practice worldwide, and is primarily
attributed to aging, obesity, alcohol use, hypertension, diabetes,
obstructive sleep apnea, valvular heart disease and heart failure
(1). Due to the increasing
morbidity and mortality rates associated with AF, it has become a
serious public health problem worldwide. According to the 2019
Global Burden of Disease Study, there were ~59.7 million cases of
AF worldwide (2), resulting in
~315,000 disease-associated mortalities and 8.39 million
disability-adjusted life-years in 2019 (3).
Research suggests that triggering (abnormal pacing
or ectopic electrical activity) and maintenance (reentry, single or
multiple wavelet hypothesis) disorders promote the occurrence and
development of AF, and atrial remodeling is the pathological basis
(4). Atrial remodeling is
characterized by electrical and structural remodeling, which
manifest as shortened action potential duration and atrial
fibrosis, respectively (5).
Although previous research has explored the molecular mechanism
underlying atrial remodeling, the pathogenesis of AF is not fully
understood, and the efficacy of current therapies for AF is
suboptimal (6). Several non-coding
RNAs have been reported to serve important roles in structural
remodeling, electrical remodeling and autonomic nerve remodeling in
AF, which has opened up novel prospects for the development of
therapies and improving the prognosis of patients with AF (7–9).
Circular RNAs (circRNAs/circs) are single-stranded,
covalently closed non-coding RNAs without 5′end caps or 3′poly(A)
tails. This molecular characteristic makes them more stable than
linear RNAs and ideal biomarkers for diseases (10). Although the diagnostic, prognostic
and potential therapeutic values of circRNAs in other diseases,
such as cancer (11), acute
ischemic stroke (12) and
non-alcoholic fatty liver disease (13), have been intensively researched
over the past decade, their regulatory role in AF remains to be
explored. Previous studies have reported that circRNA expression is
dysregulated in patients with AF (14,15).
Furthermore, several circRNAs have been reported to be involved in
atrial fibrosis. For example, circRNA_010567 and circRNA_000203
have been reported to promote cardiac fibrosis by sponging microRNA
(miRNA/miR)-141 and miR-26b-5p, respectively (16,17).
However, there is still a lack of understanding in terms of the
regulatory function and underlying mechanism of circRNAs in AF.
Our previous study used RNA sequencing to assess
differences in circRNA expression profiles between 7 patients with
AF and 7 patients without AF. The results revealed that 280
circRNAs were differentially expressed with a fold change of
>1.5 (18). As non-valvular AF
is the most common type of AF (96.1% in a national cross-sectional
epidemiological study of China from July 2020 to September 2021)
(19), the present study aimed to
assess the prognostic value of the candidate circRNAs in
non-valvular AF. However, as the availability of atrial tissues
from patients with non-valvular AF is limited (few patients with
non-valvular AF patients ever undergo surgery, making their cardiac
tissue rare for researchers), candidate dysregulated circRNAs in
valvular AF were first determined and, subsequently, their
potential as a prognostic biomarker in non-valvular AF was
assessed.
The present study aimed to evaluate the prognostic
role of hsa_circ_0000118, a upregulated circRNA and plasma-stable
circRNA, in non-valvular AF, and to explore its role in cardiac
fibroblasts and potential action mechanisms. The present study may
hold significance for screening circRNAs with clinical prognostic
value in AF, and elucidating its function and regulatory mechanism
in atrial structural remodeling.
Materials and methods
Subjects and sample collection
The present study was a retrospective study.
Patients with valvular heart disease with and without AF were
consecutively recruited at Southwest Hospital of Army Medical
University (Chongqing, China) between January 2014 and December
2017. In our previous study, a total of 7 pairs of atrial tissues
from age- and sex-matched patients with and without AF were
subjected to Hiseq/Proton RNA sequencing, as described previously
(18). A total of 34 pairs of age-
and sex-matched atrial tissues from patients with and without AF
(average age, 49.2 years; male: 28.6%) were used to validate
candidate circRNA expression by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) between July 2021 and August
2021 (18). Briefly, the inclusion
criteria were as follows: i) Diagnosis of valvular heart disease;
and ii) recent diagnosis of AF (AF group) or no AF (non-AF group).
AF was diagnosed according to the guidelines for the management of
AF from the European Society of Cardiology (20). The exclusion criteria were as
follows: i) Presence of other arrhythmias; and ii) comorbid
cardiovascular disease, including coronary heart disease,
congenital heart disease or myocardial bridge. All atrial tissues
were obtained during valve replacement or valvuloplasty, and were
frozen in liquid nitrogen at −196°C immediately after
resection.
To assess the possibilities of the candidate
dysregulated circRNA as a biomarker in non-valvular AF, venous
blood collected from 221 continuously recruited newly diagnosed
patients with non-valvular AF (average age: 68.3 years; male:
52.5%) at Southwest Hospital of Army Medical University between
January 2014 and December 2017 was used. The following inclusion
criteria were used: i) Recent diagnosis of non-valvular AF [AF was
diagnosed according to the guidelines for the management of AF from
the European Society of Cardiology (20)]; and ii) age ≥18 years. Furthermore,
the exclusion criteria for patients with non-valvular AF included:
i) Diagnosis of structural heart disease; ii) moderate-to-severe
mitral stenosis; iii) malignant tumor; iv) prosthetic valve
replacement; v) sepsis; vi) hyperthyroidism; vii) history of drug
abuse; and viii) undergoing ablation. Venous blood was collected
from patients with AF within 24 h of admission without any
treatment, and the anticoagulant ethylenediaminetetraacetic acid
was added. Subsequently, the collected blood was centrifuged at 400
× g for 15 min at 4°C within 2 h after drawing. The upper plasma
fraction was aspirated and divided into 500 µl aliquots into
cryopreservation tubes for long-term storage in a −80°C
refrigerator. The expression levels of the 12 dysregulated circRNAs
in 29 plasma samples (mean age, 70.3 years, male: 58.6%) and those
of hsa_circ_0000118 in 192 plasma samples (average age: 68.0 years,
male: 51.6%) were examined between September 2021 and October
2021.
The baseline clinical characteristics of patients
were collected from electronic medical records according to the
designed questionnaire, which included age, sex, lifestyle, medical
history, and examination results. The primary end points of the
present study were stroke and all-cause death, and these data were
collected retrospectively by a trained investigator based on
medical records and via telephone. Stroke was defined as an
ischemic event diagnosed by CT or magnetic resonance imaging with
clinical symptoms such as neurological dysfunction lasting for
>24 h. All-cause death was defined as death from any cause. If
the patient died, the cause and time of death were documented.
Written informed consent was obtained from all
included patients. The study protocol, including tissue and blood
sample utilization procedures, was approved by the Ethics Committee
of Southwest Hospital, Army Medical University (approval no.
KY2020231; Chongqing, China). All procedures involving human
participants were performed in accordance with The Declaration of
Helsinki.
circRNA selection
In the present study, the candidate circRNAs were
first selected based on previous RNA sequencing (18), and the screening criteria for
circRNAs were as follows: i) Fold change of >2, with P<0.05
and a false discovery rate of <0.05; ii) counts were >0 in at
least 8 atrial tissue samples for sequencing; and iii) the host
genes of the circRNAs were associated with possible mechanisms of
AF, such as apoptosis, fibrosis, energy metabolism and inflammation
(21), which were identified
through circbank (circbank.cn/) and GeneCards (genecards.org/).
