Introduction
The human Pin1 (EC 5.2.1.8) was identified in 1996
by yeast two-hybrid screens as a protein that binds and inhibits
the toxicity of never-in-mitosis A (NIMA), a fungal mitotic kinase
(1). Human Pin1 contains 163 amino
acid residues and consists of a substrate-recognition WW domain in
the N-terminal and a C-terminal catalytic PPIase domain (2).
Subsequent studies found that Pin1 specifically
binds and isomerizes pSer/Thr-Pro motifs in a large and defined
subset of phosphoproteins, which are usually the substrates of
proline-directed protein kinases. Proline-directed phosphorylation
plays an essential role in normal, as well as in malignant, cell
proliferation (3–5).
Functionally, the phosphorylation-dependent
cis/trans-isomerization mediated by Pin1 has a profound
impact on cell events. Such a conformational change regulates the
activities of its substrates including catalysis, protein-protein
interaction, subcellular localization, protein dephosphorylation
and stability (5–7). For instance, Pin1 is abnormally
overexpressed in a range of human cancers, including lung, breast,
colon and prostate cancers, and is considered a biomarker of poor
prognosis (8–12).
Extensive studies on Pin1 have identified over 50
proteins as biological substrates of Pin1. These proteins include
cell cycle-regulated proteins such as Raf-1, Cdc25, cyclin D1,
cyclin E, GTP-binding protein Rab4, anti-apoptotic protein Bcl-2,
transcription factors c-Jun, β-catenin, NF-κB, p53, c-Myc, p73,
c-fos, and Alzheimer's disease-related proteins APP and Tau
(13–18).
Pin1 regulates the cancer suppressor protein p53 via
its WW domain by promoting p53's stability in response to DNA
damage induced by genotoxic drugs, UV light and ionizing radiation
(19,20). Pin1 is capable of interacting with a
number of pSer/Thr-Pro motifs in p53, including pSer33, pSer46,
pThr81 and pSer315 (21–23). Binding of Pin1 mediates a p53
conformational change, which in turn enhances interaction between
p53 and the checkpoint kinase 2 (Chk2) and subsequent p53
phosphorylation at Ser20. Interaction between p53 and Chk2 protects
p53 from Mdm2-mediated ubiquitination, nuclear export and
degradation. Accumulation of p53 in turn enhances its
transcriptional activity towards the cell cycle inhibitor p21. This
elevation eventually leads to a cell cycle checkpoint arrest in
response to DNA damage (24,25).
Therefore, Pin1 and p53 are considered to be
involved in the regulation of cancer progression. In the present
study, we conducted a quantitative investigation on 110 esophageal
cancer specimens and matching normal esophageal tissue to examine
the levels of Pin1 expression in esophageal cancer and normal
esophageal specimens by means of immunohistochemistry and reverse
transcription polymerase chain reaction (RT-PCR) to ascertain the
effect of Pin1 on esophageal cancer pathogenesis and
development.
Materials and methods
Patients and tissues
Prior informed consent for the following studies was
obtained from all patients. A total of 110 cancer specimens and
matching normal esophageal tissues were obtained in consultation
with the surgeon and the pathologist at the Xiamen Hospital of
Traditional Chinese Medicine, Xiamen, China, between September 2004
and November 2008. The tissues were obtained immediately following
surgery. The tissues for RNA isolation were snap-frozen and stored
at −80˚C, and those for immunohistochemistry were fixed with 10%
formalin and embedded in paraffin wax. The clinicopathological
characteristics of the subjects are shown in Table I.
 | Table IClinicopathological characteristics of
110 esophageal cancer patients. |
Table I
Clinicopathological characteristics of
110 esophageal cancer patients.
