Introduction
In Japan, one-third of all mortalities are
cancer-related (1). The incidence
of lung, colorectal and breast cancer is increasing in Japan as
well as worldwide (1). Esophageal
carcinoma has a lower incidence than other types of cancer, but
5-fluorouracil (5-FU) and cisplatin (CDDP)-based chemoradiotherapy
results in moderately high response and survival rates relative to
other types of cancer. In fact, the complete response and 5-year
survival rates following 5-FU and CDDP-based chemoradiotherapy have
been reported to be 58 and 29%, respectively, among Japanese
esophageal carcinoma patients (2).
However, chemotherapy remains ineffective in certain patients.
Therefore, identifying the factors that affect sensitivity to 5-FU
and CDDP is necessary for enhancing the clinical outcome of
chemotherapy for esophageal carcinoma.
Certain factors affecting sensitivity to 5-FU or
CDDP have previously been revealed, including the molecular
mechanisms involved in the cellular kinetics and dynamics of 5-FU
and CDDP. For example, overexpression of the ABC transporter
superfamily C5 (ABCC5/MRP5) decreases cellular accumulation of
5-FU, resulting in resistance to 5-FU (3). In addition, dihydropyrimidine
dehydrogenase (DPYD), a 5-FU metabolizing enzyme, has been
correlated with clinical response to 5-FU-based chemotherapy among
colon cancer patients (4,5). The cytotoxic effects of CDDP are also
attenuated by ERCC1, a DNA repair-related enzyme associated with
restoration of DNA damage induced by chemotherapeutic agents or UV
rays (6–8). However, there is little information
concerning whether the levels of these molecules are predictive of
sensitivity to 5-FU or CDDP in esophageal carcinoma.
In the present study, sensitivity to 5-FU and CDDP
and mRNA levels of 35 genes, including drug transporters, DNA
repair enzymes and metabolic enzymes, were evaluated in 5 human
esophageal carcinoma cell lines. Based on these findings, factors
affecting the sensitivity of esophageal carcinoma cells to 5-FU and
CDDP were examined.
Materials and methods
Chemicals
5-FU was obtained from Sigma-Aldrich Chemical Co.
(St. Louis, MO, USA). CDDP was purchased from Wako Pure Chemical
Industries, Ltd. (Osaka, Japan). Gimeracil and MK571 were purchased
from Toronto Research Chemicals, Inc. (Toronto, ON, Canada) and
Cayman Chemical Company (Ann Arbor, MI, USA), respectively.
2-(4-Iodophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium
salt (WST-1) and 1-methoxy-5-methylphenazinium methylsulfate were
purchased from Dojindo Laboratories (Kumamoto, Japan).
Cell culture
The human esophageal adenocarcinoma cell line OE33
was purchased from DS Pharma Biomedical Co., Ltd. (Osaka, Japan)
and the squamous carcinoma cell lines KYSE30, KYSE70, KYSE140 and
KYSE150 (9) were obtained from
Health Science Research Resources Bank (Osaka, Japan). OE33 and the
other cell lines were maintained in RPMI-1640 medium (Invitrogen
Corp., Carlsbad, CA, USA) and Dulbecco’s modified Eagle’s medium
(Invitrogen), respectively, supplemented with 10% heat-inactivated
fetal bovine serum (lot no. 1335770 and 348777, Invitrogen). Cells
were cultured in an atmosphere of 95% air and 5% CO2 at
37°C and subcultured every 3 or 4 days at a density of
1×106 cells/25 cm2 culture flask. The number
of passages for OE33, KYSE30, KYSE70, KYSE140 and KYSE150 cells was
15–25, 15–28, 15–26, 21–31 and 19–31, respectively.
Growth rate of esophageal carcinoma cell
lines
The growth rate of esophageal carcinoma cells was
evaluated with a WST-1 assay utilizing succinate dehydrogenase
activity. Cells were seeded onto a 96-well plate (Corning Inc.,
Corning, NY, USA) at a density of 5×103 cells/well/100
μl and cultured in an atmosphere of 95% air and 5%
CO2 at 37°C. After 0, 6, 12, 18, 24, 36, 48, 72 and 96
h, the culture medium was exchanged for 110 μl of medium
containing WST-1 reagent solution (10 μl WST-1 solution and
100 μl culture medium), and 3 h later the absorbance was
determined using a micro-plate reader at 450 nm with a reference
wavelength of 620 nm (SpectraFluor™, Tecan Group Ltd., Männedorf,
Switzerland). The doubling time for cell growth was calculated from
the logarithmic phase of a growth curve (10) as follows: Doubling time =
(t1 - t0) ×
log102/(log10N1 -
log10N0). N0 and N1 are
the number of cells (% of day 0) at t1 and
t0, respectively.
Growth inhibitory activity assay
Cells were seeded onto 96-well plates (Corning Inc.)
at a density of 5×103 cells/well/100 μl on day 0.
After incubation for 24 h, the culture medium was exchanged for one
containing 5-FU or CDDP at various concentrations (day 1). On day
4, a WST-1 assay was performed as described above.
The effects of gimeracil and MK571 on the growth
inhibitory effects of 5-FU and CDDP were also evaluated by WST-1
assay. Cells were incubated for 24 h as described above and the
culture medium was exchanged for one containing 5-FU or CDDP at
various concentrations with or without gimeracil (100 μM) or
MK571 (50 μM). Following incubation for 72 h at 37°C, the
culture medium was replaced with a medium containing WST-1 and the
absorbance was measured.
The 50% growth inhibitory concentrations
(IC50) were calculated according to the sigmoid
inhibitory effect model: E = Emax × [1 -
Cγ/(Cγ + IC50γ)], using
the nonlinear least-squares fitting method (Solver,
Microsoft® Excel). E and Emax represent the
surviving fraction (% of control) and its maximum, respectively. C
and γ are the drug concentration in the medium and the sigmoidicity
factor, respectively. Relative sensitivity was calculated as
follows: Relative sensitivity = IC50 (without gimeracil
or MK571)/IC50 (with gimeracil or MK571).
