Introduction
Colorectal cancer (CRC) is the third most common
type of malignant neoplasm and the third leading cause of
cancer-related mortalities worldwide (1). Conventional pathological diagnosis is
traumatic, with the majority of cases being detected in the later
stage. Early detection of CRC has puzzled clinicians and scientists
for years. Even with annual fecal occult-blood testing, which has
decreased the 13-year cumulative mortality rate from CRC by 33%
(2), the outcome remains poor in
patients with advanced disease. Only CRC diagnosed at an early
stage is likely to be cured by surgical resection. Genetic
alterations present in CRC, including those in APC, K-ras or p53,
do not demonstrate a confirmed correlation between their mutation
rate and clinical stage (3).
Therefore, a reliable, sensitive and specific molecular diagnostic
test for CRC is highly desirable from a clinical perspective.
Accumulating evidence suggests that microRNAs
(miRNAs) play a crucial role in the tumorigenesis and prognosis of
cancer (4–7). miRNAs are non-coding, single-stranded
RNAs of 18–25 nucleotides in length, which are able to regulate
gene expression by inhibiting translation or decreasing stability
of target mRNAs. Over the past few years, interest in the
identification, detection and utilization of miRNA molecules has
expanded rapidly. Bioinformatic analysis suggests that up to 30% of
human genes may be regulated by miRNAs, despite the fact that they
only constitute approximately 1% of the human genome (8,9).
Changes in miRNA expression have been observed in a
variety of human tumors, including breast (5), prostate (6), hepatocellular (7), colorectal (10) and oral cancer (11), chronic lymphocytic leukemia
(12) and bladder cancer (13) (Table
I). miRNAs are involved oncogenesis, disease progression,
invasion and metastasis, and are associated with patient prognosis
(4,14). Studies have confirmed that the
expression of miR-21 and miR-31 is upregulated in CRC patients
(15). miR-21 has been demonstrated
to accelerate tumorigenesis by targeting tumor suppressor genes,
including phosphatase and tensin homolog (PTEN), tropomyosin 1
(TPM1) and programmed cell death 4 (PDCD4) (7,16,17).
Ultimately, this increases tumor cell growth, migration and
invasion.
 | Table IMean fold change of analyzed miRNAs in
colorectal tumor samples. |
Table I
Mean fold change of analyzed miRNAs in
colorectal tumor samples.
| miRNA | Mean fold change | Chromosome
localization | Correlation with
cancer in previous studies (Refs.) | Putative targets
(Refs.) |
|---|
| miR-21 | 1.5 | 17q23.2 | ↑CRC (10,15),
↑CLL (12), ↑breast cancer
(5), ↑pancreatic cancer (32), ↑HCC (7) | PTEN (5,7), TPM1
(16), PDCD4 (17), TIAM1 (18) |
| miR-31 | 33.5 | 9p21.3 | ↑CRC (10,15,29),
↑oral cancer (11) | FOXC-2, FOXC-3
(15), SATB2 (24), TIAM1 (18) |
| miR-96 | 2.2 | 7q32.2 | ↑CRC (15), ↑bladder cancer (13), (26)
↑prostate carcinoma (6),
↓pancreatic cancer (26) | K-ras (26), CHES1 (15) |
| miR-135b | 17.9 | 1q32.1 | ↑CRC (15,19),
↑prostate cancer (33) | MSH2 (15), APC (19) |
Recent studies have demonstrated that miR-21 and
miR-31 are positive regulators of colon carcinoma cells with
migratory and invasive properties. As the direct target gene of
miR-21 and miR-31, the T lymphoma and metastasis gene 1 (TIAM1) has
been found to regulate the migration and invasion of colon
carcinoma cells (18). Bandrés
et al (15) examined the
expression of 156 mature miRNAs using real-time PCR in 15 CRC cell
lines as well as 12 matched pairs of tumoral and non-tumoral
adjacent tissues. These authors found that miR-31, miR-96 and
miR-135b were upregulated in both cell lines and tissues and the
expression level of miR-31 was correlated with the clinical stage
of CRC. Another study confirmed that there was a considerable
upregulation of miR-135b in CRC (19). The same study also revealed that the
two miR-135, isoforms a and b, correlated with low APC mRNA levels,
suggesting that miR-135a and miR-135b contribute to the
pathogenesis of CRC.
Accordingly, we predicted that miR-21, miR-31,
miR-96 and miR-135b may function as micro-oncogenes in colorectal
carcinogenesis and regulate CRC development and progression.
In the present study, we developed a highly
sensitive and specific quantitative real-time PCR (qPCR) method
based on SYBR-Green I. This was used to detect the expression of
miR-21, miR-31, miR-96 and miR-135b and to analyze their
correlation to the clinicopathological parameters of CRC. An
understanding of the miRNA association with CRC clinicopathological
features is essential to gain an insight into the miRNA involvement
in colorectal carcinogenesis. This method is less costly than the
method based on the TaqMan probe assay. Therefore, it may become
more widely utilized in conventional laboratories, as well as used
for the analysis of numerous samples.
