Introduction
Chronic myeloid leukemia (CML) is a clonal malignant
disorder of a pluripotent hematopoietic stem cells and is
characterized by the presence of the Philadelphia chromosome (Ph)
in over 90% of cases (1). Ph is a
product of reciprocal translocation between the long arms of
chromosomes 9 and 22. This rearrangement combines the ABL1
proto-oncogene on chromosome 9 with the breakpoint cluster region
(BCR gene) on chromosome 22. However, variant complex chromosomal
translocations involving one or more chromosomes in addition to 9
and 22 are detected in 2–10% of CML cases. It is generally accepted
that the clinical, prognostic and hematological features of CML
with variant translocations are not distinct from those with the
typical t(9;22) translocation (1).
The development of new fluorescence in situ
hybridization (FISH) techniques has led to the identification of
unexpected deletions adjacent to the translocation breakpoint on
the derivative chromosome 9 [der(9)] in 10–15% of CML patients with classic
Ph-positive status, and in as many as 30–40% of patients with
variant Ph translocations (2).
These deletions are thought to occur simultaneously as the Ph
translocation rather than as a secondary event, and may involve the
loss of sequences from chromosome 9, chromosome 22 or both
(3). However, the deletions on
der(9) are associated with a
shorter duration of the chronic phase and a poor response to
interferon and imatinib mesylate (4–5).
Imatinib mesylate (Glivec, formerly known as STI571)
was specifically designed to inhibit the tyrosine kinase activity
of the BCR/ABL protein and other tyrosine kinases, including cABL,
c-KIT and platelet-derived growth factor receptor (PDGFR). Glivec
inactivates downstream signaling by binding to an active site of
the tyrosine kinase, halting cell proliferation and inducing
apoptosis (6). Imatinib therapy has
demonstrated high efficacy, achieving a complete or major
cytogenetic response, i.e., a reduction to 0–34% Ph-positive cells.
This positive effect was observed in cases with a simple t(9;22)
translocation combined with complex translocations resulting in
BCR/ABL gene fusion, as well as in cases with clonal evolution
(7–8).
In this study, we report a novel Ph
chromosome-positive CML case with an absence of the BCR/ABL fusion
gene on der(9) and a new complex
rearrangement formed by chromosomes 11 and 20 as well as 9 and 22.
This unusual translocation has been characterized by FISH and
array-proven multicolor banding (aMCB), the latter being extremely
useful in characterizing breakpoint regions in detail. Underlying
mechanisms and prognostic factors are discussed.
Materials and methods
Case report
A 55-year-old female was diagnosed as suffering from
CML in the chronic phase in September 2007. The patient had a white
blood cell count (WBC) of 3.190×109/l with 46.7%
neutrophils, 48.6% lymphocytes, 1.3% eosinophiles and 3.4%
basophiles. The platelet count was 398×109/l and the
hemoglobin level was 10.3 g/dl. The patient was treated with
imatinib mesylate at a dose of 400 mg/day for a period of 16
months. During that period the patient showed no symptoms. Later
the patient was lost during follow-up.
Cytogenetic analysis
Chromosome analysis using GTG-banding was performed
according to standard procedures (9). A total of 20 metaphases derived from
the unstimulated bone marrow of the patient were analyzed.
Karyotypes were described according to the international system for
human cytogenetic nomenclature (10).
Molecular cytogenetics
FISH was performed using an LSI BCR/ABL dual-color
dual-fusion translocation probe (Abbott Molecular/Vysis, Abbott
Park, IL, USA) and a whole chromosome painting (WCP) probe for
chromosomes 9, 11, 20 and 22 (MetaSystems, Altlussheim, Germany),
was applied according to the manufacturer’s instructions (11). aMCB sets based on
microdissection-derived region-specific libraries for chromosomes
9, 11, 20 and 22 were applied as previously described (12–13). A
total of 20 metaphase spreads were analyzed using a fluorescence
microscope (Axio Imager.Z1 mot, Zeiss, Germany) equipped with
appropriate filter sets to distinguish between a maximum of five
fluorochromes and the counterstain DAPI
(4′,6-diamino-2-phenylindole). Image capturing and processing were
carried out using an ISIS imaging system (MetaSystems) for the MCB
evaluation.