Real-time reverse transcription
(RT)-PCR
The mRNA expression levels were measured by
real-time RT-PCR. Cells were seeded at a density of
2×106 cells/60 mm culture dish and 48 h later, total RNA
was extracted from the cells with a GenEluteTM Mammalian Total RNA
Miniprep kit (Sigma-Aldrich). Total RNA (1 μg) was used for
RT with a PrimeScriptTM RT reagent kit (Takara Bio, Inc., Shiga,
Japan) and a thermal cycler (i-Cycler, Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The RT reaction was conducted in 40 μl
reaction buffer at 37°C for 15 min and terminated by heating at
85°C for 5 sec followed by cooling at 4°C.
Real-time PCR was performed with a 7500 Real-time
PCR system (Applied Biosystems, Carlsbad, CA, USA) and SYBR Premix
Ex Taq™ (Takara Bio, Inc.). The primer sequences are shown in
Table I. PCR was performed at 95°C
for 10 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34
sec. Dissociation was initiated at 95°C for 15 sec followed by 60°C
for 1 min and 95°C for 15 sec. To compare the relative expression
of target mRNA levels between the cell lines, the comparative Ct
method was used, as previously described (10); β-actin (ACTB) was used as an
internal standard. Samples were prepared in duplicate and three
independent sample sets were analyzed.
 | Table I.Sequences of oligonucleotide primers
designed for real-time PCR. |
Table I.
Sequences of oligonucleotide primers
designed for real-time PCR.
| Function and
gene | Forward
(5′–3′) | Reverse
(5′–3′) | Reference |
|---|
| ACTB |
TCATGAAGTGTGACGTGGACATC |
TGCATCCTGTCGGCAATG | 10 |
| Transport | | | |
| SLC22A1 |
TCTTCCATCGTCACTGAGTTCAAC |
AGAAGCCCGCATTCAAACAG | 10 |
| SLC22A2 |
TCTACTCTGCCCTGGTTGAATTC |
ATGCAGCCCAAGGGTAACG | 10 |
| SLC22A3 |
TAGCCCCATTTCTGCTCTTTC |
AGATGGATGCCAGGATACCAA | 10 |
| SLC23A2 |
TCTTTGTGCTTGGATTTTCGAT |
ACGTTCAACACTTGATCGATTC | 23 |
| SLC31A1 |
ACAAGTCAGCATTCGCTACAATTC |
TTGCAGGAGGTGAGGAAAGC | 9 |
| ABCB1 |
TTCCTTCACCCAGGCAATG |
ATGAGTTTATGTGCCACCAAGTAG | a |
| ABCC1 |
CAGTGACCTCTGGTCCTTAAACAA |
TTGGCGCATTCCTTCTTCC | 24 |
| ABCC2 |
ACTTGTGACATCGGTAGCATGGA |
AAGAGGCAGTTTGTGAGGGATGA | a |
| ABCC3 |
GTCCGCAGAATGGACTTGAT |
TCACCACTTGGGGATCATTT | 25 |
| ABCC4 |
GCTCAGGTTGCCTATGTGCT |
CGGTTACATTTCCTCCTCCA | 25 |
| ABCC5 |
CGAAGGGTTGTGTGGATCTT |
GTTTCACCATGAAGGCTGGT | a |
| ABCC6 |
TGTCGCTCTTTGGAAAATCC |
AGGAACACTGCGAAGCTCAT | 25 |
| ABCG2 |
TGACGGTGAGAGAAAACTTAC |
TGCCACTTTATCCAGACCT | 26 |
| ATP7A |
AGATACTGGGACACTGGAGAAA |
AGGTCATCCCTTCCACTTTCA | 10 |
| ATP7B |
TGATTTATAACCTGGTTGGGATACC |
ATGAGAGCACCACAGACACAGA | 10 |
| DNA repair | | | |
| ERCC1 |
TACAAGGCCTATGAGCAGAAACCA |
TCTCTTGATGCGGCGATGAG | a |
| ERCC2 |
CTGGAGGTGACCAAACTCATCTA |
CCTGCTTCTCATAGAAGTTGAGC | 27 |
| ERCC3 |
TATCCCAGGACACACAGGAAAT |
TCACCTTGAAGCTATAACCTTGA | a |
| XPA |
TGCGGCGAGCAGTAAGAAG |
TCATGGCCACACATAGTACAAGTC | a |
| Rad51 |
TGGGAACTGCAACTCATCTGG |
GCGCTCCTCTCTCCAGCAG | 28 |
| BRCA1 |
ACAGCTGTGTGGTGCTTCTGTG |
CATTGTCCTCTGTCCAGGCATC | 29 |
| BRCA2 |
TGAAGAGCAGTTAAGAGCCTTGAA |
ACGGTTGTGACATCCCTTGATAAA | a |
| HMGB1 |
CAAGCGAACAGCAGGGTTAG |
CAGATTGAGTCATTTGCTCCTCTTA | a |
| HMGB2 |
TGAACATCGCCCAAAGATCA |
TCAGACCACATTTCACCCAATT | a |
| MLH1 |
GATTACCCCTTCTGATTGACA |
ACTGAGGCTTTCAAAACA | 30 |
| MSH2 |
CAGTATATTGGAGAATCGCA |
AGGGCATTTGTTTCACC | 30 |
| PMS2 |
AGTCAGCGTGCAGCAGTTATT |
GACCATTTTGGCATACTCCTTCT | a |
| RPP25 |
AGAATGGTGGACAGTGGGATT |
TACTTCAGGTGCTCTTCGTGAATG | a |
| Metabolism | | | |
| GSTP1 |
CTGCGCATGCTGCTGGCAGATC |
TTGGACTGGTACAGGGTGAGGTC | 31 |
| GCLC |
GGCAAGATACCTTTATGACCAGTT |
TGCAGCACTCAAAGCCATAA | 32 |
| GCLM |
TGACTGCATTTGCTAAACAATTTGA |
CGTGCGCTTGAATGTCAGG | 33 |
| TYSM |
GCCTCGGTGTGCCTTTCA |
CCCGTGATGTGCGCAAT | 34 |
| DPYD |
AATGATTCGAAGAGCTTTTGAAGC |
GTTCCCCGGATGATTCTGG | 35 |
| UMPS |
TAGTGTTTTGGAAACTGTTGAGGTT |
CTTGCCTCCCTGCTCTCTGT | 36 |
| MTHFR |
CGGGTTAATTACCACCTTGTCAA |
GCATTCGGCTGCAGTTCA | 36 |
Statistical analyses
Data are shown as the mean ± standard deviation
(SD). Comparisons between 2 and among 3 or more groups were
performed with Student’s unpaired t-test and repeated one-way
analysis of variance (ANOVA) followed by Scheffe’s F test,
respectively. P<0.05 (two-tailed) was considered to indicate a
statistically significant result. The correlation analysis was
performed using Pearson’s correlation coefficient (r).