| Parameters | No. of patients
(N) |
|---|
| Patients | 110 |
| Lymph node |
| Positive | 65 |
| Negative | 45 |
| Differentiation |
| Low | 30 |
| Middle | 69 |
| High | 11 |
| Stage |
| I | 31 |
| II | 40 |
| III + IV | 39 |
| TNM
classification |
| T1 | 8 |
| T2 | 67 |
| T3 | 11 |
| T4 | 24 |
| T1+T2 | 75 |
| T3+T4 | 35 |
| Gender |
| Female | 18 |
| Male | 92 |
| Age |
| <49 | 17 |
| 50–59 | 18 |
| 60–69 | 46 |
| >70 | 29 |
Immunohistochemistry
Sections (4 μm) of formalin-fixed, paraffin-embedded
tissue samples were cut with a microtome and dried overnight at
37˚C on a silanized slide. The protocol for the
avidin-biotin-peroxidase complex method was followed for each
sample. Samples were deparaffinized in xylene at room temperature
for 30 min, rehydrated with graded ethanol and washed in
phosphate-buffered saline (PBS). The samples were then laced in 10
mM citrated buffer (pH 6.0) and boiled in a microwave for 10 min
for epitope retrieval. Endogenous peroxidase activity was quenched
by incubating tissue sections in 3% H2O2 for
10 min. The primary antibodies mouse Pin1 (Abnova Corp., Taipei,
Taiwan) and mouse p53 (Santa Cruz Biotechnology, Santa Cruz, CA,
USA) were used overnight at 4˚C, at dilutions of 1:300 and 1:200,
respectively. The slides were washed and biotinylated goat
anti-mouse secondary antibody (Santa Cruz Biotechnology) was
applied for 1 h. Following rinsing in PBS, the slides were treated
using the peroxidase-labeled Vectastain Elite ABC kit, and the
peroxidase was subsequently developed with a diaminobenzidine kit
and counterstained with hematoxylin. Goat serum was used for the
negative controls instead of the primary antibody for Pin1 and
p53.
Specimens of immunohistochemical staining for Pin1
and p53 were evaluated in a semi-quantitative manner, which
considers the intensity of the staining and the percentage of cells
stained at each intensity. In each case, the intensity (weak,
moderate or strong) and pattern (incomplete or complete) of nuclear
and cytoplasmic staining, and the percentage of neoplastic
immunoreactive cells (cut-off of 10%) were evaluated. Tumors were
scored as: score 0, no appreciable staining or staining in <5%
of neoplastic cells; score 1+, tumors with faint/barely appreciable
incomplete nuclear and cytoplasmic staining in 5–25% of neoplastic
cells; score 2+, tumors with weak to moderate complete nuclear and
cytoplasmic staining or containing 25–50% of neoplastic cells with
moderate incomplete basolateral nuclear and cytoplasmic
immunostaining; score 3+, strong immunoreactivity of the entire
nucleus and cytoplasm in >50% of neoplastic cells or containing
>50% neoplastic cells with strong basolateral incomplete nuclear
and cytoplasmic immunoreactivity. Tumors were classified as 0 or
1+, ‘negative’ and 2+ or 3+, ‘positive’.
Primers
Primer sequences were designed for the qRT assay
using Primer Premier Analysis Software, version 5.0. To avoid
possible amplification of contaminating genomic DNA, primers were
designed so that each PCR product covered at least one intron. The
primer sequences utilized were: Pin1 forward,
5′-TGATCAACGGCTACATCCAG-3′; reverse, 5′-CAAACGAGGCGTCTTCAAAT-3′;
and GAPDH forward, 5′-CATGACAACTTTGGTATCGTG-3′; reverse,
5′-GTGTCGCTGTTGAAGTCAGA-3′. GAPDH was amplified as an internal
reference housekeeping gene for assessing the status of the mRNA
sample.
Real-time RT-PCR assay
Total cellular RNA from tissue specimens was
isolated using a TRIzol reagent (Gibco BRL Life Technologies, Grand
Island, NY, USA) according to the manufacturer's instructions. The
total RNA (3 μg) was reversed transcribed using Moloney murine
leukemia virus reverse transcriptase and random primers (Promega,
Madison, WI, USA).