Subsequently, circRNAs that met the inclusion criteria were further
verified to be stable in the plasma of 29 patients with
non-valvular AF by RT-qPCR (Fig.
S1). The candidate circRNA (hsa_circ_0000118) that was stably
expressed in all plasma samples was selected for further assessment
of its expression in 34 pairs of atrial tissues from patients with
and without AF, and the association between its expression in
plasma samples from 192 patients with AF and prognosis.
Cell culture
The mouse cardiac fibroblast cell line (cat. no.
340098) was purchased from BeNa Culture Collection; Beijing Beina
Chunglian Institute of Biotechnology. The cells were cultured in
RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin solution (Beyotime Institute of
Biotechnology). All cells were cultured at 37°C in a 5%
CO2 incubator.
Plasmid construction and cell
transfection
The mouse homologous circRNA of hsa_circ_0000118,
mmu_circ_0010297, was identified using the circBase database
(circbase.org/) and amplified by PCR and subcloned into the pLC
5-ciR vector to form the pLC 5-ciR-C10297 overexpression vector by
Guangzhou Gisai Biotechnology Co., Ltd. The empty plasmid vector
pLC 5-ciR was used as a control. Sanger sequencing was used to
demonstrate whether the plasmid was constructed successfully.
Furthermore, five small interfering RNAs (siRNAs/sis) were
synthesized for mmu_circ_0010297 silencing, along with a
non-targeting siRNA negative control (siNC), and miR-34a-5p mimics
were prepared for overexpression, together with a non-targeting
mimic NC (Shanghai GenePharma Co., Ltd.). The sequences of the
siRNAs and miR-34a-5p mimics are listed in Table SI. The plasmid (2 µg), siRNAs,
mimics (2.5 µg), or NC (all 2.5 µg) were transfected for 6 h at
room temperature into cells using Lipofectamine™ 2000 (Invitrogen™;
Thermo Fisher Scientific, Inc.) in 6-well plates according to the
manufacturer's instructions. RNA was extracted after 48 h of
transfection and protein was extracted after 72 h.
RT-qPCR
Total RNA was extracted from tissues, plasma and
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The Primer Script™ RT Reagent kit with gDNA Eraser (Takara
Biotechnology Co., Ltd.) was used for cDNA synthesis for circRNAs
and mRNAs, while stem-loop RT-qPCR using the miRNA RT Reagent Kit
(Sangon Biotech Co., Ltd.) was used for miRNAs. RT was performed as
follows: 2 min at 42°C followed by holding at 4°C, and 15 min at
37°C followed by 5 sec at 85°C and final holding at 4°C for the
second step, while the stem-loop method utilizes a protocol of 30
min at 16°C, 30 min at 37°C, 5 min at 85°C and subsequent
maintenance at 4°C. Subsequently, cDNA was amplified using
SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.).
The thermocycling conditions consisted of an initial step at 95°C
for 30 sec, followed by 40 cycles of denaturation at 95°C for 5 sec
and annealing/extension at 60°C for 30 sec. All experiments were
performed according to the manufacturer's instructions. The primer
sequences used in the experiments are listed in Tables SII and SIII. The relative expression of genes
was calculated using the 2−ΔΔCq method (22). β-actin was used as the internal
reference for circRNAs and mRNAs, while U6 was used as the internal
reference for miRNAs.
RNase R treatment
circRNA expression was assessed using quantification
of circRNAs and their linear parent genes in RNA samples treated
with and without RNase R enzyme. RNase R enzyme is an exonuclease
that can digest linear RNA molecules without degrading circRNAs due
to their covalently closed circular structure (23). The same batch of total RNA was
equally divided into two groups, in which one group was treated
with 8 U RNase R, 8 ng RNA and 2 µl 10X RNase R buffer, and the
other group was not treated with RNase R. In both groups, RNase
R-free water was added to 20 µl, and samples were digested at 37°C
for 30 min. Subsequently, the stability of both hsa_circ_0000118
(circRNA) and MANIA2 (linear mRNA) was evaluated by RT-qPCR as
aforementioned.
Cell Counting Kit-8 (CCK-8) assay
Cardiac fibroblast cells were seeded at a density of
3,000 cells per well into 96-well cell culture plates. Cell
proliferation was detected on days 1, 2, 3, 4 and 5 after
transfection using CCK-8 reagent (Dojindo Laboratories, Inc.). A
mixture of 100 µl complete medium and 10 µl CCK-8 reagent was added
to each well, and the absorbance of each sample was measured at a
wavelength of 450 nm after incubation for 2 h in an incubator. The
growth curves of cardiac fibroblasts were drawn based on the
average absorbance of the three wells at each time point.
Transwell cell migration assay
After 48 h of transfection, cardiac fibroblasts were
resuspended and diluted to 1×105 cells/ml using FBS-free
medium (RPMI-1640 culture medium, HyClone; Cytiva). A total of 100
µl cell suspension (containing 10,000 cells) was inoculated into
the upper chamber of an insert (8-µm pore size; MilliporeSigma),
and 500 µl medium (1640 culture medium, HyClone; Cytiva) containing
10% FBS (Gibco; Thermo Fisher Scientific, Inc.) was added to the
lower chamber. After incubation at 37°C for 12 h, cells that
migrated were fixed with 100% anhydrous methanol for 30 min at room
temperature, stained with 0.1% crystal violet for 15 min at room
temperature and washed in distilled water. Subsequently, cell
migration was assessed under a fluorescence microscope (ECLIPSE
55I; Nikon) after drying. Cells were manually counted in five
fields of view, selected from four quadrants and the central area
of the chamber.
Western blotting
Total protein was extracted from cells using RIPA
(Beyotime Institute of Biotechnology) containing 1/10 volume of 100
mM PMSF (Beyotime Institute of Biotechnology). The protein
concentration was detected using a bicinchoninic acid protein
concentration determination kit (Beyotime Institute of
Biotechnology). A total of 30 µg denatured protein was loaded and
separated on SDS-PAGE gels (8%), which were prepared according to
the molecular weight of the protein. Subsequently, the proteins
were transferred to a PVDF membrane (Bio-Rad Laboratories, Inc.),
which was blocked with 5% skimmed milk on a shaker at room
temperature for 1 h. Subsequently, the PVDF membrane was placed in
diluted primary antibodies (1:1,000) overnight at 4°C. The primary
antibodies used in the present study included: Collagen type I α1
chain (Col1a1; cat. no. ab270993; Abcam), collagen type III α1
chain (Col3a1; A0817; ABclonal Biotech Co., Ltd.), Smad4 (A5657;
ABclonal Biotech Co., Ltd.) and β-actin (ab8227; Abcam). The next
day, after three 15-min washes in 20 mM PBS with 0.1% Tween 20
(PBST), the PVDF membrane was incubated with secondary antibodies
(HRP-conjugated Goat anti-Rabbit IgG; 1:2,000; cat. no. AS014;
ABclonal Biotech Co., Ltd.) for 1 h at room temperature and then
washed three times with PBST before detection. Equal volumes of
Developer A and B (Super ECL Plus Western Blotting Substrate,
Bioground Biotech Co., Ltd.) were mixed to prepare the working
solution. The washed PVDF membrane was saturated with this solution
and detected in a gel imaging system (Bio-Rad). The densitometric
analysis was performed using ImageJ (version 1.54f; National
Institutes of Health).