Reverse transcription-polymerase chain
reaction (RT-PCR) for BCR/ABL fusion transcripts
RT-PCR was carried out as previously described
(14).
Results
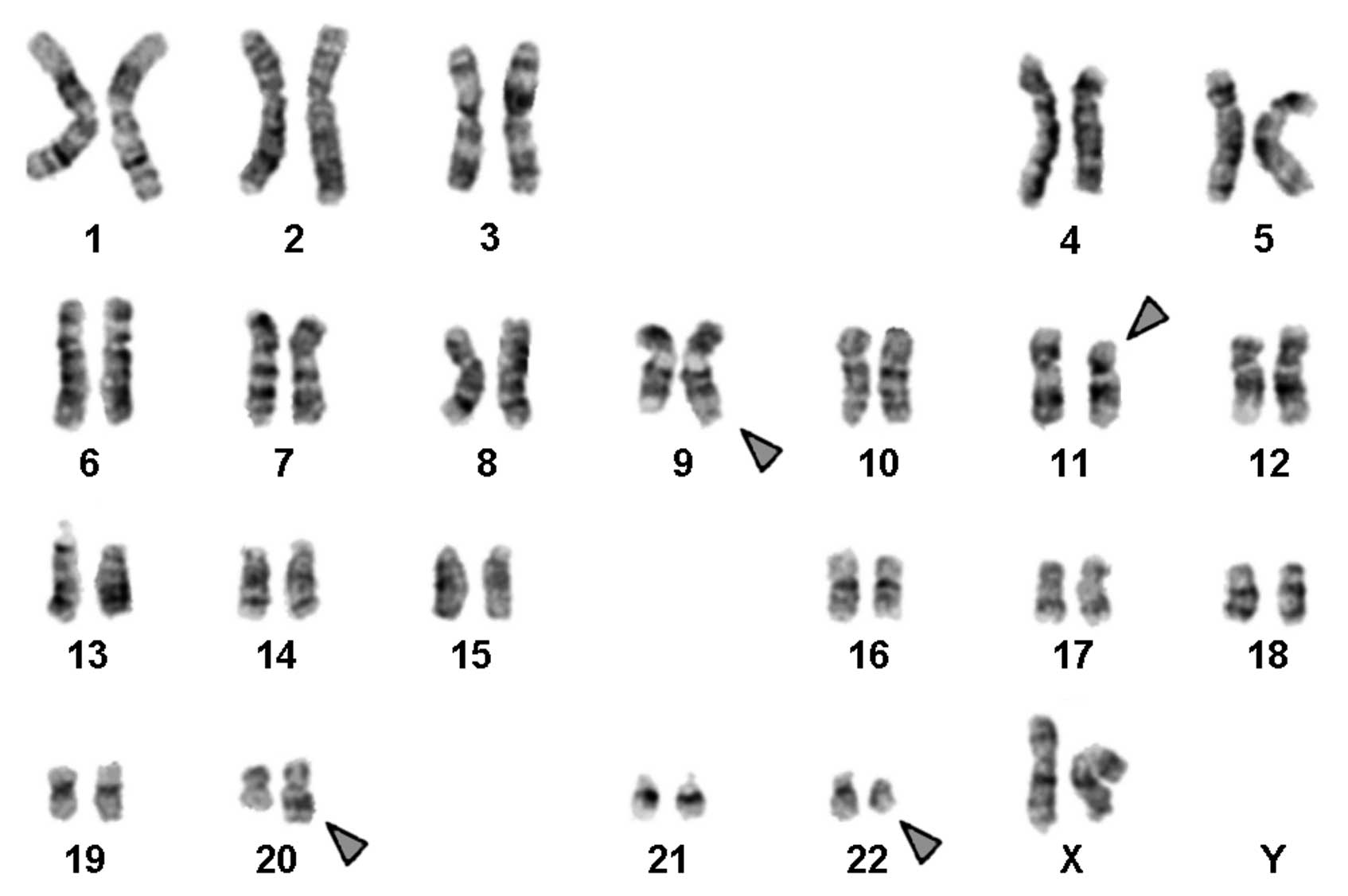
Karyotyping was performed following the chemotherapy
treatment. A complex karyotype 46,XX,t(9;11;20;22) was determined
by GTG-banding (Fig. 1) and further
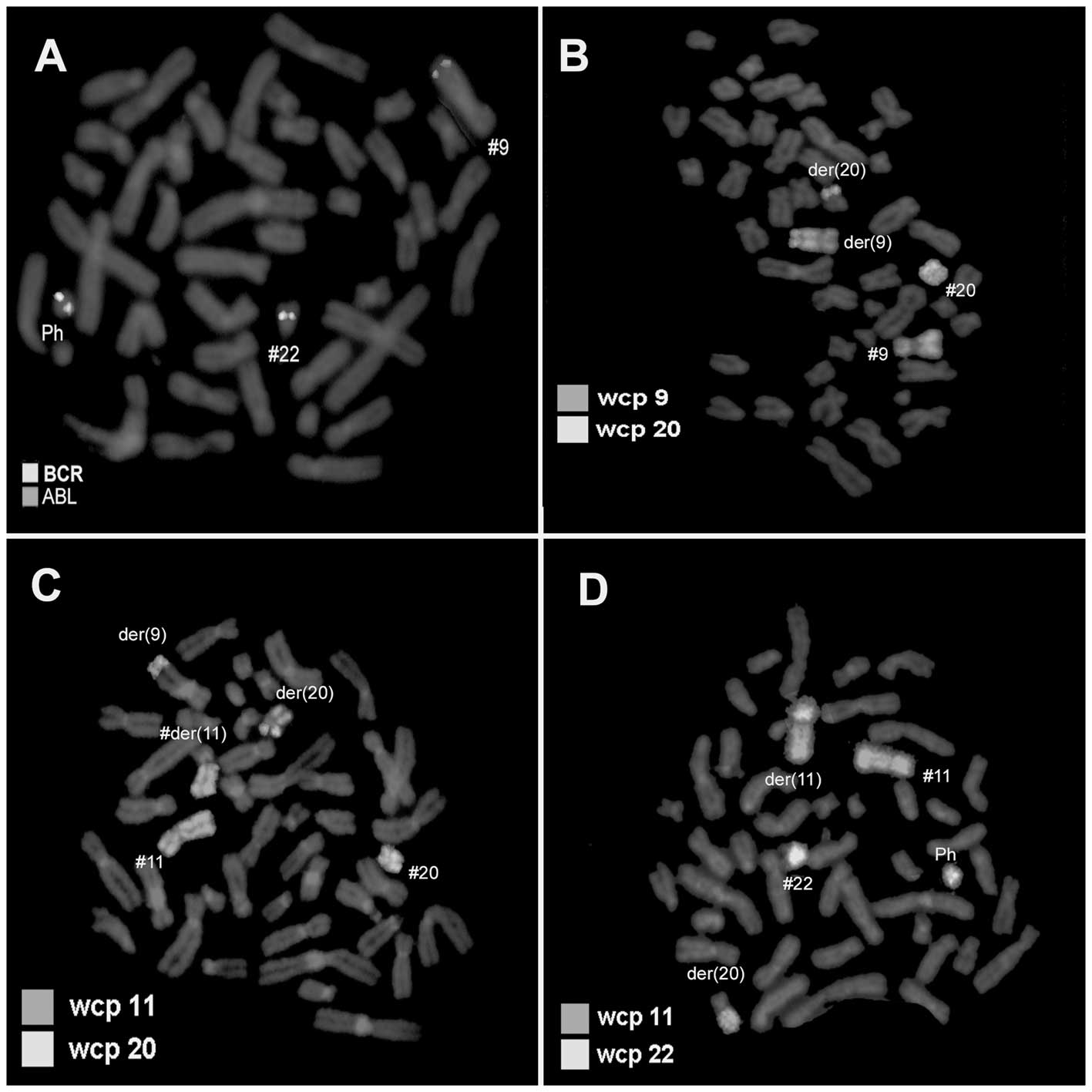
specified by molecular cytogenetic studies (Figs. 2 and 3). Dual-color FISH using a probe specific
for BCR and ABL revealed typical Ph status and the BCR/ABL fusion
gene on der(22), while the BCR/ABL
fusion gene on der(9) was not
observed (Fig. 2A). Dual-color FISH
using WCP-specific probes was performed to confirm the
rearrangement (Fig. 2B–D). Thus,
chromosomes 9, 11, 20 and 22 were found to be involved in the
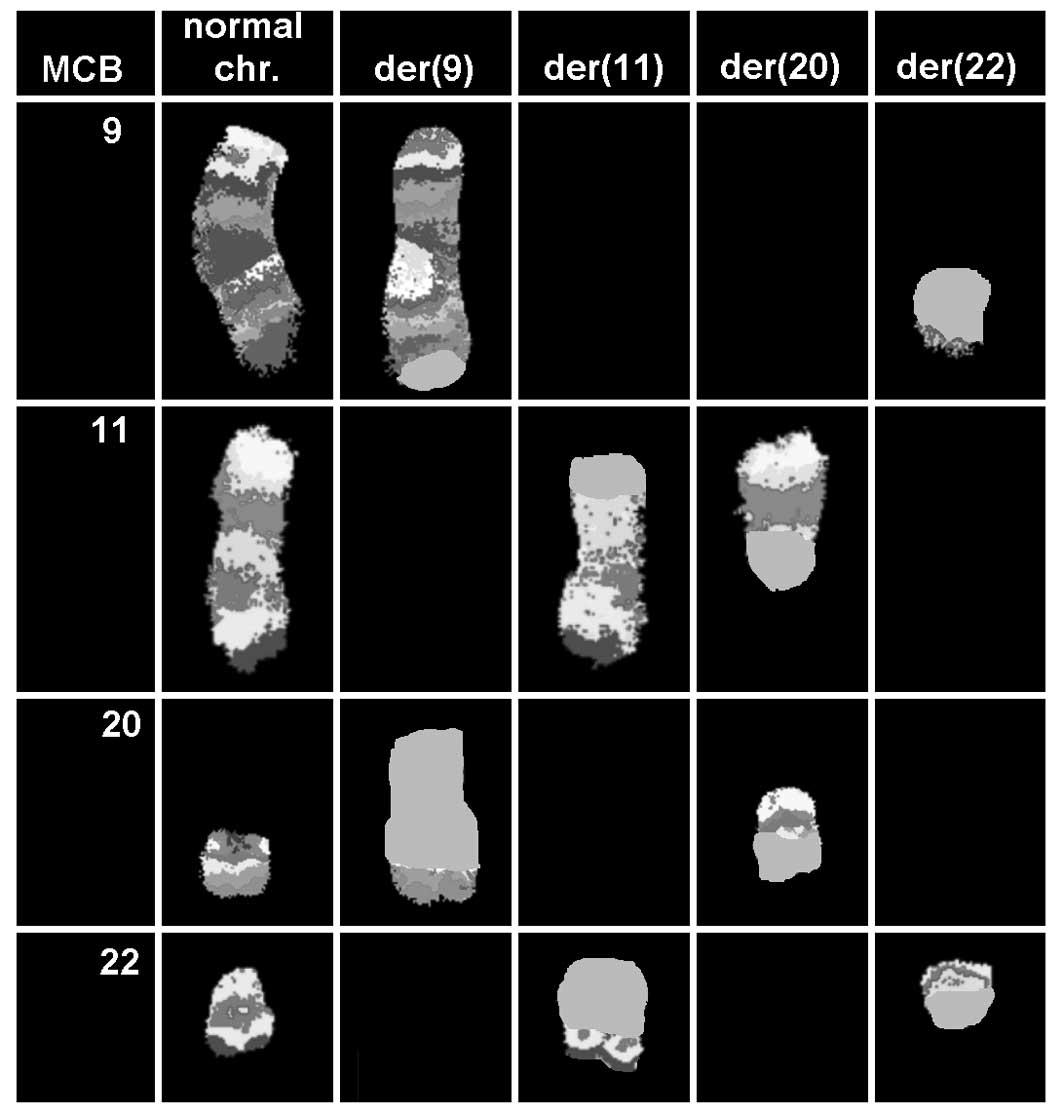
karyotypic changes. aMCB using probes for the corresponding
chromosomes was applied as previously reported (13). The presence of a complex