Materials and methods
Study population and tissue sample
preparation
In the present study, we recruited 52 patients with
CRC, who were diagnosed and received surgery at the China-Japan
Friendship Hospital, Beijing, China, between October 2009 and
December 2010. After obtaining written informed consent from all
subjects or their guardians and approval from the Beijing Hospital
Institutional Review Committee, resected tumors and the
corresponding adjacent normal mucosa tissues were obtained from
surgically treated patients with CRC. Samples were immediately
stored in liquid nitrogen until RNA extraction was performed.
Generally, normal mucosal tissue was obtained from a region >10
cm away from the cancer tissue. The eligible tumors had to result
from primary CRC without prior preoperative radiotherapy or
chemotherapy treatment. The study group comprised 26 males and 26
females, with a median age of 62 years, range 36–89. The TNM
classification was in accordance with the American Joint Committee
on Cancer (AJCC) TNM staging. Patient details, including gender,
tumor location, TNM stage, grade and lymphovascular invasion, are
shown in Table II.
 | Table IICorrelation between miR-21, miR-31,
miR-96 and miR-135b relative expression levels and
clinicopathological features in CRC patients. |
Table II
Correlation between miR-21, miR-31,
miR-96 and miR-135b relative expression levels and
clinicopathological features in CRC patients.
| | miR-21 | miR-31a | miR-96b | miR-135ba |
|---|
| |
|
|
|
|
|---|
| n | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value | Mean ± SD | P-value |
|---|
| Tissue |
| Cancer | 52 | 4.803±3.037 | 0.003 | 0.804±0.851 | 0.003 | 1.242±0.961 | 0.013 | 1.167±1.186 |
<0.001 |
| Normal | 52 | 3.192±2.242 | | 0.024±0.030 | | 0.553±0.811 | | 0.065±0.074 | |
| Gender |
| Male | 26 | 4.603±3.150 | 0.4 | 0.644±0.846 | 0.528 | 1.153±0.857 | 0.286 | 1.161±1.153 | 0.808 |
| Female | 26 | 5.003±2.969 | | 0.399±0.628 | | 1.334±1.069 | | 1.239±1.510 | |
| Localization |
| Right side | 17 | 4.330±3.344 | 0.067 | 0.852±1.093 | 0.493 | 1.119±0.990 | 0.628 | 0.883±0.849 | 0.2 |
| Left side | 35 | 5.033±2.900 | | 0.448±0.671 | | 1.625±2.126 | | 1.486±1.661 | |
| Lymph node |
| Positive | 32 | 6.560±2.805 | 0.032 | 0.498±0.757 | 0.529 | 1.358±1.039 | 0.829 | 1.319±1.332 | 0.127 |
| Negative | 20 | 4.191±3.416 | | 0.714±0.976 | | 1.062±0.819 | | 0.932±0.898 | |
|
Differentiation |
| High | 11 | 5.925±4.546 | 0.348 | 0.816±1.198 | 0.968 | 0.975±0.828 | 0.12 | 1.058±1.194 | 0.194 |
| Moderate | 31 | 4.847±2.634 | | 0.427±0.593 | | 1.452±0.995 | | 1.353±1.261 | |
| Low | 10 | 3.433±1.600 | | 0.414±0.494 | | 0.855±0.865 | | 0.702±0.849 | |
| Stage |
| II | 19 | 3.782±1.460 | 0.048 | 0.462±0.454 | 0.76 | 1.055±0.751 | 0.063 | 1.206±1.098 | 0.029 |
| III | 24 | 4.272±2.120 | | 0.697±0.970 | | 1.156±0.932 | | 1.002±1.508 | |
| IV | 6 | 5.828±2.948 | | 0.145±0.107 | | 2.298±1.283 | | 2.826±1.903 | |
| Liver
metastasis |
| Yes | 7 | 5.076±3.113 | 0.778 | 0.131±0.104 | 0.29 | 2.387±1.195 | 0.006 | 2.560±1.875 | 0.013 |
| No | 45 | 4.760±3.059 | | 0.655±0.890 | | 1.060±0.792 | | 0.946±0.883 | |
Total RNAs of the tumor tissues and the
corresponding normal tissues were isolated using the mirVana miRNA
Isolation kit (Ambion, Carlsbad, CA, USA), according to the
manufacturer’s instructions. RNA concentration and purity were
analyzed using a UV spectrophotometer (A260/A280 >2.0; A260/A230
>1.8).
Preparation and quantification of
standards
Reliable standards are essential in qPCR analysis.
Therefore, we selected a cloned circular plasmid, which is more
stable than PCR products (20). The
four miRNAs (miR-21, miR-31, miR-96 and miR-135b) and U6 snRNA from
the HT29 cells were reverse-transcribed. The target miRNAs were
then amplified according to the procedures described below.