Luciferase reporter assay
Using miRanda
(cbio.mskcc.org/miRNA2003/miranda.html), miRDB (mirdb.org/) and
RNA22 (https://cm.jefferson.edu/rna22/) databases to screen
the potential candidate target miRNAs of mmu_circ_0010297.
Wild-type mmu_circ_0010297 sequences that were predicted to bind
miR-34a-5p and the corresponding mutant mmu_circ_0010297 sequences
were constructed and inserted into the pmirGLO Dual-Luciferase
vector (Promega Corporation) by Shanghai GenePharma Co., Ltd.
(Fig. S2). The constructed
plasmids were co-transfected with miR-34a-5p mimics or negative
control mimics into mouse cardiac fibroblasts using Lipofectamine™
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). A
Dual-Luciferase reporter gene detection kit (cat. number: RG027,
Beyotime Institute of Biotechnology) was used to detect the Firefly
and Renilla luciferase activities after transfection for 48
h. The Firefly luciferase activity was normalized to the Renilla
luciferase activity for each sample.
RNA immunoprecipitation (RIP)
The EZ-Magna RIP Kit (MilliporeSigma) was employed
to perform RIP according to the manufacturer's protocol. Briefly,
500 µl cell lysates (cell lysis buffer for Western and IP, Beyotime
Institute of Biotechnology) of cardiac fibroblasts were incubated
with 50 µl protein A/G beads coupled with anti-IgG or
anti-argonaute 2 (5 µg, Ago2, ab186733; Abcam) overnight at 4°C.
The beads were collected using a magnetic stand and washed six
times with RIP Wash Buffer (MilliporeSigma). The RNA extraction was
performed by adding 150 µl of prepared Proteinase K Buffer to the
washed beads, followed by incubation at 55 °C for 30 min with
vortexing every 5 min. After magnetic separation, the supernatant
was transferred to a new tube and mixed with 250 µl of RIP Wash
Buffer. Then, 400 µl of RNA extraction buffer (composed of phenol :
chloroform : isoamyl alcohol in a ratio of 125 : 24 : 1) was added,
and the mixture was vortexed and centrifuged at 19,100 × g for 10
min at room temperature. The upper aqueous phase (350 µl) was
collected, combined with 400 µl chloroform, vortexed, and
centrifuged again under the same conditions. Subsequently, 300 µl
aqueous layer was transferred to a new tube, and RNA was
precipitated by adding 50 µl Salt Solution I (MilliporeSigma), 15
µl of Salt Solution II (MilliporeSigma), 5 µl of Precipitation
Enhancer (MilliporeSigma), and 850 µl of anhydrous ethanol,
followed by overnight storage at −80 °C. The sample was centrifuged
at 19,100 × g for 30 min at 4 °C, washed with 1 ml 80% ethanol, and
centrifuged at 19,100 × g for 15 min at 4°C before air-drying. The
immunoprecipitated RNA was finally dissolved in 10 µl nuclease-free
water and was subjected to RT-qPCR analysis as described
before.
Statistical analysis
All experiments were performed independently with ≥3
biological replicates. Continuous variables are presented as the
mean ± standard deviation or median (interquartile range), and
comparisons between two groups were made using the unpaired t-test
if the data were normally distributed or the Mann-Whitney U test if
the data were not normally distributed. Normality was assessed
using the Shapiro-Wilk test. One-way ANOVA was used for comparisons
among three groups if the data were normally distributed, and the
Sidak's post hoc test was used for correction for multiple
comparisons. The Kruskal Wallis test was used for comparison among
three groups if the data were not normally distributed, and Dunn's
post hoc test was used for multiple comparisons. The results of the
CCK-8 assay were analyzed by two-way ANOVA, and the comparison
between groups was corrected using the Sidak method. Categorical
variables are presented as n (%).
For the prognostic analysis, the relative expression
levels of hsa_circ_0000118 were transformed into binary variables,
and X-tile software version 3.6.1 (Yale School of Medicine) was
used to determine the best cut-off value. Survival analysis was
performed using the Kaplan-Meier method with log-rank test for
comparison. The multivariate Cox proportional hazards model was
used to analyze the association between hsa_circ_0000118 and stroke
and death outcomes by adjusting for potential confounding variables
with P<0.1 in the univariate Cox proportional hazards model.
A two-sided P<0.05 was considered to indicate a
statistically significant difference. All analyses were performed
using SPSS 19.0 (IBM Corp.), R 3.5.2 (The R Foundation for
Statistical Computing) and GraphPad Prism 9.0 (Dotmatics).
Results
Molecular characteristics of
hsa_circ_0000118
A total of 12 dysregulated circRNAs (six upregulated
and six downregulated) were selected based on previous RNA
sequencing results, according to the predefined screening criteria.
To identify the circRNAs that could be the potential biomarkers for
AF prognosis, the expression levels of these 12 candidate circRNAs
were examined in plasma samples from 29 patients with AF.
hsa_circ_0000118 was the only stably expressed circRNA in all 29
plasma samples (Table SIV;
Fig. S1). Subsequently,
hsa_circ_0000118 expression was validated in atrial tissues from 34
patients with AF and 34 patients without AF using RT-qPCR and the
outward primer of hsa_circ_0000118 (Table SII). Consistent with the
sequencing results, it was demonstrated that hsa_circ_0000118
expression was significantly upregulated in atrial tissue of
valvular heart disease patients with AF compared with those without
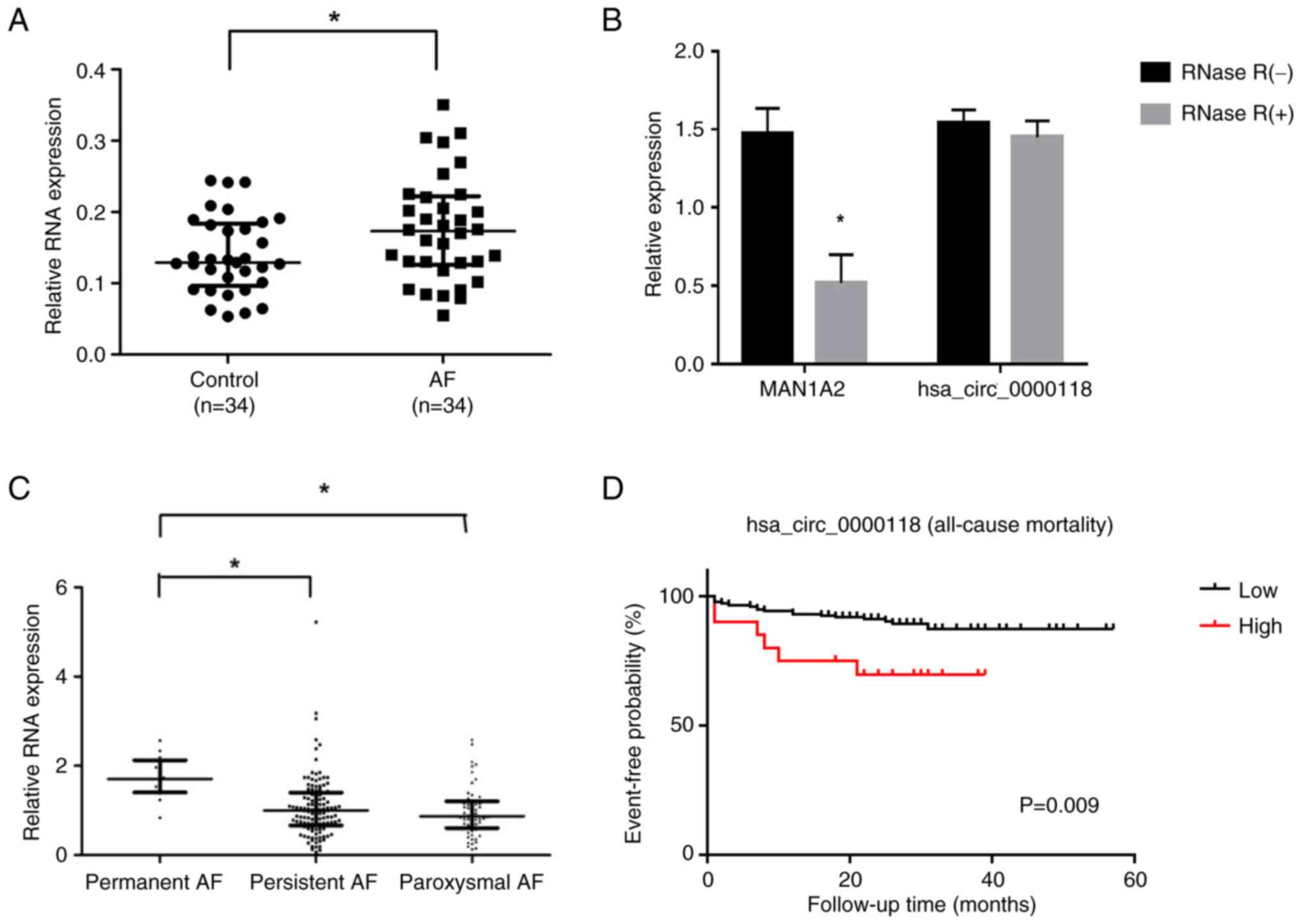
AF (Fig. 1A). Therefore,
hsa_circ_0000118 was selected as the candidate circRNA to assess
its predictive effect in AF prognosis and its underlying molecular
mechanism.