translocation among the four chromosomes was confirmed (Fig. 3), and the final karyotype obtained
was determined as
46,XX,t(9;11;20;22)(q34;p11.2;q11.21;q11)[20].
RT-PCR analysis of the fusion transcript revealed a
band corresponding to the b2a2 transcript (data not shown).
Discussion
We described a novel Ph-positive CML case with a new
complex variant translocation
t(9;11;20;22)(q34;p11.2;q11.21;q11)[20]. To the best of our
knowledge, this translocation has not been observed in CML
previously (15).
In 2–10% of Ph chromosome CML cases, complex
trans-locations have been reported in addition to those involving
chromosomes 9 and 22 (1). At
present, it appears that in such rearrangements any other
chromosome may be involved. However, it has been suggested that the
distribution of chromosomes and breakpoints is non-random with the
chromosomal bands most susceptible to breakage being 1p36, 3p21,
5q31, 6p21, 9q22, 10q22, 11q13, 12p13, 17p13, 17q21, 17q25, 19q13,
21q22, 22q12 and 22q13 (1). The
breakpoints 11p11.2 and 20q11.21 have not yet been reported in
variant Ph rearrangement (1).
A possible candidate in 11p11.2 is the cluster of
differentiation 82 (KAI1/CD82), a human protein encoded by the CD82
gene, originally identified as a putative metastasis suppressor
gene (16). KAI1/CD82 is widely
expressed in human tissues, and downregulation of this gene is
associated with the metastatic phenotype of several malignancies,
including carcinomas of the prostate, lung, colon, pancreas,
stomach, liver and bladder. Downregulation of KAI1/CD82 is
associated with the acquisition of high metastatic properties in
Dunning rat prostate cancers (17).
The transfer of the KAI1/CD82 gene into mammary cancer cells
suppresses their metastatic potential but does not affect primary
tumor growth (18).
The breakpoint 20q11.21 was again reported in
patients with B-cell precursor acute lymphoblastic leukemia
(BCP-ALL) (19,20). The KIF3B gene mapped at position
20q11.21 (19) may be involved in
this translocation. It encodes kinesin-like protein KIF3B in
humans. KIF3A and KIF3B form a heterodimer that functions as a
microtubule-based fast anterograde translocator of membranous
organelles (21). The head domain
of KIF3A/B, containing the ATPase activity, binds to a microtubule
and the tail domain binds to KAP3 (21,22).
Further studies are required to establish which
genes, if any, are involved in complex Ph translocations. Notably,
in the present case, Ph is associated with a deletion in the
derivative chromosome 9. In 2000, Sinclair et al(2) reported deletions on der(9)t(9;22) including 3′ BCR and 5′ ABL1 in
10–15% of CML patients and suggested that these findings had
adverse prognostic significance. Certain investigators have
suggested an association between ASS deletion and resistance to
therapy (23). Kreil et
al(24) demonstrated that
derivative chromosome 9 deletions had a heterogeneous prognostic
effect on prognosis. Only deletions spanning the ABL1/BCR
breakpoint were associated with an adverse prognosis. In this
study, we only identified the deletion of ABL1/BCR, and not that of
the ASS gene.