Following these procedures, the products were cloned into a pMD-18T
vector (Takara, Otsu, Shiga, Japan), the ligated fragments were
transformed into DH5α competent cells and the transformed cultures
were spread onto lysogeny broth plates containing ampicillin (75
μg/ml). Clones were screened by PCR reactions and positive clones
were selected and processed for plasmid isolation. The purity and
concentration of the plasmids were accurately quantified using a
Qubit™ dsDNA BR Assay kit (Invitrogen, Ltd., Inchinnan Business
Park, UK). The exact sequence of the inserted plasmids was analyzed
and confirmed by sequencing with RV-M universal primers. We
obtained a copy number based upon the molecular weight of the
plasmid and insert (21). The
plasmids were then diluted in 1× Tris-EDTA (TE) to 1010
copy/μl. To maximize accuracy, dilutions were performed over a
range of copy numbers that included the amount of target mRNA
expected in the experimental RNA samples. Thus, serial 10-fold
dilutions from 1010 to 100 copy/μl of the
plasmids were used as standards. Plots of the logarithm of the
template concentration versus the Ct were graphed, and the PCR
efficiency was calculated using the equation: E = 10(−1/slope)
(22).
Reverse transcription and real-time
PCR
Stem-loop real-time RT-PCR was used to analyze the
expression of miRNA. We designed miR-21, miR-31, miR-96 and
miR-135b stem-loop RT primers and amplification primers according
to the method developed by Chen et al (23). cDNAs were synthesized from the total
RNA using unique stem-loop RT primers. The sequences were as
follows: 5′-GTCGTATCCAGTGCAGG GTCCGAGGTATTCGCACTGGATACGACTCAACA-3′
(miR-21), 5′-GTCGTATCCAGTGCAGGGTCCGAGGT ATTCGCACTGGATACGACAGCTAT-3′
(miR-31), 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACT GGATACGACAGCAAA-3′
(miR-96) and 5′-GTCGT ATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATAC
GACTCACAT-3′ (miR-135b). Reverse transcriptase reactions contained
the following reagents: 10 ng RNA sample, 60 nM stem-loop RT
primer, 1× RT buffer, 0.25 mM each of dNTP, 4 U/μl M-MLV reverse
transcriptase (Promega, Madison, WI, USA) and 0.4 U/μl RNase
inhibitor (Takara). Reactions (10 μl) were incubated in a GenAmp
PCR System 9700 (Applied Biosystems, Foster City, CA, USA) at 16°C
for 30 min, 42°C for 30 min and 85°C for 5 min. The samples were
then held at 4°C. Real-time PCR was performed using the Thermal
Cycler Dice real-time system TP800 (Takara). The universal reverse
primer for the four miRNAs was 5′-CAGTGCAGGGTCC GAGGT-3′. The
specific forward primers were as follows:
5′-GCCCGCTAGCTTATCAGACTGATG-3′ (miR-21), 5′-GCCGCAGGCAAGATGCTGGC-3′
(miR-31), 5′-GCCC GCTTTGGCACTAGCACATT-3′ (miR-96) and 5′-GCCCG
CTATGGCTTTCATTCCT-3′ (miR-135b). The 25 μl PCR reaction mixture
included 1× SYBR premix Ex Taq mix (Takara), 2 μl RT products and
10 nM of each forward and reverse primer. Reactions were incubated
in a 96-well plate at 95°C for 30 sec, followed by 45 cycles of
95°C for 15 sec and 60°C for 21 sec. Dissociation from 65 to 95°C
was conducted to confirm the specificity of the amplification
products. The threshold cycle data were determined using second
derivative max settings. U6 was used as an internal control to
normalize the level of target miRNAs. The stem-loop RT primers and
amplification primers of U6 were obtained from Ribobio Co., Ltd.,
Guangzhou, China).
Normalization and data analysis
In the present study, we applied a quantification
method using U6 as an internal reference to analyze the expression
of the miRNAs. The cycle number at the threshold level of the
log-based fluorescence is defined as the Ct value. According to the
principle that the Ct value is inversely proportional to the
logarithm of the initial copy number, the copy number of the target
miRNA can be accurately and quantitatively calculated based on the
construction of a standard curve. For each sample, the normalized
expression of the miRNAs was calculated according to the equation
(24): En = Copy (target)/Copy
(reference); where En is the normalized expression of the miRNA in
either tumor tissue or normal mucosal tissue, copy (target) is the
copy number of the target miRNA by comparing it to the
corresponding standard curve, and copy (reference) is the copy
number of U6 by comparing it with the corresponding standard curve.
The fold change of the miRNA expression level in tumor tissue was
calculated as: FC = ET/EN; where
ET is the normalized expression in the tumor tissue and
EN is the normalized expression in the normal mucosal
tissue.
Statistical analysis
An independent sample t-test was used to compare the
differential expression between the tumor tissues and the normal
mucosal tissues. Statistical differences between the
clinicopathological parameters and the miRNA levels were evaluated
using non-parametric tests. The Mann-Whitney U test was used
between 2 groups and the Kruskall-Wallis test was used between 3 or
more groups. P<0.05 was considered to indicate a statistically
significant difference. All calculations were performed using SPSS
version 16.0 software.