Using circbank, it was demonstrated that
hsa_circ_000018 was derived from exons 2–5 of the mannosidase α
class 1A member 2 (MAN1A2) gene and was located on the sense strand
of chromosome 1 (data not shown). The resistance of
hsa_circ_0000118 to digestion by the RNase R exonuclease confirmed
that it was circular (Fig. 1B).
Furthermore, a mouse homologous circRNA (mmu_circ_0010297) of
hsa_circ_0000118 was identified using the circBase database
(Fig. S3).
Upregulation of hsa_circ_0000118
expression is associated with poor prognosis in AF
To assess the clinical value of hsa_circ_0000118 and
its potential as a prognostic biomarker, the association between
its plasma levels and clinical stages and prognosis of AF was
analyzed in a cohort study. A total of 192 patients with newly
diagnosed AF were enrolled, with a median age of 68 years. A total
of 51.6% of the patients were male, and 15 were lost during
follow-up (Table I). Among the 192
patients with AF, 60 (31.3%) had paroxysmal AF, 120 (62.5%) had
persistent AF and 12 (6.3%) had permanent AF. The plasma levels of
hsa_circ_0000118 were highest in patients with permanent AF, who
had significantly higher levels than patients with persistent and
paroxysmal AF (Fig. 1C). During a
median follow-up of 26 months, the incidence rates of stroke and
all-cause death were 5.93 and 5.76 per 100 person-years,
respectively (data not shown). Patients with higher levels of
hsa_circ_0000118 had a significantly higher mortality rate than
those with lower levels (P=0.009; Fig.
1D). Multivariate Cox proportional hazards regression analysis
did not demonstrate that hsa_circ_0000118 was associated with the
risk of stroke [hazard ratio (HR), 0.69; 95% CI, 0.28–1.68;
P=0.412], whereas elevated levels of hsa_circ_0000118 were
associated with a 5.0-fold increased risk of all-cause death (HR,
5.02; 95% CI, 1.94–12.98; P<0.001; data not shown).
 | Table I.Baseline demographic and clinical
data of 192 patients with AF. |
Table I.
Baseline demographic and clinical
data of 192 patients with AF.
| Characteristic | No. (%) |
|---|
| Age, years |
|
|
<65 | 66 (34.4) |
|
65-74 | 75 (39.1) |
|
≥75 | 51 (26.6) |
| Male | 99 (51.6) |
| Education |
|
| Junior
high school and below | 159 (82.8) |
| High
school and above | 33 (17.2) |
| Family annual per
capita income, 10,000 yuan/year |
|
|
<2.5 | 91 (47.4) |
|
≥2.5 | 101 (52.6) |
| Smoking | 63 (32.8) |
| Drinking | 160 (83.3) |
| BMI,
kg/m2 |
|
|
<18.5 | 14 (7.3) |
|
18.5–23.9 | 96 (50.0) |
|
≥24 | 82 (42.7) |
| Type of AF |
|
|
Paroxysmal AF | 60 (31.3) |
|
Persistent AF | 120 (62.5) |
|
Permanent AF | 12 (6.3) |
| Comorbidities |
|
|
Hypertension | 107 (55.7) |
|
Diabetes | 38 (19.8) |
|
Coronary heart disease | 87 (45.3) |
|
Myocardiopathy | 21 (10.9) |
| Heart
failure | 65 (33.9) |
| TIA or
stroke | 28 (14.6) |
|
Vascular disease | 7 (3.6) |
| Drug therapy |
|
|
Antiarrhythmic drugs | 90 (46.9) |
|
ACEI | 65 (33.9) |
|
ARB | 19 (9.9) |
| β
blockers | 75 (39.1) |
|
Warfarin | 64 (33.3) |
|
Statins | 98 (51.0) |
| Electrocardiogram
parameters |
|
| Left
atrial diameter, mm |
|
|
≤37 | 33 (17.2) |
|
>37 | 159 (82.8) |
| LVEF,
% |
|
|
<50 | 39 (20.3) |
|
≥50 | 153 (79.7) |
|
CHA2DS2-VASc
score ≥2 | 147 (76.6) |
hsa_circ_0000118 promotes the
migration of cardiac fibroblasts and increases the expression
levels of collagen I and collagen III
As hsa_circ_0000118 was demonstrated to be
associated with the prognosis of patients with AF, its effect on
atrial structural remodeling was evaluated, of which the main
manifestation is cardiac fibrosis (6–8).
Furthermore, the effects of mmu_circ_0010297 on proliferation,
migration and the expression levels of extracellular matrix-related
proteins in cardiac fibroblasts were assessed to investigate the
potential role of hsa_circ_0000118 in atrial fibrosis. Gain- and
loss-of-function cell models were established in cardiac
fibroblasts, and 2/5 siRNAs (si3 and si5) were demonstrated to be
effective in knocking down mmu_circ_0010297 expression without
markedly affecting the linear form of MAN1A2 (Figs. 2A and S4). Furthermore, it was revealed that
overexpression of mmu_circ_0010297 did not affect the proliferation
rate of cardiac fibroblasts; however, it significantly promoted
their migration compared with the empty vector group (Fig. 2B and C). Subsequently, the RT-qPCR
results revealed that overexpression of mmu_circ_0010297 had no
marked effect on the mRNA expression levels of Acta2 and Vimentin
(Vim) (markers of the transformation of cardiac fibroblasts into
cardiac myofibroblasts), but significantly increased the expression
levels of collagen I and collagen III (main component of the
extracellular matrix) (Fig. 3A)
(24,25). Compared with those in the control
group, the protein levels of collagen I and collagen III in the
mmu_circ_0010297 overexpression group were also significantly
increased (Fig. 3C). By contrast,
downregulation of mmu_circ_0010297 did not affect the proliferation
rate of the cardiac fibroblasts; however, it significantly
decreased their migration rate (Fig.