Results
Growth rates of esophageal carcinoma cell
lines
Table II shows the
cell growth doubling times for the 5 esophageal carcinoma cell
lines. Doubling times for the cells varied from 20 to 25 h,
revealing a significant difference between lines. KYSE30 cells
(20.1±1.41 h) had the shortest doubling time and OE33 cells
(25.0±0.90 h) the longest.
 | Table II.Doubling times of esophageal
carcinoma cell lines. |
Table II.
Doubling times of esophageal
carcinoma cell lines.
| Cell line | Doubling time, mean
± SD (h) |
|---|
| OE33 | 25.0±0.90 |
| KYSE30 | 20.1±1.41 |
| KYSE70 | 21.8±0.51 |
| KYSE140 | 23.3±1.07 |
| KYSE150 | 20.6±0.53 |
Sensitivity of esophageal carcinoma cell
lines to 5-FU and CDDP
The IC50 values for 5-FU were markedly
different among the cell lines (0.524–30.2 μM); the OE33
cells showed the highest sensitivity to 5-FU and the KYSE30 cells
the lowest sensitivity (Table III).
In the case of CDDP, the IC50 values were also
substantially different among the cell lines (2.17–19.5 μM).
The rank order of sensitivity to CDDP was comparable to that for
5-FU.
 | Table III.IC50 values for 5-FU and
CDDP in esophageal carcinoma cell lines. |
Table III.
IC50 values for 5-FU and
CDDP in esophageal carcinoma cell lines.
| IC50
value, mean ± SD (μM)
|
|---|
| Cell line | 5-FU | CDDP |
|---|
| OE33 | 0.524±0.08 | 2.17±0.33 |
| KYSE30 | 30.2±8.29 | 19.5±3.67 |
| KYSE70 | 13.1±13.3 | 5.27±0.36 |
| KYSE140 | 1.88±0.38 | 3.09±0.67 |
| KYSE150 | 4.75±1.46 | 14.0±1.02 |
Correlation analysis of factors affecting
drug sensitivity
The level of mRNA expression differed among the
esophageal carcinoma cell lines (Table
IV). The correlations between the IC50 values and
the mRNA levels of the 35 different genes were analyzed (Table V). SLC22A3 mRNA was not detected in
any cells, with the exception of the OE33 cell line. ABCC6 mRNA
expression was not observed in KYSE30 and KYSE70 cells.
 | Table IV.Expression levels of mRNA in
esophageal carcinoma cell lines. |
Table IV.
Expression levels of mRNA in
esophageal carcinoma cell lines.
| Expression ratio,
mean ± SD (2−ΔCt×10−4)
|
|---|
| Function and
gene | OE33 | KYSE30 | KYSE70 | KYSE140 | KYSE150 |
|---|
| Transport | | | | | |
| SLC22A1 | 0.11±0.03 | 0.07±0.02 | 0.01±0.003 | 0.03±0.02 | 0.12±0.07 |