Results
Sensitivity and specificity of detecting
miRNAs
Standard curves were created by plotting the input
copy number of a standard plasmid DNA, which contained 62–65 bp of
the miR-21, miR-31, miR-96, miR-135b or U6 and the Ct value. Each
of the plasmid DNAs were confirmed using sequencing to guarantee
the authenticity of the PCR products. Using serial 10-fold
dilutions of the plasmid standard, a wide linear range of
101–108 copies for miR-21,
102–109 copies for miR-31,
102–108 copies for miR-96 and miR-135b, and
101–108 copies for U6 snRNA, was detected.
Dissociation curves of each miRNA are shown as single,
sharply-defined narrow peaks, indicating that specific, homogeneous
PCR products were produced (Fig.
1).
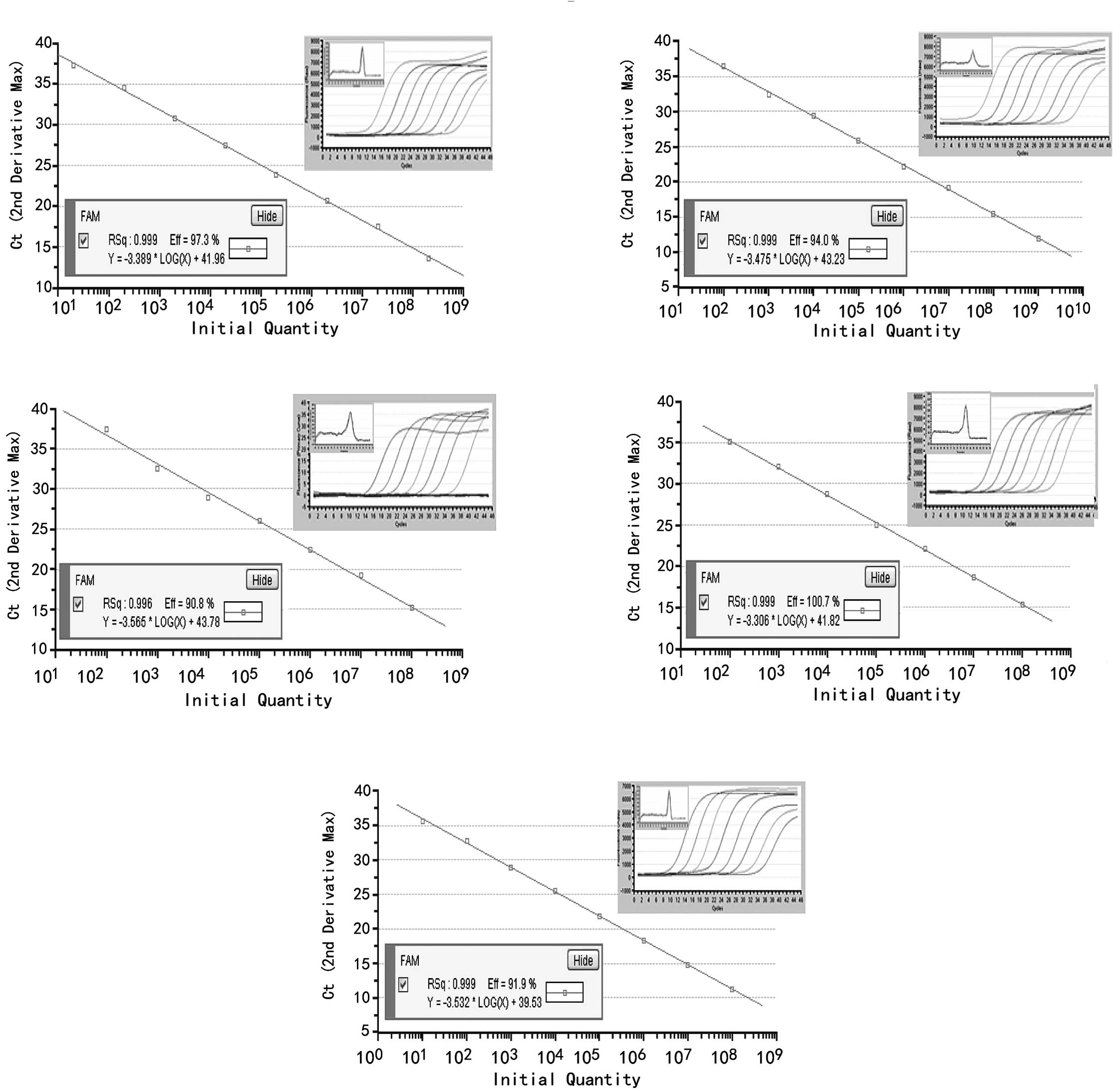 | Figure 1Standard curves were created with
10-fold serially diluted plasmid DNA containing (A) miR-21, (B)
miR-31, (C) miR-96, (D) miR-135b or (E) U6 by SYBR-Green I
real-time PCR. The Ct values obtained from the real-time PCR assays