2D and E). In addition, knockdown of mmu_circ_0010297 resulted
in significantly decreased expression levels of type I and III
collagen at both the mRNA and protein levels (Fig. 3B and D).
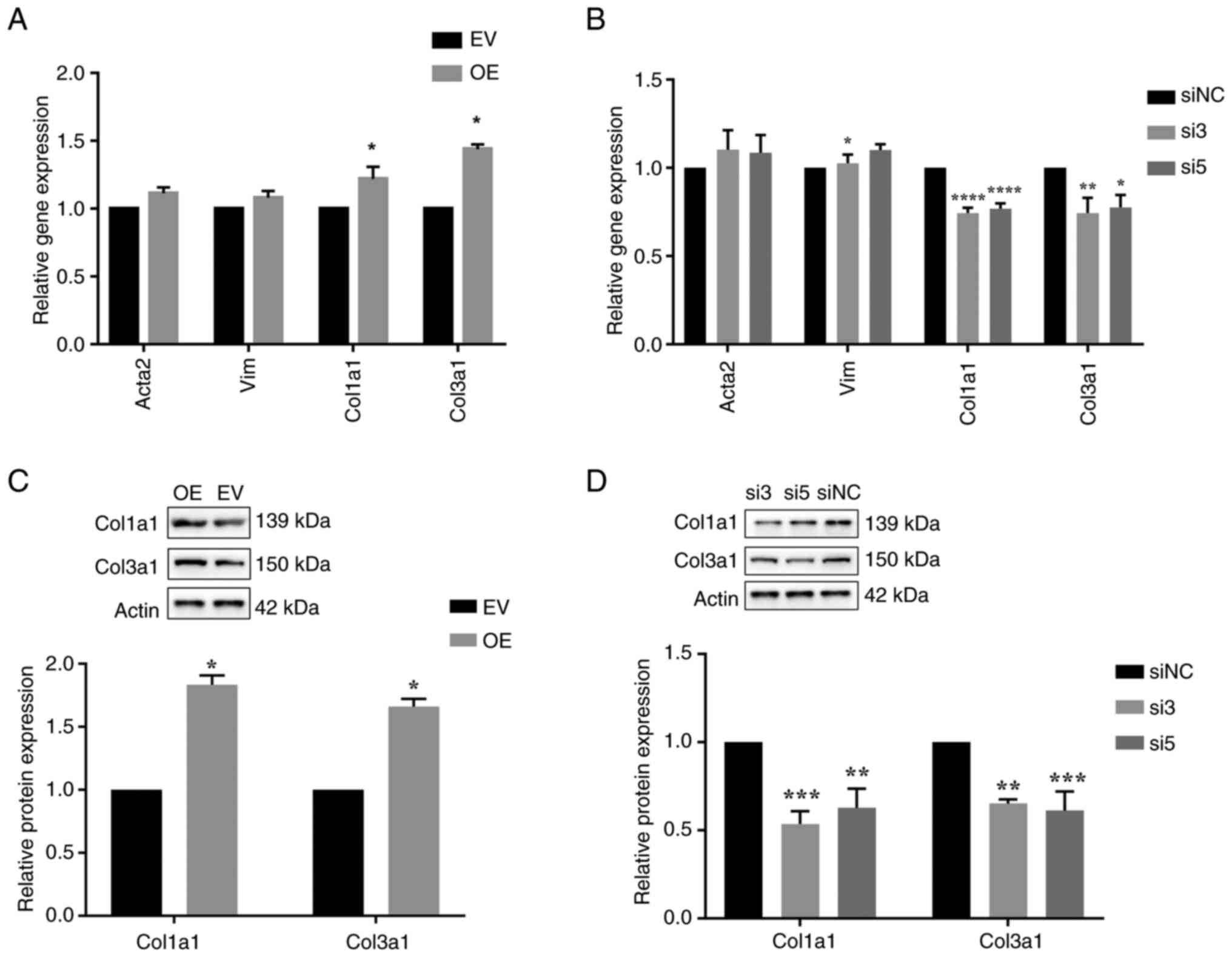 | Figure 3.Effect of mmu_circ_0010297
overexpression and knockdown on differentiation into myofibroblasts
and collagen expression. (A) Relative Acta2, Vim, Col1a1 and Col3a1
mRNA expression of cardiac fibroblasts after transfection with
mmu_circ_0010297 or EV (unpaired t-test). (B) Relative Acta2, Vim,
Col1a1 and Col3a1 mRNA expression of cardiac fibroblasts after
transfection with mmu_circ_0010297 siRNAs or siNC (one-way ANOVA
followed by the Sidak method). (C) Relative Col1a1 and Col3a1
protein expression of cardiac fibroblasts after transfection with
mmu_circ_0010297 OE or EV (unpaired t-test). (D) Relative Col1a1
and Col3a1 protein expression of cardiac fibroblasts after
transfection with mmu_circ_0010297 siRNAs or siNC (one-way ANOVA
followed by the Sidak method). *P<0.05, **P<0.01,
***P<0.001, ****P<0.0001 vs. EV or siNC. circ, circular RNA;
Col1a1, collagen type I α1 chain; Col3a1, collagen type III α1
chain; EV, empty vector; NC, negative control; OE, overexpression;
siRNA/si, small interfering RNA; Vim, Vimentin. |
Taken together, the aforementioned findings
suggested that mmu_circ_0010297 could increase the migration rate
and collagen synthesis of cardiac fibroblasts in atrial
fibrosis.
hsa_circ_0000118 participates in
fibrosis of cardiac fibroblasts by sponging miR-34-5p to regulate
Smad4
The present study assessed whether hsa_circ_0000118
may act as a natural miRNA sponge to prevent miRNAs from binding to
their target mRNAs in atrial fibrosis. Possible candidate target
miRNAs were screened in the miR databases miRanda, miRDB and RNA22,
revealing 10 candidate target miRNAs of mmu_circ_0010297 (data not
shown). Subsequently, the present study evaluated whether the
expression levels of the 10 candidate target miRNAs were altered
when mmu_circ_0010297 was overexpressed or knocked down in cardiac
fibroblasts. The results demonstrated that when mmu_circ_0010297
was overexpressed, 7 out of 10 targeted miRNAs were significantly
downregulated; however, when mmu_circ_0010297 was knocked down,
three miRNAs, miR-34a-5p, miR-6987-5p, and miR-6387, were
significantly upregulated. Notably, only miR-34a-5p and miR-6387
showed significant changes following both overexpression and
knockdown conditions of mmu_circ_0010297 (Fig. 4A and B). The RIP assay further
revealed that, compared with the IgG antibodies, the Ago2
antibodies could coimmunoprecipitate mmu_circ_0010297 and
miR-34a-5p, but not miR-6387 (Fig.
4C). Therefore, mimics of miR-34a-5p were designed, which could
significantly upregulate the expression levels of miR-34a-5p in
mouse cardiac fibroblasts (Fig.