Real-time PCR was carried out with an Opticon 2
Real-Time Thermocycler (two colors) detection system (MJ Research,
Canada). The PCR reaction solution (25 μl) contained cDNA from 250
ng of total RNA, 1.5 mM MgCl2, 0.2 mM dNTP, 0.6 μM of
each primer, 1 unit Taq polymerase (Takara, Otsu, Shiga, Japan),
0.625 μl 1X SYBR-Green1 (Molecular Probes, Eugene, OR, USA;
dilution 1:1000) and 2.5 μl 10X AmpliTaq buffer for the final
volume. PCR was performed under the following conditions: samples
were initially denatured by heating at 94˚C for 5 min, followed by
40 cycles of denaturation at 94˚C for 30 sec and annealing at 58˚C
for 30 sec for the two genes, and extension at 72˚C for 1 min. Each
assay was performed at least three times to verify the results.
The standard curve for quantifying mRNA copy number
was established by amplifying six aliquots of templates with known
copy numbers (2.1×103–2.1×108 copies). cDNA
was synthesized as follows: RT-PCR on the sample RNA was performed,
electrophoresis was run on a 2% agarose gel, the Pin1 cDNA and
GAPDH cDNA were ligased separately into pCR II-TOPO cloning vectors
(Invitrogen, San Diego, CA, USA), the cDNA clones were transformed
into Escherichia coli DH5-α cells and the cultures were
expanded as previously described (9). Plasmids containing the target gene
were purified and quantified for use in the qRT setup. To confirm
that the inserted PCR product size was correct, plasmids were
digested with specific restriction enzymes, and the cDNA clone PCR