were plotted against the initial plasmid DNA copy number. The
curves demonstrated a wide linear range:
101–108 copies for miR-21,
102–109 copies for miR-31,
102–108 copies for miR-96 and miR-135b and
101–108 copies for U6 snRNA. Correlation
coefficients were >0.996, and the melting-curves of miR-21,
miR-31, miR-96, miR-135b and U6 are shown as a single,
sharply-narrow peak, indicating that pure, homogeneous qPCR
products were produced. |
Expression of miRNAs in the tumor and
corresponding normal tissues
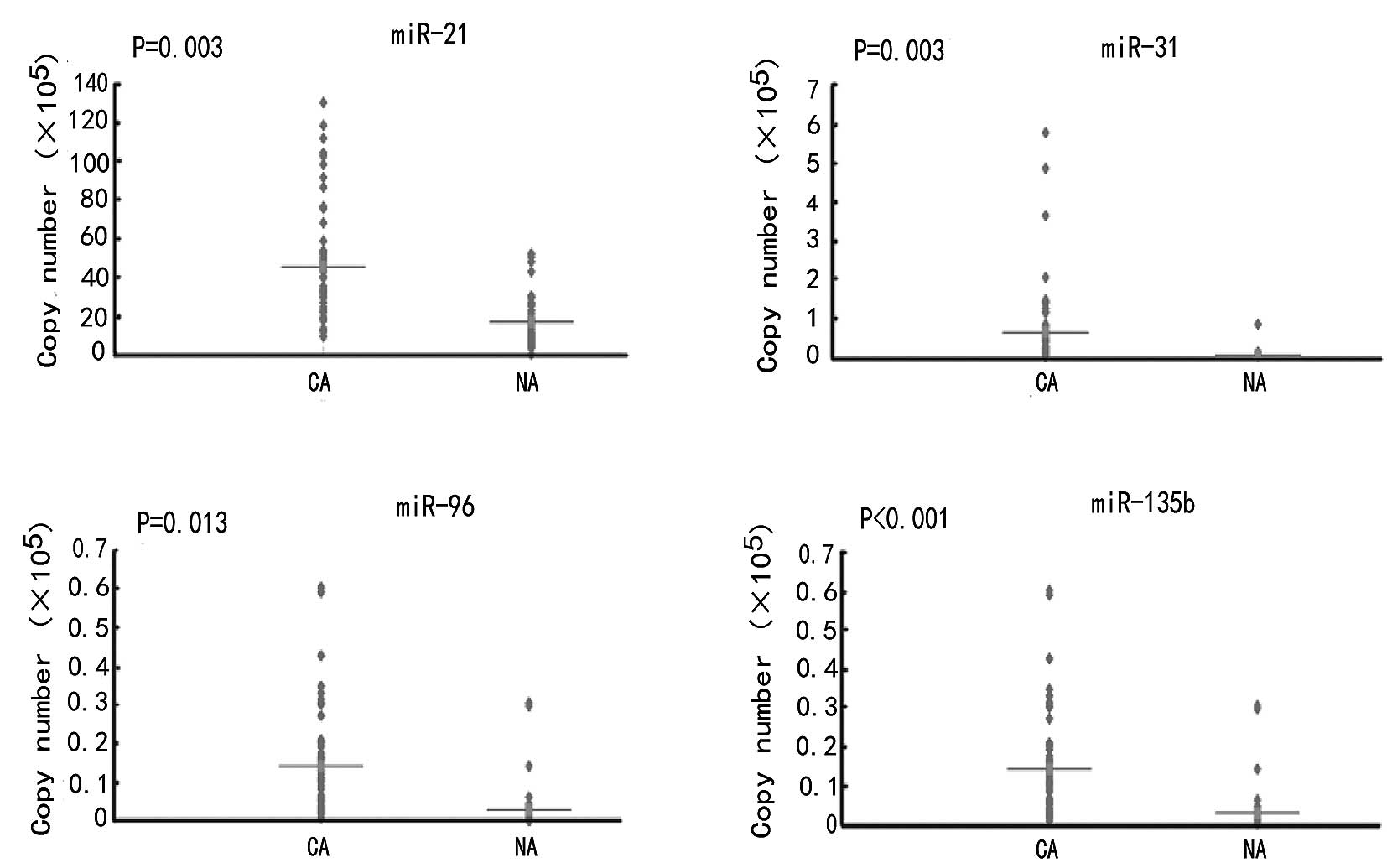
The expression levels of all analyzed miRNAs were
significantly different between the tumor and normal adjacent
tissues. Expression levels of miR-21, miR-31, miR-96 and miR-135b
were upregulated in CRC tissues by 1.5, 33.5, 2.2 and 17.9 times,
respectively, compared to the normal tissues (P<0.05) (Table I; Fig.
2). The mean fold changes of the miRNAs analyzed in the
colorectal tumor samples and their possible correlation with cancer
and putative targets were detected (Table I).