| SLC22A2 | 0.49 ±0.15 | 0.16±0.03 | 0.23±0.03 | 0.87±0.45 | 0.47±0.36 |
| SLC22A3 | 36.3±9.24 | ND | ND | ND | ND |
| SLC23A2 | 76.1±13.8 | 36.6±6.39 | 59.8±4.66 | 61.1±43.8 | 92.9±64.0 |
| SLC31A1 | 125±26.6 | 131±10.2 | 179±23.4 | 252±135 | 244±147 |
| ABCB1 | 0.53±0.14 | 0.16±0.03 | 0.25±0.06 | 0.54±0.20 | 0.79±0.59 |
| ABCC1 | 74.7±11.3 | 37.0±3.83 | 246±32.9 | 123±75.6 | 67.8±36.6 |
| ABCC2 | 0.05±0.01 | 2.57±0.89 | 1.36±0.07 | 0.38±0.16 | 0.38±0.23 |
| ABCC3 | 80.5±15.1 | 10.2±2.64 | 60.7±8.62 | 40.6±23.3 | 123±86.7 |
| ABCC4 | 26.1±2.17 | 28.9±1.95 | 52.1±5.53 | 189±105 | 92.5±51.3 |
| ABCC5 | 9.06±1.30 | 62.14±17.0 | 65.38±8.60 | 22.92±10.6 | 26.76±19.2 |
| ABCC6 | 1.79±0.12 | ND | ND | 0.05±0.05 | 0.04±0.04 |
| ABCG2 | 6.25±1.29 | 4.64±0.21 | 1.66±0.34 | 2.47±0.69 | 33.1±18.3 |
| ATP7A | 8.66±1.27 | 7.99±0.70 | 4.68±1.20 | 6.09±3.61 | 15.4±7.98 |
| ATP7B | 1.91±0.25 | 1.89±0.73 | 2.21±0.47 | 5.72±4.77 | 3.37±2.69 |
| DNA repair | | | | | |
| ERCC1 | 219±66.1 | 143±35.8 | 96.3±13.2 | 241±131 | 296±175 |
| ERCC2 | 43.9±4.57 | 35.1±8.01 | 21.0±2.52 | 47.9±22.4 | 52.2±22.6 |
| ERCC3 | 82.3±11.5 | 79.3±19.4 | 57.4±6.31 | 134±97.9 | 185±104 |
| XPA | 91.4±16.0 | 107±16.7 | 193±14.8 | 461±290 | 283±164 |
| Rad51 | 3.84±1.03 | 2.64±0.37 | 3.67±1.09 | 6.09±3.07 | 4.72±1.75 |
| BRCA1 | 90.5±15.6 | 61.1±2.46 | 65.3±4.56 | 188±116 | 151±78.8 |
| BRCA2 | 110±20.4 | 111±3.99 | 43.5±3.74 | 261±165 | 204±108 |
| HMGB1 | 35.2±7.29 | 35.9±1.84 | 51.4±3.98 | 79.9±47.8 | 37.6±19.1 |
| HMGB2 | 509±87.3 | 1340±150 | 1343±129 | 1980±947 | 1367±679 |
| MLH1 | 27.5±4.55 | 21.3±1.61 | 23.9±1.83 | 28.2±18.4 | 70.9±36.8 |
| MSH2 | 185±39.8 | 540±38.6 | 331±23.5 | 338±154 | 272±114 |
| PMS2 | 25.3±3.22 | 34.2±4.24 | 77.1±12.8 | 123±81.6 | 67.0±42.8 |
| RPP25 | 27.5±4.35 | 6.32±0.99 | 0.13±0.03 | 74.9±59.9 | 0.74±0.47 |
| Metabolism | | | | | |
| GSTP1 | 2444 ±425 | 2926±644 | 3421±380 | 5784±3549 | 7249±3978 |
| GCLC | 5.21±0.51 | 3.87±1.15 | 45.0±4.30 | 10.8±4.41 | 9.05±5.39 |
| GCLM | 8.38±2.61 | 31.34±4.30 | 75.1±10.8 | 33.0±15.4 | 32.1±22.2 |
| TYMS | 54.0±11.6 | 163±3.10 | 2511±136 | 81.8±32.9 | 215.6±102 |
| DPYD | 5.81±2.03 | 62.4±6.50 | 0.82±0.29 | 1.23±0.87 | 12.4±9.82 |
| UMPS | 85.6±17.4 | 69.0±3.85 | 162±20.6 | 183±108 | 165±73.2 |
| MTHFR | 5.78±1.85 | 10.0±2.39 | 18.6±2.72 | 25.6±23.2 | 23.7±15.3 |