In conclusion, we report a novel case of a
Ph-positive CML in the chronic phase with an absence of the ABL/BCR
fusion gene on der(9) and a new
complex variant Ph translocation involving the four chromosomal
regions 9q34, 11p11.2, 20q11.21 and 22q11. Notably, the patient
concerned showed a good response to imatinib despite loss during
follow-up.
Acknowledgements
We thank Professor I. Othman, the
Director General of the Atomic Energy Commission of Syria (AECS)
and Dr N. Mirali, Head of the Molecular Biology and Biotechnology
Department for their assistance. This study was supported by the
AECS, and in parts by the Stefan-Morsch-Stiftung,
Monika-Kutzner-Stiftung and the DAAD (D/07/09624).
References
|
1.
|
Johansson B, Fioretos T and Mitelman F:
Cytogenetic and molecular genetic evolution of chronic myeloid
leukemia. Acta Haematol. 107:76–94. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Sinclair PB, Nacheva EP, Leversha M,
Telford N, Chang J, Reid A, Bench A, Champion K, Huntly B and Green
AR: Large deletions at the t(9;22) breakpoint are common and may
identify a poor-prognosis subgroup of patients with chronic myeloid
leukemia. Blood. 95:738–743. 2000.PubMed/NCBI
|
|
3.
|
Huntly BJP, Bench A and Green AR: Double
jeopardy from a single translocation: deletions of the derivative
chromosome 9 in chronic myeloid leukemia. Blood. 102:1160–1168.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Reid AG, Huntly BJ, Grace C, Green AR and
Nacheva EP: Survival implications of molecular heterogeneity in
variant Philadelphia-positive chronic myeloid leukaemia. Br J
Haematol. 121:419–427. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Bennour A, Sennana H, Laatiri MA, Elloumi
M, Khelif A and Saad A: Molecular cytogenetic characterization of
variant Philadelphia translocations in chronic myeloid leukemia:
genesis and deletion of derivative chromosome 9. Cancer Genet
Cytogenet. 194:30–37. 2009. View Article : Google Scholar
|
|
6.
|
Griffen J: The biology of signal
transduction inhibition: basic science to novel therapies. Semin
Oncol. 28:3–8. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kantarjian H, Sawyers C, Hochhaus A, et
al: International STI571 CML Study Group: Hematologic and
cytogenetic responses to imatinib mesylate in chronic myelogenous
leukemia. N Engl J Med. 346:645–652. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Cortes JE, Talpaz M, Giles F, O’Brien S,
Rios MB, Shan J, Garcia-Manero G, Faderl S, Thomas DA, Wierda W,
Ferrajoli A, Jeha S and Kantarjian HM: Prognostic significance of
cytogenetic clonal evolution in patients with chronic myelogenous
leukemia on imatinib mesylate therapy. Blood. 101:3794–3800. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Claussen U, Michel S, Mühlig P, Westermann
M, Grummt UW, Kromeyer-Hauschild K and Liehr T: Demystifying
chromosome preparation and the implications for the concept of
chromosome condensation during mitosis. Cytogenet Genome Res.
98:136–146. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Shaffer L, Slovak M and Cambell L: ISCN
(2009): An International System for Human Cytogenetic Nomenclature.