products were then run on gel electrophoresis. Finally, the two
plasmids were sequenced by another biology company (Invitrogen) to
confirm the sequencing results.
Statistical analysis
Statistical analysis was carried out using the Kappa
analysis. Results were considered significant when P<0.05.
Results
This study set out to examine the protein expression
level of Pin1 and p53. A total of 110 esophageal cancer specimens
and matching normal esophageal tissues were analyzed by
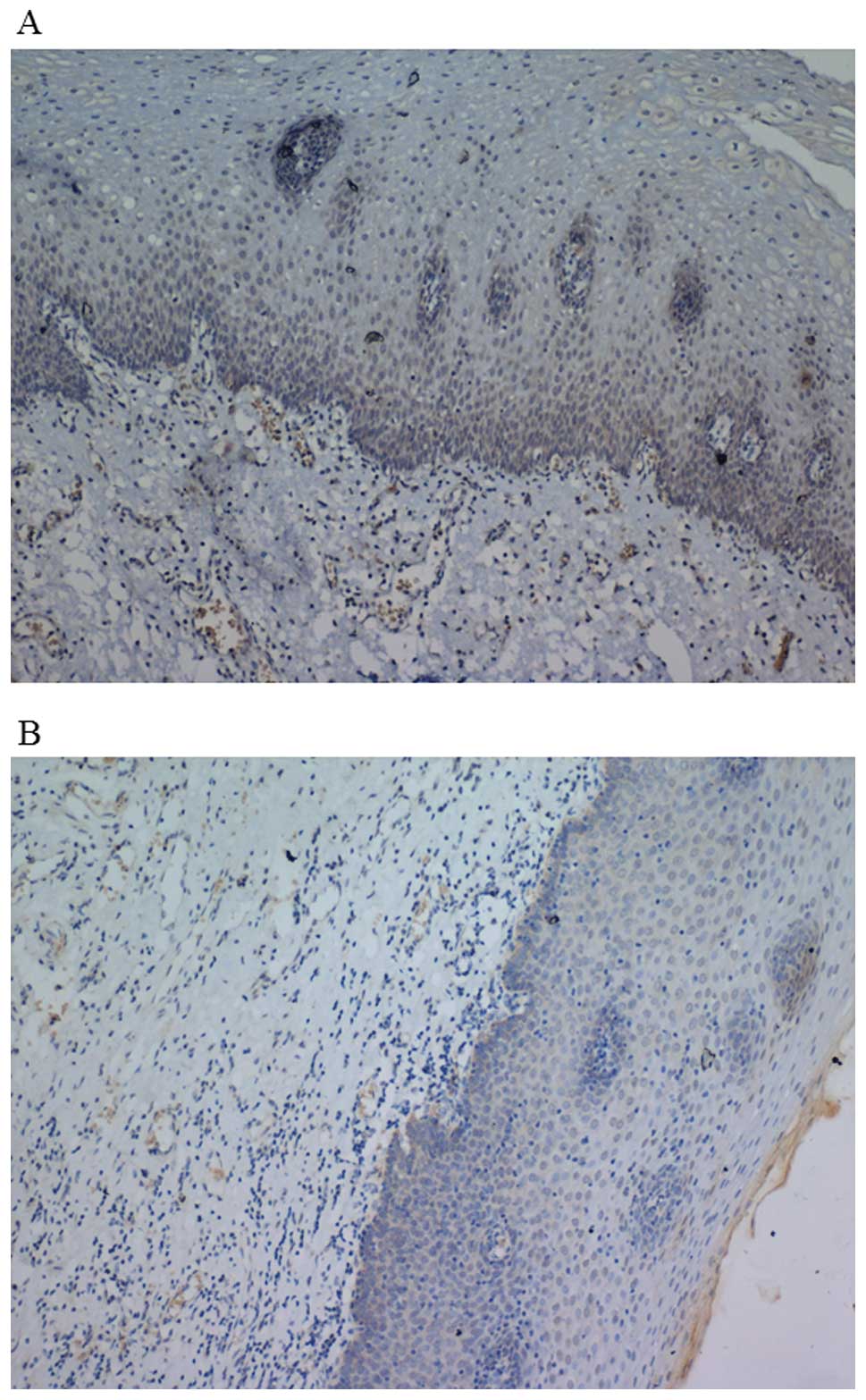
immunohistochemistry. Fig. 1 shows
representative cancer tissue sections stained for the Pin1 and p53
proteins. A number of clinicopathological categories were used to
evaluate the expression of the targets and their potential
correlation with the status of the disease. As shown in Table II, the protein level of Pin1 was
significantly higher in cancer tissue than in normal tissue. A
favorable correlation was found between Pin1 expression and the
stage of the disease. However, there significant correlation was
observed between Pin1 expression and histological type or the level
of differentiation of the cancer. On the other hand, the results
indicated that p53 was expressed more often in cancer tissue than
in normal tissue.
 | Table IIThe correlation between Pin1 and p53
protein expression and clinicopathological characteristics. |
Table II
The correlation between Pin1 and p53
protein expression and clinicopathological characteristics.