 | Table V.Pearson’s correlation coefficient
between IC50 values for 5-FU or CDDP and mRNA expression
level. |
Table V.
Pearson’s correlation coefficient
between IC50 values for 5-FU or CDDP and mRNA expression
level.
| Pearson’s
correlation coefficient (r)
|
|---|
| Function and
gene | 5-FU | CDDP |
|---|
| Transport | | |
| SLC22A1 | −0.189 | 0.333 |
| SLC22A2 | −0.764 | −0.574 |
| SLC22A3 | ND | ND |
| SLC23A2 | −0.790 | −0.302 |
| SLC31A1 | −0.477 | −0.132 |
| ABCB1 | −0.788 | −0.215 |
| ABCC1 | −0.150 | −0.530 |
| ABCC2 | 0.992b | 0.706 |
| ABCC3 | −0.659 | −0.179 |
| ABCC4 | −0.470 | −0.315 |
| ABCC5 | 0.573 | 0.234 |
| ABCC6 | ND | ND |
| ABCG2 | −0.244 | 0.398 |
| ATP7A | −0.199 | 0.451 |
| ATP7B | −0.485 | −0.314 |
| DNA repair | | |
| ERCC1 | −0.638 | −0.041 |
| ERCC2 | −0.533 | 0.019 |
| ERCC3 | −0.439 | 0.187 |
| XPA | −0.463 | −0.284 |
| Rad51 | −0.756 | −0.523 |
| BRCA1 | −0.653 | −0.274 |
| BRCA2 | −0.455 | −0.049 |
| HMGB1 | −0.341 | −0.507 |
| HMGB2 | 0.010 | 0.125 |
| MLH1 | −0.369 | 0.269 |
| MSH2 | 0.913a | 0.719 |
| PMS2 | −0.363 | −0.365 |
| RPP25 | −0.486 | −0.561 |
| Metabolism | | |
| GSTP1 | −0.401 | 0.121 |
| GCLC | 0.032 | −0.321 |
| GCLM | 0.287 | −0.011 |
| TYMS | 0.163 | −0.211 |
| DPYD | 0.881 | 0.863 |
| UMPS | −0.522 | −0.379 |
| MTHFR | −0.319 | −0.074 |
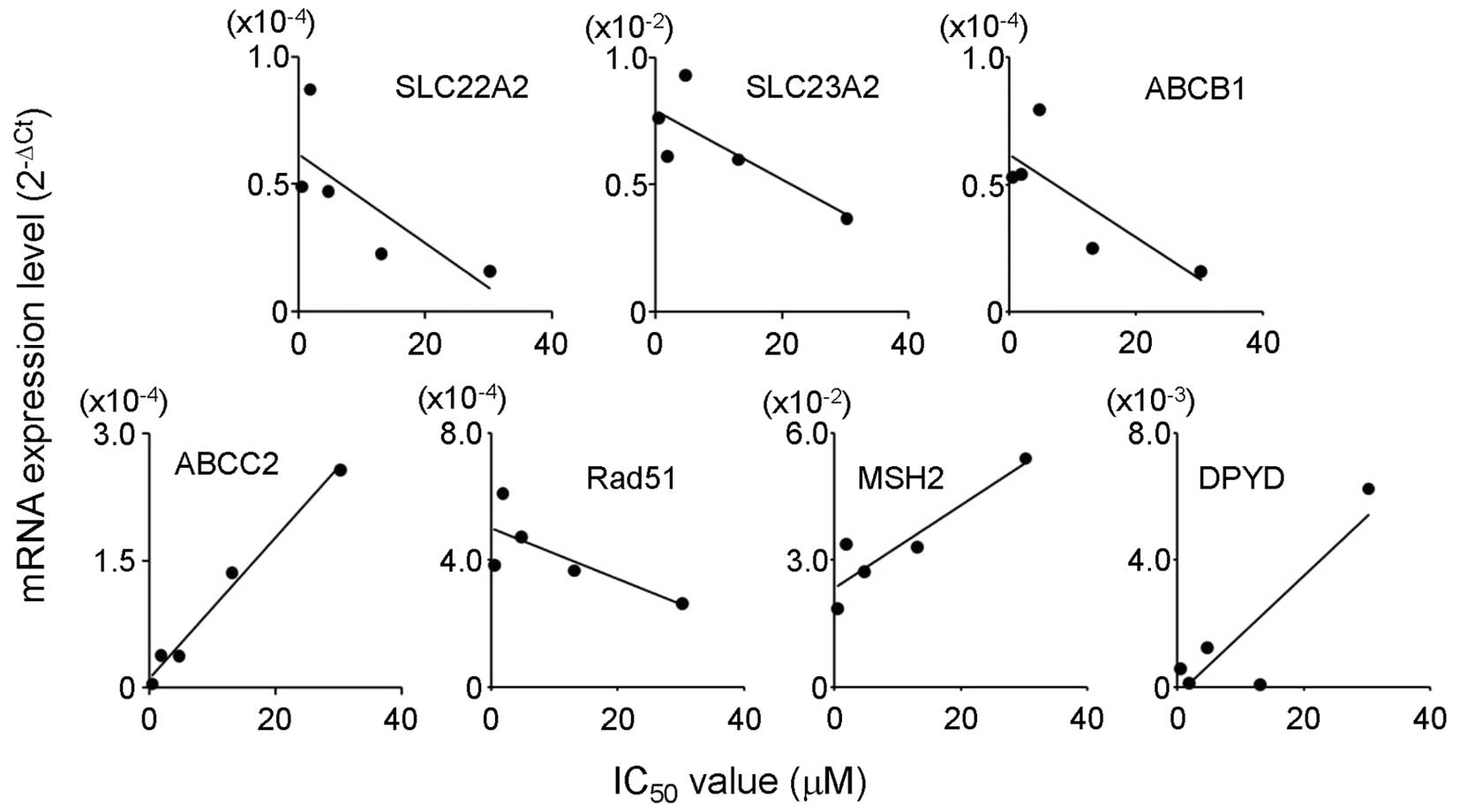
The mRNA levels of SLC22A2, SLC23A2, ABCB1 and Rad51
showed a strong negative correlation (r<−0.7) with the
IC50 values for 5-FU. ABCC2, MSH2 and DPYD were
positively correlated with the IC50 values for 5-FU
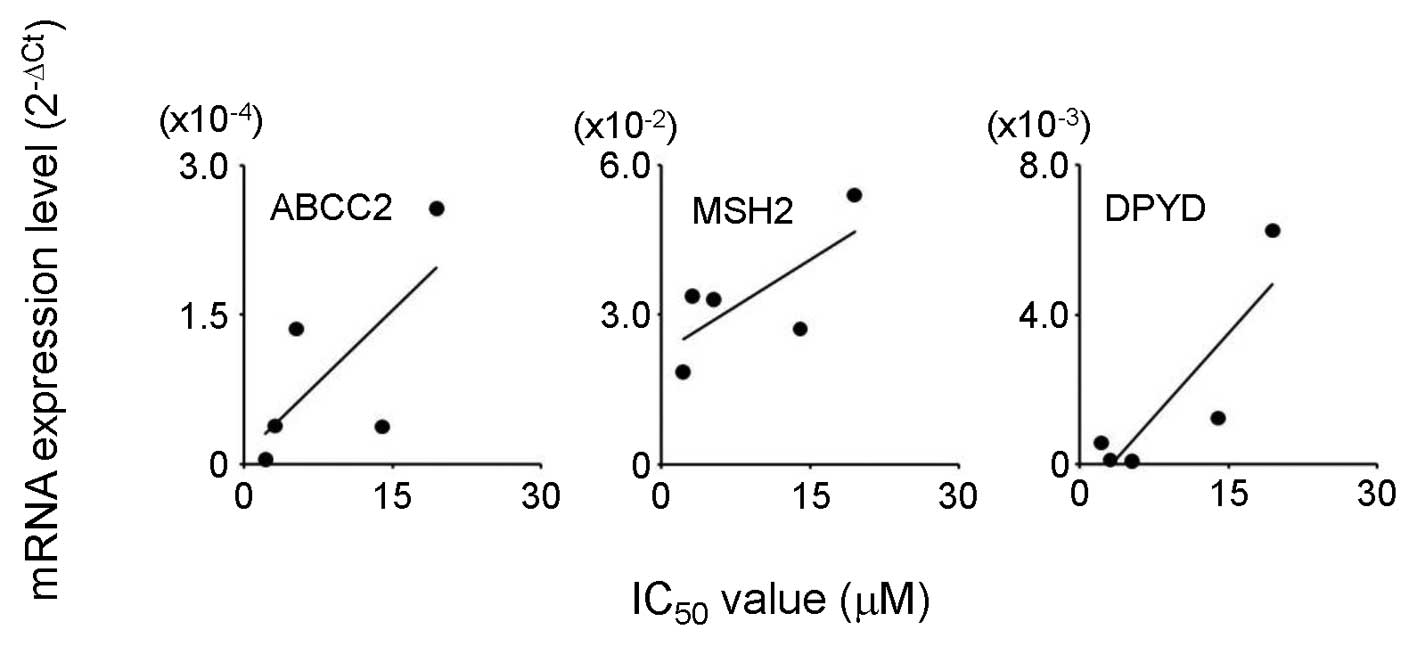
(r>0.7; Table V and Fig. 1). In the case of CDDP, a high
positive correlation coefficient (r>0.7) was found between the
IC50 values and ABCC2, MSH2 and DPYD mRNA expression
(Table V and Fig. 2).
Effects of gimeracil and MK571 on
sensitivity of esophageal carcinoma cell lines to 5-FU and
CDDP
The sensitivity of KYSE30 cells to 5-FU was enhanced
by gimeracil, but in the other cell lines gimeracil had no
observable effect (Table VI). In
addition, gimeracil showed a tendency to decrease the sensitivity
of all the cell lines to CDDP.
 | Table VI.Relative sensitivity of the
esophageal carcinoma cell lines to 5-FU or CDDP with or without
gimeracil. |
Table VI.