S5). The luciferase reporter assay demonstrated that the
luciferase activity of wild-type mmu_circ_0010297 was significantly
decreased after overexpression of miR-34a-5p, whereas that of
mutant mmu_circ_0010297 did not change, which confirmed that
miR-34a-5p was the target gene of mmu_circ_0010297 (Fig. 4D).
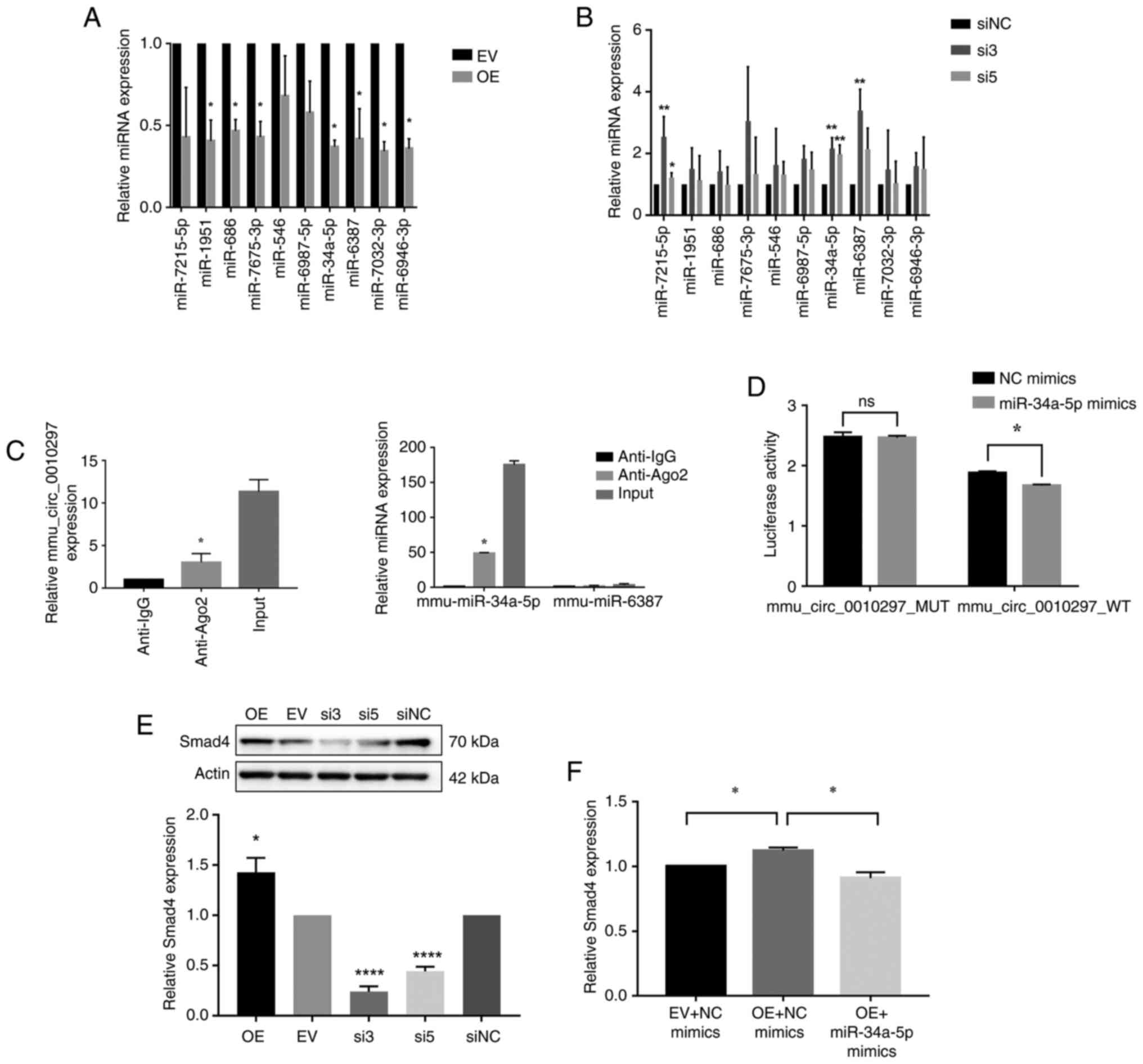 | Figure 4.Examination of the specific mechanism
of mmu_circ_0010297 in regulating collagen production. Expression
of the candidate miRNAs after (A) overexpression of
mmu_circ_0010297 (unpaired t-test) or (B) knockdown of
mmu_circ_0010297 (one-way ANOVA and the Sidak method). (C)
mmu_circ_0010297 and miR-34a-5p levels were determined using
reverse transcription-quantitative PCR after Ago2 or IgG RNA
immunoprecipitation assays (unpaired t-test, as only the Ago2 and
IgG groups were compared). (D) Dual-luciferase reporter assay
(unpaired t-test). (E) Regulatory relationship between Smad4 and
mmu_circ_0010297. For the comparison of OE and EV, an unpaired
t-test was used. For the comparison of si3, si5 and siNC, one-way
ANOVA and the Sidak method were used. (F) Relative expression
levels of Smad4 in cardiac fibroblasts were determined by RT-qPCR
after co-transfection with mmu_circ_0010297 or control EV and
miRNA-34a-5p mimics or NC mimics (one-way ANOVA and the Sidak
method). *P<0.05, **P<0.01, ****P<0.0001 vs. EV, siNC or
Anti-IgG. Ago2, argonaute 2; circ, circular RNA; EV, empty vector;
miRNA/miR, microRNA; MUT, mutant; NC, negative control; OE,
overexpression; si, small interfering RNA; WT, wild-type; RT-q,
reverse transcription-quantitative; ns, not significant. |
Previous studies have reported that Smad4, a
fibrosis-related gene, is the target of miR-34a-5p (26,27).
The present study demonstrated that overexpression of
mmu_circ_0010297 significantly increased the expression levels of
Smad4 compared with those in the empty vector group, while
knockdown of mmu_circ_0010297 had the opposite effect (Fig. 4E). A rescue experiment revealed
that although mmu_circ_irc_0010297 overexpression increased Smad4,
co-overexpression of miR-34a-5p attenuated this effect, which
suggested that mmu_circ_0010297 participated in collagen production
by sponging miR-34a-5p to regulate Smad4 (Fig. 4F).
The aforementioned data indicated that
mmu_circ_0010297 regulated atrial fibrosis by sponging miR-34a-5p
to reverse its inhibitory effect on Smad4, thereby participating in
the structural remodeling in AF.
Discussion
The present study identified that hsa_circ_0000118
expression was significantly upregulated in atrial tissues in AF.
High circulating plasma levels of hsa_circ_0000118 were associated
with poor prognosis of AF. Mechanistically, hsa_circ_0000118 could
promote the migration of cardiac fibroblasts and the expression of
collagen I and collagen III by acting as a competing endogenous RNA
of miR-34a-5p to relieve its suppressive effect on Smad4 in cardiac
fibroblasts.
circRNAs are a class of highly conserved non-coding
RNAs with good stability due to their unique closed loop structure,
making them ideal biomarkers (10,28).