Among the 52 CRC tissues, 35 (67.3%), 47 (90.4%), 42
(80.8%) and 48 (92.3%) tumors demonstrated overexpression of
miR-21, miR-31, miR-96 and miR-135b, respectively. The average
levels of miRNA expression in the tumor and normal tissues are
shown in Table II. We also
examined the frequency of the combined upregulated expression of
the four miRNAs and revealed that there were 31 cases (31/52,
59.6%) where all four miRNAs were upregulated and 49 cases (49/52,
94.2%) where at least one of the miRNAs was upregulated (data not
shown).
Correlation between miRNAs and clinical
parameters
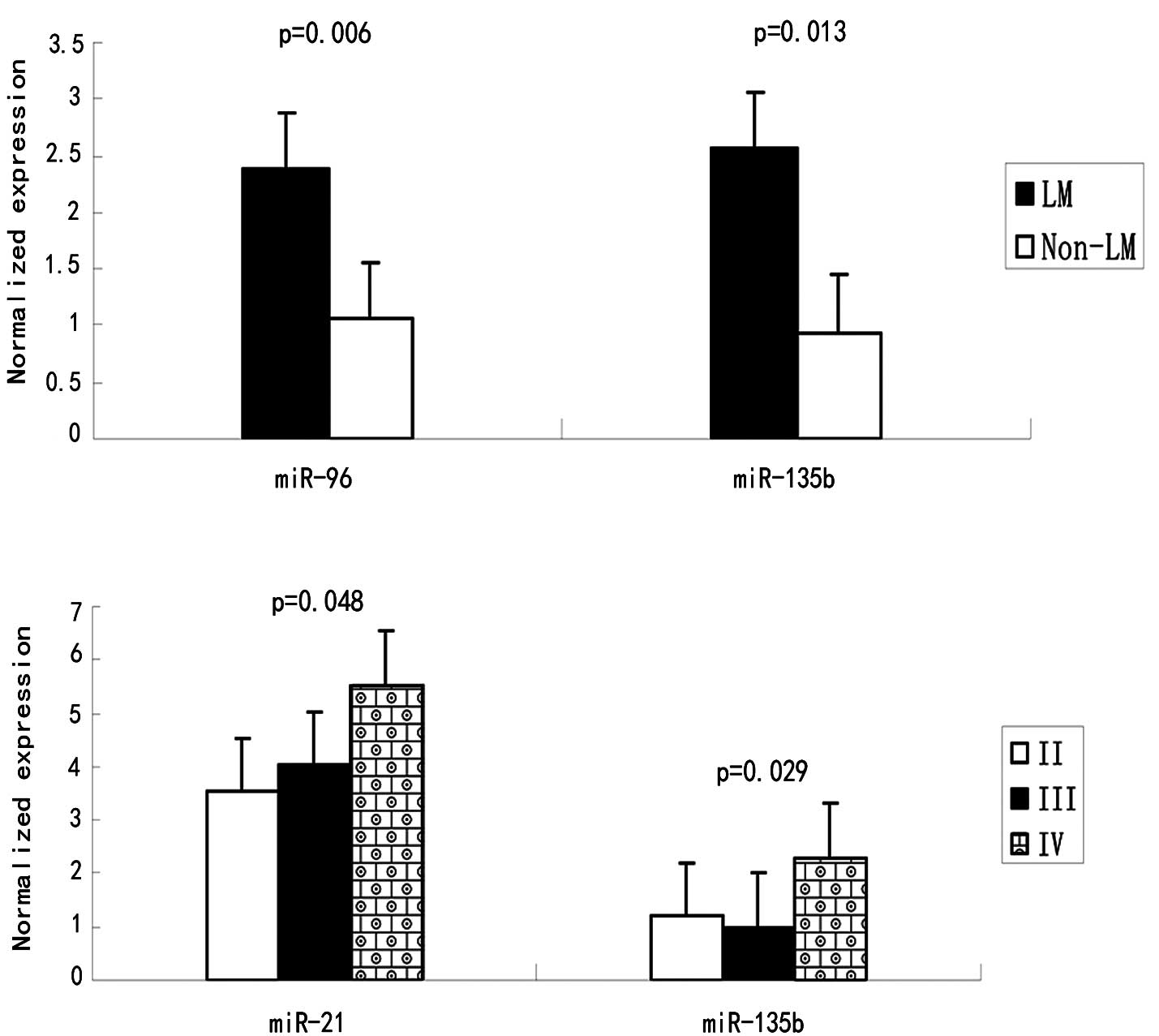
miR-96 and miR-135b were found to be associated with
liver metastasis (p=0.006 and p=0.013, respectively) (Table II and Fig. 3). miR-21 expression is associated
with lymph node metastasis (P=0.032) and clinical stage (P=0.048).
However, no correlations were observed between miR-31 and gender,
localization, lymph node metastasis, differentiation or clinical
stage. Notably, the miR-31 expression was higher in stage III
compared to II, but decreased in stage IV. A possible explanation
for this phenomenon may involve the stress response. Although the
mean expression levels of miR-96 increased progressively with the
disease stage, a statistical significance was not achieved.