| | Pin1 staining
(IHC) | p53 staining
(IHC) |
|---|
| |
|
|
|---|
| Parameters | N | Positive | Negative | P-value | Positive | Negative | P-value |
|---|
| 110 | 74 | 36 | | 63 | | |
| Lymph node |
| Positive | 65 | 54 | 11 | | 40 | 25 | |
| Negative | 45 | 20 | 25 | 0.000 | 23 | 22 | 0.277 |
| Differentiation |
| Low | 30 | 24 | 6 | | 22 | 8 | |
| Middle | 69 | 45 | 24 | | 36 | 33 | |
| High | 11 | 5 | 6 | 0.095 | 5 | 6 | 0.104 |
| Stage |
| I | 31 | 18 | 13 | | 20 | 11 | |
| II | 40 | 34 | 6 | | 22 | 18 | |
| III + IV | 39 | 29 | 10 | 0.038 | 21 | 18 | 0.626 |
| TNM
classification |
| T1 | 8 | 7 | 1 | | 7 | 1 | |
| T2 | 67 | 55 | 12 | | 35 | 32 | |
| T3 | 11 | 8 | 3 | | 7 | 4 | |
| T4 | 24 | 19 | 5 | 0.847 | 14 | 10 | 0.275 |
| T1+T2 | 75 | 62 | 13 | | 42 | 33 | |
| T3+T4 | 35 | 27 | 8 | 0.492 | 21 | 14 | 0.693 |
| Gender |
| Female | 18 | 12 | 6 | | 12 | 6 | |
| Male | 92 | 70 | 22 | 0.484 | 50 | 42 | 0.260 |
| Age |
| <49 | 17 | 12 | 5 | | 11 | 6 | |
| 50–59 | 18 | 12 | 6 | | 11 | 7 | |
| 60–69 | 46 | 28 | 18 | | 26 | 20 | |
| >70 | 29 | 18 | 11 | 0.868 | 15 | 14 | 0.833 |
To further examine the expression pattern of Pin1 in
esophageal cancer tissues, we investigated the mRNA expression of
Pin1 by quantitative real-time-PCR (q-RT). Total cellular RNA from
tissue specimens was extracted for RT-PCR analysis. Among the total
RNA extractions from 110 esophageal cancer specimens, only the RNA
extractions from 40 specimens qualified for further RT-PCR
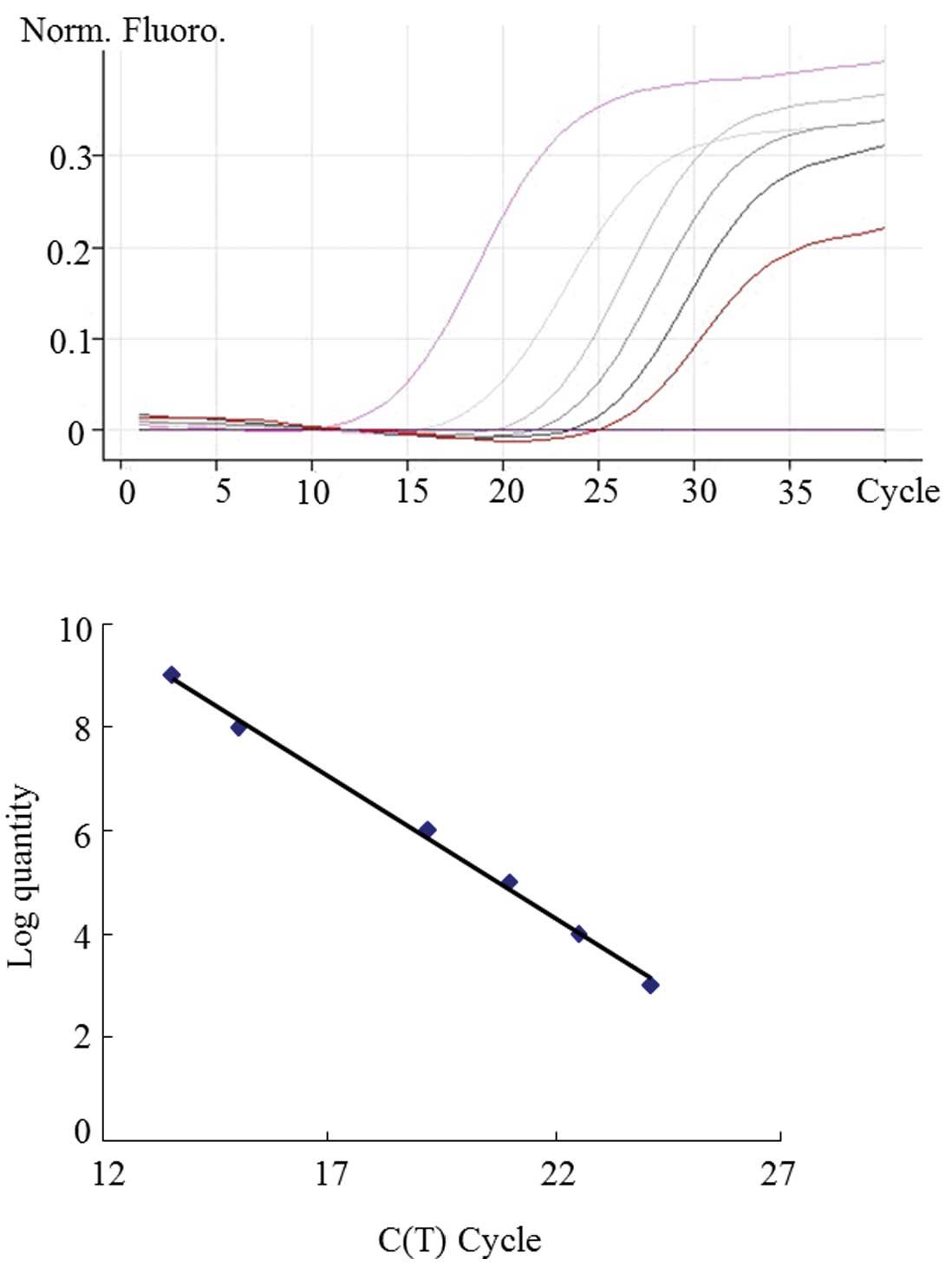
analysis. We generated standard curves using known quantities of
Pin1 prepared and confirmed by sequencing in our laboratory. A
representative graph of the curves obtained from serial dilutions
of Pin1 is shown in Fig. 2. The
Pin1 mRNA was identified as ‘overexpressed’ if, when calibrated
against GAPDH, the tumor had a ≥1.8 expression ratio compared to
normal tissue. The level of mRNA was then compared to various