Relative sensitivity of the
esophageal carcinoma cell lines to 5-FU or CDDP with or without
gimeracil.
| Relative
sensitivity, mean ± SD (fold)
|
|---|
| Cell line | 5-FU | CDDP |
|---|
| OE33 | 1.10±0.37 | 0.579±0.06 |
| KYSE30 | 2.30±0.13 | 0.710±0.03 |
| KYSE70 | 1.16±0.19 | 0.687±0.05 |
| KYSE140 | 0.989±0.15 | 0.691±0.10 |
| KYSE150 | 1.19±0.16 | 0.788±0.25 |
MK571 had no observable effect on the KYSE30,
KYSE140 and KYSE150 cells (Table
VII). However, the sensitivity of KYSE70 cells to 5-FU was
substantially accelerated by the presence of MK571, and the
sensitivity of OE33 cells to 5-FU was markedly decreased. However,
MK571 showed a tendency to decrease sensitivity to CDDP, with the
exception of the KYSE30 and KYSE150 cell lines.
 | Table VII.Relative sensitivity of the
esophageal carcinoma cell lines to 5-FU or CDDP with or without
MK571. |
Table VII.
Relative sensitivity of the
esophageal carcinoma cell lines to 5-FU or CDDP with or without
MK571.
| Relative
sensitivity, mean ± SD (fold)
|
|---|
| Cell line | 5-FU | CDDP |
|---|
| OE33 | 0.0680±0.01 | 0.852±0.28 |
| KYSE30 | 0.961±0.06 | 0.974±0.09 |
| KYSE70 | 2.36±1.36 | 0.617±0.06 |
| KYSE140 | 0.813±0.16 | 0.803±0.04 |
| KYSE150 | 0.731±0.11 | 1.08±0.26 |
Discussion
Combination chemotherapy with 5-FU and CDDP is known
to be effective against esophageal carcinoma. However, it remains
ineffective in certain patients, and the causes for this have not
been clarified. The aim of the present study was to examine the
factors affecting the sensitivity of esophageal carcinoma cells to
5-FU and CDDP.
The sensitivity of the 5 different esophageal
carcinoma cell lines to 5-FU and CDDP differed (Table III). OE33, an adenocarcinoma cell
line, showed a high sensitivity to 5-FU and CDDP, whereas the
squamous cell carcinoma KYSE30 cells showed low sensitivity to 5-FU
and CDDP. In addition, OE33 cells had the longest doubling time (an
index of cell growth) of all the cell lines and KYSE30 cells the
shortest (Table II), resulting in a
trend for lower sensitivity to chemo-therapeutic agents among cells
with higher growth activity. These findings suggest that
sensitivity to 5-FU and CDDP was influenced by the growth activity
of cells, although cytotoxic agents such as 5-FU and CDDP are known
to be more toxic in cells with higher growth activity. In order to
resolve this discrepancy, further studies concerning the
correlation between cell growth and sensitivity to 5-FU or CDDP
should be performed.
The correlations between sensitivity to 5-FU and
CDDP and the mRNA levels of the 35 genes were then examined. The
levels of target mRNA expression differed among the cell lines
(Table IV). The mRNA levels of
ABCC2, MSH2 and DPYD were positively correlated with the
IC50 values of 5-FU (r>0.7; Fig. 1 and Table V). By contrast, a negative
correlation between the IC50 values of 5-FU and the mRNA
levels of SLC22A2, SLC23A2, ABCB1 and Rad51 was observed. In the
light of the biological roles of these genes, the negative
correlation between SLC22A2 and SLC23A2 mRNA expression and
sensitivity was considered to be noteworthy. SLC22A2 encodes an
organic cation transporter which is responsible for cell uptake of
various drugs, including CDDP (11,12). A
colon carcinoma cell line exhibiting resistance to 5-FU has been
reported to show lower expression of SLC23A2 mRNA than its parent
cells (13). ABCC2, MSH2 and DPYD
are known to act in detoxifying mechanisms; they are an efflux
transporter, DNA repair-related protein and metabolic enzyme,
respectively. Although ABCB1 is a known efflux transporter that
contributes to drug resistance, the cytotoxicity of 5-FU was not
influenced by the expression of ABCB1 (14). In addition, the overexpression of
DNA-repair related proteins, including Rad51, has been reported to
contribute to resistance to DNA damaging agents (15). Although the present findings showing
a negative correlation between IC50 values and ABCB1 and
Rad51 mRNA expression levels conflict with previous findings, they
may indicate that ABCB1 and Rad51 have no significant impact on
sensitivity.
In the case of CDDP, a positive correlation
(r>0.7) between the IC50 values and the mRNA levels
of ABCC2, MSH2 and DPYD was identified. The findings for ABCC2 and
MSH2 are supported by their functions; the export of CDDP from
cells (16) and repair of DNA
damaged by CDDP (17), respectively
(Table V and Fig. 2). In addition, proliferating cell
nuclear antigen-normalized mRNA expression of DPYD has previously
been reported to be associated with sensitivity to CDDP in lung
cancer tissues (18). Although the
correlation between CDDP and DPYD has not been investigated in
detail, these previous results may support the present findings.
The mRNA levels of ABCC2, MSH2 and DPYD correlated well with
sensitivity to both 5-FU and CDDP, suggesting that these are potent
predictive factors for 5-FU and CDDP-based chemotherapy in
esophageal carcinoma patients.
Finally, the roles of ABCC2 and DPYD in sensitivity
to 5-FU and CDDP were examined, since the knock-down of MSH2 in
SW460 and HeLa cells has been reported to have no influence on
sensitivity to 5-FU (19). In the
present study, 100 μM gimeracil, which showed sufficient
inhibition of DPYD (20), enhanced
5-FU sensitivity in the KYSE30 cell line (Table VI), which had the highest level of
DPYD mRNA expression of all the cell lines tested (Table IV). The present findings support
those of Ando et al(21);
that is, DPYD was a predictor of sensitivity to 5-FU. Apart from
the correlation analysis, gimeracil decreased sensitivity to CDDP
in all cell lines (Table VI),
implying that DPYD activity may be required for the cytotoxic
effect of CDDP. Further investigations are required to resolve this
contradiction. The concomitant administration of 50 μM
MK571, a representative ABCC2 inhibitor (22), was found to decrease the sensitivity
of OE33 and KYSE150 cells to 5-FU. In addition, the growth
inhibitory activity of CDDP was decreased in KYSE30 and KYSE150
cell lines (Table VII). These
findings conflict with the function of ABCC2 function as an efflux
transporter, and further investigations are required to clarify
this situation.