Discussion
miRNAs are emerging as major contributors in normal
and diseased cell processes and have been demonstrated to be
involved in oncogenesis, disease progression, invasion and
metastasis (14). To date, a number
of different approaches to quantify miRNAs have been described,
including northern blotting (25,26),
microarrays (15), bead-based
hybridization (27), modified
invader assays (28) and real-time
PCR. Among these approaches, real-time PCR is a more quantitative
and sensitive method when compared to other high-throughput assays.
Real-time PCR based on TaqMan probes and SYBR-Green I is the most
commonly used method for miRNA detection. TaqMan probes, which are
designed to hybridize to an internal stretch of the amplicon, are
more specific. However, with this method, one miRNA must match a
single and unique probe, thereby increasing the overall cost. In
contrast, real-time RT-PCR that uses SYBR-Green I is a more
cost-effective method. It should be considered that SYBR-Green I is
not able to discriminate between different PCR products.
Additionally, it binds to all dsDNA, including non-specific
products, such as primer dimers. Thus, melting point analysis must
be performed to monitor the homogeneity of qPCR products when using
the SYBR-Green I method.
Although altered expression levels of numerous
miRNAs have been identified in human cancers, limited information
is available regarding their physiological and pathological roles.
In the present study, we established a specific and sensitive
SYBR-Green I real-time RT-PCR method to detect miRNA expression.
The melting-curves of miR-21, miR-31, miR-96, miR-135b and U6 were
each shown as a single, sharply-defined melting curve with a narrow
peak, indicating that pure, homogeneous qPCR products were produced
(Fig. 1).
In our study, we analyzed 52 cases of CRC tissues
and corresponding normal mucosal tissue, including 7 cases that
were liver metastasis-positive and 45 cases that were liver
metastasis-negative, in order to identify the expression levels of
miR-21, miR-31, miR-96 and miR-135b. The expression levels of the
four miRNAs were significantly higher in the tumor tissues than in
the matched normal musocal tissues (P<0.05), which supports
previous studies (10,15,19,29).
Furthermore, we identified that the expression of miR-21 was not
only associated with lymph node metastasis, but also with the
clinical stage; this also corresponds with a previous study
(10). Bandrés et al
(15) examined 12 matched pairs of
tumoral and non-tumoral adjacent tissues and revealed that miR-31
was correlated with stage of CRC (P=0.028). However, we did not
observe this correlation, nor did we find any other significant
link between miR-31 and the clinical features examined. Slaby et
al (10) examined 29 primary
colorectal carcinomas and 6 non-tumoral adjacent tissue specimens
using real-time PCR and also found that miR-21 and miR-31 were
upregulated in CRC patients. In our study, miR-21 was correlated
with the clinical stage (P=0.032), but miR-31 did not significantly
change in the different clinical stages. This discrepancy may
partly be due to differences in the specimens analyzed.
We also found that miR-96 and miR-135b were
upregulated in the CRC samples and were correlated with liver
metastasis (P=0.006 and P=0.013, respectively). To the best of our
knowledge, this is the first study to describe the relationship
between miR-96 and miR-135b in CRC patients with liver metastasis.
We found no correlation between miR-96 and miR-135b and other
clinicopathological characteristics such as gender, localization,
lymph node metastasis, differentiation or clinical stage (Table II).
Further studies are required to determine the
interactions of miR-21, miR-31, miR-96 and miR-135b with their
potential targets. The genes targeted by miR-21 have been under
extensive study. The PTEN tumor suppressor gene was first selected
as a potential miR-21 target in hepatocellular cancer based on its
well-characterized role in tumor biology (7). Following this, TPM and PDCD4 were
confirmed as functionally significant targets for miR-21 in breast
cancer (16,30). It was demonstrated that miR-21
promoted cell migration, invasion and metastasis by downregulating
PDCD4 gene expression. Studies on other cancer types have also
confirmed that miRNAs are able to promote metastasis and have
confirmed their role in tumor migration and invasion (31). miR-31 is able to directly target the
homeobox gene SATB2, which is responsible for chromatin remodeling
and regulation of gene expression in cancer-associated fibroblasts
(24).
miR-135b regulates APC expression and the Wnt
signaling pathway, suggesting its contribution to CRC pathogenesis
(19). miR-96 was upregulated in
colorectal, bladder and prostate cancer, but downregulated in
pancreatic cancer (6,13,15,26),
where it targets the K-ras oncogene and acts as a tumor suppressor
gene (26). From these results it
is evident that miRNAs may function as oncogenes or tumor
suppressors depending on the tissue type or target gene
expression.
In conclusion, we have established a sensitive and
specific assay to detect miRNA expression. miR-21, miR-31, miR-96
and miR-135b were upregulated in tumor tissues compared to adjacent
normal mucosal tissues. These miRNAs may have roles as oncogenes
during the development of CRC. miR-135b, in particular, may be
correlated with malignancy and the process of liver metastasis in
CRC.
Acknowledgements
This study was supported by the Key International
Science and Technology Cooperation Projects of China (No.