clinicopathological characteristics. In accordance with the earlier
comparison with the expression protein levels, high-level Pin1 mRNA
was significantly correlated with the presence of lymph node
metastasis and with the stage of disease. No significant
correlation was found between Pin1 mRNA expression and the
histological or differentiation types of the tumors, the size of
the tumor, or between Pin1 expression and the age or gender of the
patient. These results are shown in Table III. As shown in Table IV, no significant correlation
between the level of Pin1 protein expression and Pin1 mRNA
expression.
 | Table IIICorrelation between Pin1 mRNA
expression and clinicopathological characteristics. |
Table III
Correlation between Pin1 mRNA
expression and clinicopathological characteristics.
| | Pin1 mRNA
(q-RT) |
|---|
| |
|
|---|
| Parameters | N | Overexpression
(%) | Underexpression
(%) | P-value |
|---|
| 40 | 16 (40) | 24 (60) | |
| Diff | | | | |
| Low | 13 | 5 | 8 | |
| Middle | 18 | 7 | 11 | |
| High | 9 | 3 | 6 | 0.959 |
| Stage | | | | |
| I | 9 | 2 | 7 | |
| II | 12 | 7 | 5 | |
| III + IV | 19 | 7 | 12 | 0.164 |
| TNM
classification | | | | |
| T1 | 3 | 0 | 3 | |
| T2 | 26 | 8 | 18 | |
| T3 | 5 | 4 | 1 | |
| T4 | 6 | 2 | 4 | 0.099 |
| T1+T2 | 29 | 8 | 21 | |
| T3+T4 | 11 | 6 | 5 | 0.110 |
| Gender | | | | |
| Female | 6 | 3 | 3 | |
| Male | 34 | 11 | 23 | 0.403 |
| Age | | | | |
| <49 | 6 | 2 | 4 | |
| 50–59 | 12 | 5 | 7 | |
| 60–69 | 15 | 6 | 9 | |
| >70 | 7 | 3 | 4 | 0.985 |
 | Table IVCorrelation between Pin1 protein
expression (IHC) and mRNA expression (q-RT). |
Table IV
Correlation between Pin1 protein
expression (IHC) and mRNA expression (q-RT).
| | Pin1 protein
(IHC) |
|---|
| |
|
|---|
| N | Positive | Negative | P-value |
|---|
| Pin1 mRNA
(q-RT) | | | | |
| Positive | 24 | 19 | 5 | |
| Negative | 16 | 12 | 4 | 0.757 |
Discussion
Pin1 has been shown to play a significant role in
numerous steps of oncogenic signaling pathways (24,25).