In conclusion, the mRNA levels of SLC22A2, SLC23A2,
ABCB1, ABCC2, Rad51, MSH2 and DPYD were confirmed to be strongly
correlated with the IC50 values for 5-FU, and those of
ABCC2, MSH2 and DPYD were also confirmed to be strongly correlated
with the IC50 values for CDDP. These genes have the
potential to affect the sensitivity to 5-FU and CDDP. In addition,
the inhibition of DPYD was suggested to affect the cytotoxicity of
CDDP. These findings provide useful information for improving the
clinical outcome of chemotherapy against esophageal carcinoma.
Acknowledgements
This study was supported in part by a
grant of Strategic Research Foundation Grant-aided Project for
Private Universities from the Ministry of Education, Culture,
Sport, Science and Technology, Japan.
References
|
1.
|
National Cancer Center: Cancer statistics
in Japan: 2011, http://ganjoho.jp/public/statistics/backnumber/2011_jp.html.
Accessed June 29, 2012.
|
|
2.
|
Tahara M, Ohtsu A, Hironaka S, Boku N,
Ishikura S, Miyata Y, Ogino T and Yoshida S: Clinical impact of
criteria for complete response (CR) of primary site to treatment of
esophageal cancer. Jpn J Clin Oncol. 35:316–323. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Pratt S, Shepard RL, Kandasamy RA,
Johnston PA, Perry W III and Dantzig AH: The multidrug resistance
protein 5 (ABCC5) confers resistance to 5-fluorouracil and
transports its monophosphorylated metabolites. Mol Cancer Ther.
4:855–863. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Kornmann M, Schwabe W, Sander S, Kron M,
Sträter J, Polat S, Kettner E, Weiser HF, Baumann W, Schramm H,
Häusler P, Ott K, Behnke D, Staib L, Beger HG and Link KH:
Thymidylate synthase and dihydropyrimidine dehydrogenase mRNA
expression levels: predictors for survival in colorectal cancer
patients receiving adjuvant 5-fluorouracil. Clin Cancer Res.
9:4116–4124. 2003.
|
|
5.
|
Ciaparrone M, Quirino M, Schinzari G,
Zannoni G, Corsi DC, Vecchio FM, Cassano A, La Torre G and Barone
C: Predictive role of thymidylate synthase, dihydropyrimidine
dehydrogenase and thymidine phosphorylase expression in colorectal
cancer patients receiving adjuvant 5-fluorouracil. Oncology.
70:366–377. 2006. View Article : Google Scholar
|
|
6.
|
Larminat F and Bohr VA: Role of the human
ERCC-1 gene in gene-specific repair of cisplatin-induced DNA
damage. Nucleic Acids Res. 22:3005–3010. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Andersson BS, Sadeghi T, Siciliano MJ,
Legerski R and Murray D: Nucleotide excision repair genes as
determinants of cellular sensitivity to cyclophosphamide analogs.
Cancer Chemother Pharmacol. 38:406–416. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Hsu DS, Lan HY, Huang CH, Tai SK, Chang
SY, Tsai TL, Chang CC, Tzeng CH, Wu KJ, Kao JY and Yang MH:
Regulation of excision repair cross-complementation group 1 by
Snail contributes to cisplatin resistance in head and neck cancer.
Clin Cancer Res. 16:4561–4571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Shimada Y, Imamura M, Wagata T, Yamaguchi
N and Tobe T: Characterization of 21 newly established esophageal
cancer cell lines. Cancer. 69:277–284. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Kitada N, Takara K, Minegaki T, Itoh C,
Tsujimoto M, Sakaeda T and Yokoyama T: Factors affecting
sensitivity to antitumor platinum derivatives of human colorectal
tumor cell lines. Cancer Chemother Pharmacol. 62:577–584. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Kimura N, Masuda S, Tanihara Y, Ueo H,
Okuda M, Katsura T and Inui K: Metformin is a superior substrate
for renal organic cation transporter OCT2 rather than hepatic OCT1.
Drug Metab Pharmacokinet. 20:379–386. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Filipski KK, Loos WJ, Verweij J and
Sparreboom A: Interaction of Cisplatin with the human organic
cation transporter 2. Clin Cancer Res. 14:3875–3880. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Karasawa H, Miura K, Fujibuchi W, Ishida
K, Kaneko N, Kinouchi M, Okabe M, Ando T, Murata Y, Sasaki H,
Takami K, Yamamura A, Shibata C and Sasaki I: Down-regulation of
cIAP2 enhances 5-FU sensitivity through the apoptotic pathway in
human colon cancer cells. Cancer Sci. 100:903–913. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Takara K, Obata Y, Yoshikawa E, Kitada N,
Sakaeda T, Ohnishi N and Yokoyama T: Molecular changes to HeLa
cells on continuous exposure to cisplatin or paclitaxel. Cancer
Chemother Pharmacol. 58:785–793. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Takenaka T, Yoshino I, Kouso H, Ohba T,
Yohena T, Osoegawa A, Shoji F and Maehara Y: Combined evaluation of
Rad51 and ERCC1 expressions for sensitivity to platinum agents in
non-small cell lung cancer. Int J Cancer. 121:895–900. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Noma B, Sasaki T, Fujimoto Y, Serikawa M,
Kobayashi K, Inoue M, Itsuki H, Kamigaki M, Minami T and Chayama K:
Expression of multidrug resistance-associated protein 2 is involved
in chemotherapy resistance in human pancreatic cancer. Int J Oncol.
33:1187–1194. 2008.PubMed/NCBI
|
|
17.
|
Lan L, Hayashi T, Rabeya RM, Nakajima S,
Kanno S, Takao M, Matsunaga T, Yoshino M, Ichikawa M, Riele H,
Tsuchiya S, Tanaka K and Yasui A: Functional and physical
interactions between ERCC1 and MSH2 complexes for resistance to
cisdiamminedichloroplatinum(II) in mammalian cells. DNA Repair
(Amst). 3:135–143. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Takizawa M, Kawakami K, Obata T, Matsumoto
I, Ohta Y, Oda M, Sasaki T and Watanabe G: In vitro sensitivity to
platinum-derived drugs is associated with expression of thymidylate
synthase and dihydropyrimidine dehydrogenase in human lung cancer.