At present, there are few studies on hsa_circ_0000118 or other
MAN1A2-derived circRNA isoforms, and most of them focus on tumors
(29–32). For example, higher hsa_circ_0000118
(also referred to as ‘circMAN1A2’ in the study) levels were
reported to be associated with poor survival in patients with
gastric cancer, and a mechanistic study revealed that
hsa_circ_0000118 could suppress T-cell antitumor immunity by
inhibiting F-box and WD repeat domain containing 11-mediated
splicing factor proline and glutamine rich degradation (29). circMAN1A2 was upregulated in serum
samples of patients with malignant tumors such as nasopharyngeal,
oral, thyroid, ovarian and lung cancer, indicating that circMAN1A2
had good diagnostic value for malignant tumors (32). However, as most studies did not
provide detailed sequence or characterization data for
‘circMAN1A2’, it cannot be conclusively determined if circMAN1A2 is
identical to hsa_circ_0000118 or a distinct MAN1A2 circRNA variant.
Future studies should provide specific characteristic data of
circRNAs when reporting novel circRNAs. Previous studies on
circRNAs in AF have mainly focused on expression profiling and
searching for differentially expressed circRNAs (14,15).
However, the potential of differentially expressed circRNAs as
biomarkers and their function or mechanism in AF lack exploration.
Therefore, to the best our knowledge, the present study was the
first to reveal the prognostic value, regulatory function and
underlying mechanisms of hsa_circ_0000118 in patients with AF,
which may provide novel prospects for the treatment and prognosis
of AF.
Several studies have preliminarily explored the
roles of circRNAs in AF (33,34).
Gao et al (33) reported 92
notably dysregulated circRNAs in the plasma of patients with
persistent AF compared with patients with paroxysmal AF, five of
which were confirmed using RT-qPCR. The circRNA-miRNA-mRNA network
of the five dysregulated circRNAs was further established based on
predictions of miRNA response elements and target genes. Kyoto
Encyclopedia of Genes and Genomes analysis revealed that the five
dysregulated circRNAs were clustered in the mitogen-activated
protein kinase and TGF-β signaling pathways. Another study reported
that 20 circRNAs were upregulated and three were downregulated in
both left and right atrial appendages of patients with AF compared
with in patients without AF (34).
Furthermore, gene set enrichment analysis suggested that
hsa_circ_0000075 and hsa_circ_0082096 were related to AF
pathogenesis via the TGF-β signaling pathway (34). Both studies suggested that circRNAs
were associated with atrial fibrosis in AF; however, the exact
underlying mechanism was not confirmed.
Atrial remodeling is one of the main reasons for the
triggering and maintenance of AF, and its prominent manifestation
is cardiac fibrosis. Cardiac fibrosis is a multifactor-mediated
process characterized by excessive deposition of extracellular
matrix proteins, primarily type I and type III collagen (35). Cardiac fibroblasts are a key cell
type in the heart that can be activated and differentiated into
myofibroblasts during fibrosis, resulting in increased
proliferation, migration and collagen production capacity (25,36).
The results of the present study demonstrated that overexpression
of mmu_circ_0010297 in mouse cardiac fibroblasts did not alter
their proliferation rate but increased their migration rate.
Furthermore, overexpression of mmu_circ_0010297 did not change the
levels of Acta2 and Vim, but increased the expression levels of
Col1a1 and Col3a1, suggesting that mmu_circ_0010297 may affect the
synthesis of type I and III collagen without influencing the
transformation of cardiac fibroblasts into myofibroblasts.
Furthermore, the present study demonstrated that mmu_circ_0010297
could regulate Smad4 by targeting miR-34a-5p, thereby regulating
cardiac fibrosis and participating in atrial structural
remodeling.
However, the present study has several limitations.
First, the sample size used to test the prognostic effect of
hsa_circ_0000118 in AF was relatively small, which may explain why
no association with stroke was demonstrated. In addition, as the
patients with AF were recruited from one tertiary hospital,
selection bias may exist. Furthermore, the association between
hsa_circ_0000118 and poor prognosis was observed in patients with
non-valvular AF. Therefore, these findings cannot be extended to
other types of AF. At the time of the study, commercial human
cardiac fibroblasts were not widely available, and direct
harvesting of fibroblasts from human heart tissues was limited by
the difficulty of obtaining samples; thus, only the effect of the
homologous circRNA hsa_circ_0000118 was explored in mouse cardiac
fibroblasts, which made the extrapolation of the results of the
present study limited by species. Further experiments could employ
alternative human cardiac fibroblast cell lines to perform
luciferase reporter assays for direct verification of the
hsa_circ_0000118-miR-34-5p interaction. Finally, the findings of
the present study need to be further validated in animal models of
AF.
Despite the limitations, the results of the present
study are of importance as they provide a relatively comprehensive
exploration of the specific mechanistic role of hsa_circ_0000118 in
AF. The closed-loop structure of circRNAs makes them promising
disease biomarkers, and the findings demonstrated the prognostic
value of hsa_circ_0000118 in AF. Future studies are needed to
further explore how to quickly and accurately measure circRNA
expression for its clinical translation. By contrast, circRNA-based
vaccines or drugs have been reported to have advantages over
miRNA-based vaccines or drugs due to their stability, and the fact
that the amount of translation required for rolling-loop
translation is lower than for mRNA (37). With the maturity of circRNA
research, it is expected that circRNA-based drugs will be developed
in the future.
In conclusion, to the best of our knowledge, the
present study was the first to demonstrate that high plasma levels
of hsa_circ_0000118 were associated with poor prognosis of AF.
hsa_circ_0000118 was revealed to promote AF development by
increasing the expression levels of collagen I and collagen III,
and the migration of cardiac fibroblasts via the miR-34a-5p/Smad4
axis. The findings of the present study contribute to elucidating
the role and mechanism of hsa_circ_0000118 in AF development.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Chongqing (grant no. CSTB2024NSCQ-MSX0363) and the
National Natural Science Foundation of China (grant no.
82073649).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YL, LZ and ZZ conceived and designed the study. NW,
YZh, YZe, LY, JL, YC, ZY, XC, CL, LW and TC performed experiments.
YZh and YZe wrote the manuscript and confirm the authenticity of
all the raw data. NW, YZh and YZe performed statistical analysis.
NW, TC, YL, LZ and ZZ critically revised the manuscript. All
authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
included patients and the study was approved by the Ethics
Committee of Southwest Hospital, Army Medical University (approval
no. KY2020231; Chongqing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Morin DP, Bernard ML, Madias C, Rogers PA,
Thihalolipavan S and Estes NA III: The state of the art: Atrial
fibrillation epidemiology, prevention, and treatment. Mayo Clin
Proc. 91:1778–1810. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roth GA, Mensah GA, Johnson CO, Addolorato
G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ,
Benziger CP, et al: Global burden of cardiovascular diseases and
risk factors, 1990–2019: Update from the GBD 2019 study. J Am Coll
Cardiol. 76:2982–3021. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu S, Chen Y, Lin R, Huang W, Zhou H, Lin
Y and Xu M: Burden of atrial fibrillation and its attributable risk
factors from 1990 to 2019: An analysis of the global burden of
disease study 2019. Front Cardiovasc Med. 9:9976982022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lau DH, Linz D and Sanders P: New findings
in atrial fibrillation mechanisms. Card Electrophysiol Clin.
11:563–571. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sagris M, Vardas EP, Theofilis P,
Antonopoulos AS, Oikonomou E and Tousoulis D: Atrial Fibrillation:
Pathogenesis, predisposing factors, and genetics. Int J Mol Sci.