2006DFB31410) and the National Natural Science Foundation of China
(No. 81171028). We are grateful to the members of the Institute of
Geriatrics of the Ministry of Health for their advice and
assistance.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics. CA Cancer J Clin. 60:277–300. 2010.
|
|
2
|
Mandel JS, Bond JH, Church TR, et al:
Reducing mortality from colorectal cancer by screening for fecal
occult blood. Minnesota Colon Cancer Control Study. N Engl J Med.
328:1365–1371. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jeon CH, Lee HI, Shin IH and Park JW:
Genetic alterations of APC, K-ras, p53, MSI, and MAGE in Korean
colorectal cancer patients. Int J Colorectal Dis. 23:29–35. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Slack FJ and Weidhaas JB: MicroRNA in
cancer prognosis. N Engl J Med. 359:2720–2722. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang GL, Zhang XH, Guo GL, et al:
Clinical significance of miR-21 expression in breast cancer:
SYBR-Green I-based real-time RT-PCR study of invasive ductal
carcinoma. Oncol Rep. 21:673–679. 2009.PubMed/NCBI
|
|
6
|
Schaefer A, Jung M, Mollenkopf HJ, et al:
Diagnostic and prognostic implications of microRNA profiling in
prostate carcinoma. Int J Cancer. 126:1166–1176. 2010.PubMed/NCBI
|
|
7
|
Meng F, Henson R, Wehbe-Janek H, et al:
MicroRNA-21 regulates expression of the PTEN tumor suppressor gene
in human hepatocellular cancer. Gastroenterology. 133:647–658.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Slaby O, Svoboda M, Fabian P, et al:
Altered expression of miR-21, miR-31, miR-143 and miR-145 is
related to clinicopathologic features of colorectal cancer.
Oncology. 72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu CJ, Kao SY, Tu HF, et al: Increase of
microRNA miR-31 level in plasma could be a potential marker of oral
cancer. Oral Dis. 16:360–364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Borkhardt A, Fuchs U and Tuschl T:
MicroRNA in chronic lymphocytic leukemia. N Engl J Med.
354:524–525. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han Y, Chen J, Zhao X, et al: MicroRNA
expression signatures of bladder cancer revealed by deep
sequencing. PLoS One. 6:e182862011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aslam MI, Taylor K, Pringle JH and Jameson
JS: MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg.
96:702–710. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bandrés E, Cubedo E, Agirre X, et al:
Identification by Real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006.PubMed/NCBI
|
|
16
|
Zhu S, Si ML, Wu H and Mo YY: MicroRNA-21
targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol
Chem. 282:14328–14336. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Frankel LB, Christoffersen NR, Jacobsen A,
et al: Programmed cell death 4 (PDCD4) is an important functional
target of the microRNA miR-21 in breast cancer cells. J Biol Chem.
283:1026–1033. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cottonham CL, Kaneko S and Xu L: miR-21
and miR-31 converge on TIAM1 to regulate migration and invasion of
colon carcinoma cells. J Biol Chem. 285:35293–35302. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nagel R, le Sage C, Diosdado B, et al:
Regulation of the adenomatous polyposis coli gene by the miR-135
family in colorectal cancer. Cancer Res. 68:5795–5802. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dhanasekaran S, Doherty TM and Kenneth J:
Comparison of different standards for real-time PCR-based absolute
quantification. J Immunol Methods. 354:34–39. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Whelan JA, Russell NB and Whelan MA: A
method for the absolute quantification of cDNA using real-time PCR.
J Immunol Methods. 278:261–269. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee C, Kim J, Shin SG and Hwang S:
Absolute and relative QPCR quantification of plasmid copy number in
Escherichia coli. J Biotechnol. 123:273–280. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aprelikova O, Yu X, Palla J, et al: The
role of miR-31 and its target gene SATB2 in cancer-associated
fibroblasts. Cell Cycle. 9:4387–4398. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Michael MZ, SM OC, van Holst Pellekaan NG,
et al: Reduced accumulation of specific microRNAs in colorectal
neoplasia. Mol Cancer Res. 1:882–891. 2003.PubMed/NCBI
|
|
26
|
Yu S, Lu Z, Liu C, et al: miRNA-96
suppresses KRAS and functions as a tumor suppressor gene in
pancreatic cancer. Cancer Res. 70:6015–6025. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Allawi HT, Dahlberg JE, Olson S, et al:
Quantitation of microRNAs using a modified Invader assay. RNA.
10:1153–1161. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang CJ, Zhou ZG, Wang L, et al:
Clinicopathological significance of microRNA-31, -143 and -145
expression in colorectal cancer. Dis Markers. 26:27–34. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lu Z, Liu M, Stribinskis V, et al:
MicroRNA-21 promotes cell transformation by targeting the
programmed cell death 4 gene. Oncogene. 27:4373–4379. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang Q, Gumireddy K, Schrier M, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee EJ, Gusev Y, Jiang J, et al:
Expression profiling identifies microRNA signature in pancreatic
cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tong AW, Fulgham P, Jay C, et al: MicroRNA
profile analysis of human prostate cancers. Cancer Gene Ther.
16:206–216. 2009.PubMed/NCBI
|