Oncol Rep. 15:1533–1539. 2006.
|
|
19.
|
Pettersen HS, Visnes T, Vågbø CB, Svaasand
EK, Doseth B, Slupphaug G, Kavli B and Krokan HE: UNG-initiated
base excision repair is the major repair route for 5-fluorouracil
in DNA, but 5-fluorouracil cytotoxicity depends mainly on RNA
incorporation. Nucleic Acids Res. 39:8430–8444. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Li Y, Mizutani Y, Shiraishi T, Nakamura T,
Mikami K, Takaha N, Okihara K, Kawauchi A, Sakai T and Miki T: The
significance of the expression of dihydropyrimidine dehydrogenase
in prostate cancer. BJU Int. 99:663–668. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Ando T, Ishiguro H, Kuwabara Y, Kimura M,
Mitsui A, Sugito N, Mori R, Ogawa R, Katada T and Fujii Y:
Relationship between expression of 5-fluorouracil metabolic enzymes
and 5-fluorouracil sensitivity in esophageal carcinoma cell lines.
Dis Esophagus. 21:15–20. 2008.PubMed/NCBI
|
|
22.
|
Pedersen JM, Matsson P, Bergström CA,
Norinder U, Hoogstraate J and Artursson P: Prediction and
identification of drug interactions with the human ATP-binding
cassette transporter multidrug-resistance associated protein 2
(MRP2; ABCC2). J Med Chem. 51:3275–3287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Reidling JC, Subramanian VS, Dahhan T,
Sadat M and Said HM: Mechanisms and regulation of vitamin C uptake:
studies of the hSVCT systems in human liver epithelial cells. Am J
Physiol Gastrointest Liver Physiol. 295:1217–1227. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Nishimura M, Koeda A, Morikawa H, Satoh T,
Narimatsu S and Naito S: Comparison of inducibility of multidrug
resistance (MDR)1, multidrug resistance-associated protein (MRP)1,
and MRP2 mRNAs by prototypical microsomal enzyme inducers in
primary cultures of human and cynomolgus monkey hepatocytes. Biol
Pharm Bull. 31:2068–2072. 2008. View Article : Google Scholar
|
|
25.
|
Maubon N, Le Vee M, Fossati L, Audry M, Le
Ferrec E, Bolze S and Fardel O: Analysis of drug transporter
expression in human intestinal Caco-2 cells by real-time PCR.
Fundam Clin Pharmacol. 21:659–663. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Dauchy S, Miller F, Couraud PO, Weaver RJ,
Weksler B, Romero IA, Scherrmann JM, De Waziers I and Declèves X:
Expression and transcriptional regulation of ABC transporters and
cytochromes P450 in hCMEC/D3 human cerebral micro-vascular
endothelial cells. Biochem Pharmacol. 77:897–909. 2009. View Article : Google Scholar
|
|
27.
|
Shimizu J, Horio Y, Osada H, Hida T,
Hasegawa Y, Shimokata K, Takahashi T, Sekido Y and Yatabe Y: mRNA
expression of RRM1, ERCC1 and ERCC2 is not associated with
chemosensitivity to cisplatin, carboplatin and gemcitabine in human
lung cancer cell lines. Respirology. 13:510–517. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Sliwinski T, Krupa R, Majsterek I, Rykala
J, Kolacinska A, Morawiec Z, Drzewoski J, Zadrozny M and Blasiak J:
Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast
Cancer Res Treat. 94:105–109. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Amirrad M, Al-Mulla F, Varadharaj G, John
B, Saji T and Anim JT: BRCA1 gene expression in breast cancer in
Kuwait: correlation with prognostic parameters. Med Princ Pract.
14:67–72. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Müller A, Zielinski D, Friedrichs N,
Oberschmid B, Merkelbach-Bruse S, Schackert HK, Linnebacher M, von
Knebel Doeberitz M, Büttner R and Rüschoff J: Reduced mRNA
expression in paraffin-embedded tissue identifies MLH1-and
MSH2-deficient colorectal tumours and potential mutation carriers.
Virchows Arch. 453:9–16. 2008.PubMed/NCBI
|
|
31.
|
Veeriah S, Kautenburger T, Habermann N,
Sauer J, Dietrich H, Will F and Pool-Zobel BL: Apple flavonoids
inhibit growth of HT29 human colon cancer cells and modulate
expression of genes involved in the biotransformation of
xenobiotics. Mol Carcinog. 45:164–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Uthus EO, Reeves PG and Saari JT: Copper
deficiency decreases plasma homocysteine in rats. J Nutr.
137:1370–1374. 2007.PubMed/NCBI
|
|
33.
|
Hoang YD, Avakian AP and Luderer U:
Minimal ovarian upregulation of glutamate cysteine ligase
expression in response to suppression of glutathione by buthionine
sulfoximine. Reprod Toxicol. 21:186–196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Gustavsson B, Kaiser C, Carlsson G,
Wettergren Y, Odin E, Lindskog EB, Niyikiza C and Ma D: Molecular
determinants of efficacy for 5-FU-based treatments in advanced
colorectal cancer: mRNA expression for 18 chemotherapy-related
genes. Int J Cancer. 124:1220–1226. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Yoshinare K, Kubota T, Watanabe M, Wada N,
Nishibori H, Hasegawa H, Kitajima M, Takechi T and Fukushima M:
Gene expression in colorectal cancer and in vitro chemosensitivity
to 5-fluorouracil: a study of 88 surgical specimens. Cancer Sci.
94:633–638. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Matsubara J, Nishina T, Yamada Y, Moriwaki
T, Shimoda T, Kajiwara T, Nakajima TE, Kato K, Hamaguchi T, Shimada
Y, Okayama Y, Oka T and Shirao K: Impacts of excision repair
cross-complementing gene 1 (ERCC1), dihydropyrimidine
dehydrogenase, and epidermal growth factor receptor on the outcomes
of patients with advanced gastric cancer. Br J Cancer. 98:832–839.
2008. View Article : Google Scholar
|