23:62021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brundel B, Ai X, Hills MT, Kuipers MF, Lip
GYH and de Groot NMS: Atrial fibrillation. Nat Rev Dis Primers.
8:212022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Šustr F, Stárek Z, Souček M and Novák J:
Non-coding RNAs and cardiac arrhythmias. Adv Exp Med Biol.
1229:287–300. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeng Y, Wu N, Zhang Z, Zhong L, Li G and
Li Y: Non-coding RNA and arrhythmias: Expression, function, and
molecular mechanism. Europace. 25:1296–1308. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Franco D, Aranega A and Dominguez JN:
Non-coding RNAs and atrial fibrillation. Adv Exp Med Biol.
1229:311–325. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang X, Wan M, Min X, Chu G, Luo Y, Han
Z, Li W, Xu R, Luo J, Li W, et al: Circular RNA as biomarkers for
acute ischemic stroke: A systematic review and meta-Analysis. CNS
Neurosci Ther. 29:2086–2100. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zeng Q, Liu CH, Wu D, Jiang W, Zhang N and
Tang H: LncRNA and circRNA in patients with Non-alcoholic fatty
liver disease: A systematic review. Biomolecules. 13:5602023.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu X, Tang X, Chong H, Cao H, Fan F, Pan
J, Wang D and Zhou Q: Expression profiles of circular RNA in human
atrial fibrillation with valvular heart diseases. Front Cardiovasc
Med. 7:5979322020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Ke X, Liu J, Ma X, Liu Y, Liang
D, Wang L, Guo C and Luo Y: Characterization of circRNA-associated
ceRNA networks in patients with nonvalvular persistent atrial
fibrillation. Mol Med Rep. 19:638–650. 2019.PubMed/NCBI
|
|
16
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN,
Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al: CircRNA_000203
enhances the expression of fibrosis-associated genes by
derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac
fibroblasts. Sci Rep. 7:403422017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu N, Li J, Chen X, Xiang Y, Wu L, Li C,
Zhang H, Tong S, Zhong L and Li Y: Identification of long
non-coding RNA and circular RNA expression profiles in atrial
fibrillation. Heart Lung Circ. 29:e157–e167. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shi S, Tang Y, Zhao Q, Yan H, Yu B, Zheng
Q, Li Y, Zheng L, Yuan Y, Zhong J, et al: Prevalence and risk of
atrial fibrillation in China: A national cross-sectional
epidemiological study. Lancet Reg Health West Pac.
23:1004392022.PubMed/NCBI
|
|
20
|
European Heart Rhythm Association and
European Association for Cardio-Thoracic Surgery, . Camm AJ,
Kirchhof P, Lip GY, Schotten U, Savelieva I, Ernst S, Van Gelder
IC, Al-Attar N, et al: Guidelines for the management of atrial
fibrillation: The task force for the management of atrial
fibrillation of the european society of cardiology (ESC). Eur Heart
J. 31:2369–2429. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu Z, Ding L and Yao Y: Atrial
fibrillation: Mechanism and clinical management. Chin Med J (Engl).
136:2668–2676. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tarbit E, Singh I, Peart JN and Rose'Meyer
RB: Biomarkers for the identification of cardiac fibroblast and
myofibroblast cells. Heart Fail Rev. 24:1–15. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tallquist MD: Cardiac fibroblast
diversity. Annu Rev Physiol. 82:63–78. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qi Y, Zhao A, Yang P, Jin L and Hao C:
miR-34a-5p Attenuates EMT through targeting SMAD4 in Silica-induced
pulmonary fibrosis. J Cell Mol Med. 24:12219–12224. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xue F, Yang J, Li Q and Zhou H:
Down-regulation of microRNA-34a-5p promotes trophoblast cell
migration and invasion via targetting Smad4. Biosci Rep.
39:BSR201816312019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ and
Xu RH: Circular RNA: Metabolism, functions and interactions with
proteins. Mol Cancer. 19:1722020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shen Y, Lin J, Jiang T, Shen X, Li Y, Fu
Y, Xu P, Fang L, Chen Z, Huang H, et al: GC-derived exosomal
circMAN1A2 promotes cancer progression and suppresses T-cell
antitumour immunity by inhibiting FBXW11-mediated SFPQ degradation.
J Exp Clin Cancer Res. 44:242025. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guo R, Cui X, Li X, Zang W, Chang M, Sun
Z, Liu Z, Sun Y, Jia J and Li W: CircMAN1A2 is upregulated by
Helicobacter pylori and promotes development of gastric cancer.
Cell Death Dis. 13:4092022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dang QQ, Li PH, Wang J, Zhao JY, Zhai SN,
Zheng YJ and Yang DK: CircMAN1A2 contributes to nasopharyngeal
carcinoma progression via enhancing the ubiquitination of ATMIN
through miR-135a-3p/UBR5 axis. Hum Cell. 36:657–675. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fan CM, Wang JP, Tang YY, Zhao J, He SY,
Xiong F, Guo C, Xiang B, Zhou M, Li XL, et al: circMAN1A2 could
serve as a novel serum biomarker for malignant tumors. Cancer Sci.
110:2180–2188. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao Y, Liu Y, Fu Y, Wang Q, Liu Z, Hu R,
Yang X and Chen M: The potential regulatory role of
hsa_circ_0004104 in the persistency of atrial fibrillation by
promoting cardiac fibrosis via TGF-β pathway. BMC Cardiovasc
Disord. 21:252021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang PP, Sun J and Li W: Genome-wide
profiling reveals atrial Fibrillation-related circular RNAs in
atrial appendages. Gene. 728:1442862020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu M, López de Juan Abad B and Cheng K:
Cardiac fibrosis: Myofibroblast-mediated pathological regulation
and drug delivery strategies. Adv Drug Deliv Rev. 173:504–519.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Souders CA, Bowers SL and Baudino TA:
Cardiac fibroblast: The renaissance cell. Circ Res. 105:1164–1176.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao X, Zhong Y, Wang X, Shen J and An W:
Advances in circular RNA and its applications. Int J Med Sci.
19:975–985. 2022. View Article : Google Scholar : PubMed/NCBI
|















![Effect of mmu_circ_0010297
overexpression and knockdown on the proliferation and migration of
mouse cardiac fibroblasts. (A) Overexpression and knockdown
efficiency of mmu_circ_0010297 in cardiac fibroblasts [unpaired
t-test (left) and one-way ANOVA followed by the Sidak method
(right)]. (B) Growth curves of cardiac fibroblasts after
transfection with mmu_circ_0010297 or EV (two-way ANOVA). (C)
Migration of cardiac fibroblasts after transfection with
mmu_circ_0010297 or EV (unpaired t-test). (D) Growth curves of
cardiac fibroblasts after transfection with mmu_circ_0010297 siRNAs
or siNC (two-way ANOVA). (E) Migration of cardiac fibroblasts after
transfection with mmu_circ_0010297 siRNAs or siNC (magnification,
×100×; one-way ANOVA followed by the Sidak method). *P<0.05,
***P<0.001, ****P<0.0001 vs. EV or siNC). circ, circular RNA;
EV, empty vector; NC, negative control; OD, optical density; OE,
overexpression; siRNA/si, small interfering RNA.](/article_images/mmr/32/6/mmr-32-06-13704-g01.jpg)