S. Karger; Basel: 2009
|
|
11.
|
Al-Achkar W, Wafa A and Nweder MS: A
complex translocation t(5;9;22) in Philadelphia cells involving the
short arm of chromosome 5 in a case of chronic myelogenous
leukemia. J Exp Clin Cancer Res. 26:411–415. 2007.PubMed/NCBI
|
|
12.
|
Weise A, Mrasek K, Fickelscher I, Claussen
U, Cheung SW, Cai WW, Liehr T and Kosyakova N: Molecular definition
of high-resolution multicolor banding probes: first within the
human DNA sequence anchored FISH banding probe set. J Histochem
Cytochem. 56:487–493. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Liehr T, Heller A, Starke H, Rubtsov N,
Trifonov V, Mrasek K, Weise A, Kuechler A and Claussen U:
Microdissection based high resolution multicolor banding for all 24
human chromosomes. Int J Mol Med. 9:335–339. 2002.PubMed/NCBI
|
|
14.
|
Al-Achkar W, Wafa A, Ali BY, Manvelyan M
and Liehr T: A rare chronic myeloid leukemia case with Philadelphia
chromosome, BCR-ABL e13a3 transcript and complex translocation
involving four different chromosomes. Oncol Lett. 1:797–800.
2010.PubMed/NCBI
|
|
15.
|
Mitelman F, Johansson B and Mertens F:
Mitelman Database of Chromosome Aberrations in Cancer, 2009.
http://cgap.nci.nih.gov/Chromosomes/Mitelman.
|
|
16.
|
Dong JT, Lamb PW, Rinker-Schaeffer CW,
Vukanovic J, Ichikawa T, Isaacs JT and Barrett JC: KAI1, a
metastasis suppressor gene for prostate cancer on human chromosome
11p11.2. Science. 268:884–886. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Suzuki H, Dong JT, Gao AC, Barrett JC and
Issacs JT: Identification of the rat homologue of KAI1 and its
expression in Dunning rat prostate cancers. Prostate. 37:253–260.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Yang X, Wei LL, Tang C, Slack R, Mueller S
and Lippman ME: Overexpression of KAI1 suppresses in vitro
invasiveness and in vivo metastasis in breast cancer cells. Cancer
Res. 61:5284–5288. 2001.PubMed/NCBI
|
|
19.
|
An Q, Wright SL, Moorman AV, Parker H,
Griffiths M, Ross FM, Davies T, Harrison CJ and Strefford JC:
Heterogeneous breakpoints in patients with acute lymphoblastic
leukemia and the dic(9;20)(p11∼13;q11) show recurrent involvement
of genes at 20q11.21. Haematologica. 94:1164–1169. 2009.PubMed/NCBI
|
|
20.
|
An Q, Wright SL, Konn ZJ, Matheson E,
Minto L, Moorman AV, Parker H, Griffiths M, Ross FM, Davies T, Hall
AG, Harrison CJ, Irving JA and Strefford JC: Variable breakpoints
target PAX5 in patients with dicentric chromosomes: A model for the
basis of unbalanced translocations in cancer. Proc Natl Acad Sci
USA. 105:17050–17054. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Yamazaki H, Nakata T, Okada Y and Hirokawa
N: KIF3A/B: a heterodimeric kinesin superfamily protein that works
as a microtubule plus end-directed motor for membrane organelle
transport. J Cell Biol. 130:1387–1399. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Yamazaki H, Nakata T, Okada Y and Hirokawa
N: Cloning and characterization of KAP3: a novel kinesin
superfamily-associated protein of KIF3A/3B. Proc Natl Acad Sci USA.
93:8443–8448. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Estes DA, Lovato DM, Khawaja HM, Winter SS
and Larson RS: Genetic alterations determine chemotherapy
resistance in childhood T-ALL: modelling in stage-specific cell
lines and correlation with diagnostic patient samples. Br J
Haematol. 139:20–30. 2007. View Article : Google Scholar
|
|
24.
|
Kreil S, Pfirrmann M, Haferlach C, Waghorn
K, Chase A, Hehlmann R, Reiter A, Hochhaus A and Cross NC; German
Chronic Myelogenous Leukemia (CML) Study Group: Heterogeneous
prognostic impact of derivative chromosome 9 deletions in chronic
myelogenous leukemia. Blood. 110:1283–1290. 2007. View Article : Google Scholar : PubMed/NCBI
|