For example, Pin1 collaborates with Ras signaling to increase the
transcriptional activity of c-Jun towards cyclin D1 (24,25).
Furthermore, Pin1 is involved in the DNA damage response through
modulation of p53 function upon genotoxic stress (24,25).
Pin1 is overexpressed in human breast and oral cancers, and a high
Pin1 expression is correlated with cancer development and poor
prognosis in patients with prostate cancer (24,25).
However, whether or not there is any correlation between Pin1
expression and the clinical outcome of cancer patients remains to
be determined. To address this question, we determined Pin1
expression in 110 specimens using immunohistochemistry, followed by
quantitative real-time-PCR.
This is the first large-scale study of Pin1
expression in clinical samples of esophageal cancer. We have shown
that Pin1 is overexpressed in esophageal cancer, and that its
presence correlates with lymph node involvement and late stage
disease. We also examined tumors for a correlation between Pin1
expression and the expression of p53. In tumors with high levels of
Pin1 expression, there was also a strong likelihood for the tumors
to exhibit high levels of p53 expression. Since Pin1 is
overexpressed in tumors, and since high levels of expression
correlate with a poorer prognosis for patients, Pin1 may have the
potential to be an excellent tumor marker.
Furthermore, an elevated Pin1 expression correlates
with clinical stage. The Pin1 expression level may prove useful
during biopsies, perhaps indicating a need for further treatment
for patients with high Pin1 tumors. The strong correlation between
the Pin1 level and clinical outcome of esophageal cancer suggests
the involvement of Pin1 in the progression of the disease. We have
previously found that Pin1 is activated by oncogenic pathways via
the transcriptional factor E2F, and that Pin1 overexpression
activates multiple steps in oncogenic signaling pathways. No
significant correlation was observed between the level of Pin1
protein expression and Pin1 mRNA expression. Therefore, this result
would have suggested post-transcriptional regulation. Thus, our
study establishes the role of Pin1 as a prognostic maker for
biochemical recurrence in esophageal cancer and suggests that Pin1
is a novel target for treating patients with esophageal cancer.
Acknowledgements
This study was supported by the Scientific Research
Foundation for Returned Overseas Chinese Scholars, the State
Education Ministry (to X.W.) and the Xiamen Municipal Science and
Technology Program (3502Z20064012 to J.J.).
References
|
1
|
Lu KP, Hanes SD and Hunter T: A human
peptidyl-prolyl isomerase essential for regulation of mitosis.
Nature. 380:544–547. 1996. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ranganathan R, Lu KP, Hunter T and Noel
JP: Structural and functional analysis of the mitotic rotamase Pin1
suggests substrate recognition is phosphorylation dependent. Cell.
89:875–886. 1997. View Article : Google Scholar
|
|
3
|
Lu KP: Pinning down cell signaling, cancer
and Alzheimer's disease. Trends Biochem Sci. 29:200–209. 2004.
View Article : Google Scholar
|
|
4
|
Zhou XZ, Lu PJ, Wulf G and Lu KP:
Phosphorylation-dependent prolyl isomerization: a novel signaling
regulatory mechanism. Cell Mol Life Sci. 56:788–806. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim MR, Choi HS, Heo TH, Hwang SW and Kang
KW: Induction of vascular endothelial growth factor by
peptidyl-prolyl isomerase Pin1 in breast cancer cells. Biochem
Biophys Res Commun. 369:547–553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW
and Lu KP: PIN1 is an E2F target gene essential for Neu/Ras-induced
transformation of mammary epithelial cells. Mol Cell Biol.
22:5281–5295. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lu KP, Liou YC and Zhou XZ: Pinning down
proline-directed phosphorylation signaling. Trends Cell Biol.
12:164–172. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wulf GM, Ryo A, Wulf GG, et al: Pin1 is
overexpressed in breast cancer and cooperates with Ras signaling in
increasing the transcriptional activity of c-Jun towards cyclin D1.
EMBO J. 20:3459–3472. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bao L, Kimzey A, Sauter G, Sowadski JM, Lu
KP and Wang DG: Prevalent overexpression of prolyl isomerase Pin1
in human cancers. Am J Pathol. 164:1727–1737. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ayala G, Wang D, Wulf G, et al: The prolyl
isomerase Pin1 is a novel prognostic marker in human prostate
cancer. Cancer Res. 63:6244–6251. 2003.PubMed/NCBI
|
|
11
|
He J, Zhou F, Shao K, et al:
Overexpression of Pin1 in non-small cell lung cancer (NSCLC) and
its correlation with lymph node metastases. Lung Cancer. 56:51–58.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu KP: Prolyl isomerase Pin1 as a
molecular target for cancer diagnostics and therapeutics. Cancer
Cell. 4:175–180. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wulf G, Garg P, Liou YC, Iglehart D and Lu
KP: Modeling breast cancer in vivo and ex vivo reveals an essential
role of Pin1 in tumorigenesis. EMBO J. 23:3397–3407. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lim J and Lu KP: Pinning down
phosphorylated tau and tauopathies. Biochim Biophys Acta.
1739:311–322. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brondani V, Schefer Q, Hamy F and Klimkait
T: The peptidyl-prolyl isomerase Pin1 regulates phospho-Ser77
retinoic acid receptor alpha stability. Biochem Biophys Res Commun.
328:6–13. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liou YC, Ryo A, Huang HK, et al: Loss of
Pin1 function in the mouse causes phenotypes resembling cyclin
D1-null phenotypes. Proc Natl Acad Sci USA. 99:1335–1340. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liou YC, Sun A, Ryo A, et al: Role of the
prolyl isomerase Pin1 in protecting against age-dependent
neurodegeneration. Nature. 424:556–561. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Balastik M, Lim J, Pastorino L and Lu KP:
Pin1 in Alzheimer's disease: multiple substrates, one regulatory
mechanism? Biochim Biophys Acta. 1772:422–429. 2007.
|
|
19
|
Bulavin DV, Saito S, Hollander MC, et al:
Phosphorylation of human p53 by p38 kinase coordinates N-terminal
phosphorylation and apoptosis in response to UV radiation. EMBO J.
18:6845–6854. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Buschmann T, Potapova O, Bar-Shira A, et
al: Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is
important for p53 stabilization and transcriptional activities in
response to stress. Mol Cell Biol. 21:2743–2754. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Radhakrishnan SK and Gartel AL: CDK9
phosphorylates p53 on serine residues 33, 315 and 392. Cell Cycle.
5:519–521. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
D'Orazi G, Cecchinelli B, Bruno T, et al:
Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser
46 and mediates apoptosis. Nat Cell Biol. 4:11–19. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kurihara A, Nagoshi H, Yabuki M, Okuyama
R, Obinata M and Ikawa S: Ser46 phosphorylation of p53 is not
always sufficient to induce apoptosis: multiple mechanisms of
regulation of p53-dependent apoptosis. Genes Cells. 12:853–861.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wulf GM, Liou YC, Ryo A, Lee SW and Lu KP:
Role of Pin1 in the regulation of p53 stability and p21
transactivation, and cell cycle checkpoints in response to DNA
damage. J Biol Chem. 277:47976–47979. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ryo A, Liou YC, Lu KP and Wulf G: Prolyl
isomerase Pin1: a catalyst for oncogenesis and a potential
therapeutic target in cancer. J Cell Sci. 116:773–783. 2003.
View Article : Google Scholar : PubMed/NCBI
|