Introduction
The family with sequence similarity 83, member D
(Fam83D, also known as CHICA) is located on chromosome 20 of the
human genome (1). Fam83D contains
an uncharacterized DUF1669 domain in the N terminus. The members of
this domain family are found in all eukaryotes and are composed of
sequences derived from hypothetical eukaryotic proteins of unknown
function. Some members of this domain family are noted as being
potential phospholipases, but no evidence from literature or
sequence analysis was found to support this (2). Fam83D was identified as a putative
mitotic spindle component in a mass spectrometry study (3). Furthermore, another study revealed
that although Fam83D is primarily found in the cytoplasm during
interphase, during prophase it associates with spindle
microtubules, on which it remains throughout metaphase and anaphase
(4). The same article also revealed
that Fam83D is an interaction partner of chromokinesin KID, which
is required for the generation of polar ejection forces and
chromosome congression, and has roles in organizing the metaphase
plate (4).
As all the mitotic spindle-associated proteins are
involved in the control and regulation of cell proliferation, as
well as in carcinogenesis, we further investigated Fam83D using
in silico tools. Our results revealed that Fam83D is
coexpressed with important mitosis-related genes, including
Aurora-A, Aurora-B, Plk-1, Plk-4, Cdc20, Cdk1, Nek2, Geminin and
CENP family members. All these molecules are well-known genes that
have crucial roles in different stages of mitosis, from equal
segregation of chromosomes to production of daughter cells.
Therefore, we speculate that Fam83D is involved in mitotic
processes to regulate cell division. Moreover, our results also
demonstrated that this gene is differentially expressed in various
cancers in concordance with the previously mentioned coexpression
partners.
This is the first study concerning the correlation
between Fam83D and cancer. It is well-known that differentially
expressed genes in cancers are candidates for diagnostic and
prognostic approaches. Therefore, this article suggests that Fam83D
is a strong candidate for prognostic and diagnostic approaches and
should be investigated further.
Materials and methods
Meta-analysis of Fam83D
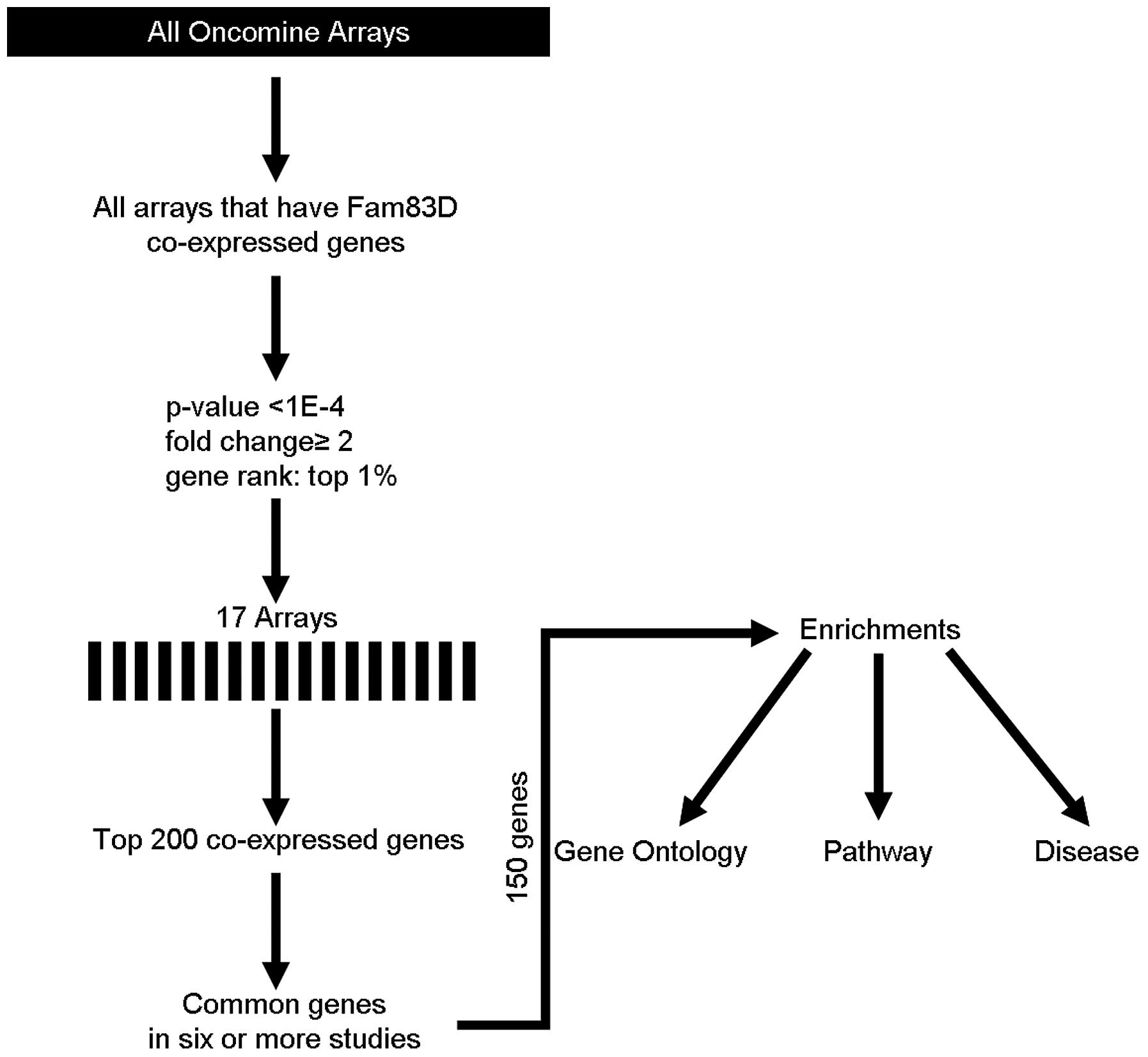
To understand the function of Fam83D, coexpression
analysis was performed using the Oncomine database (http://oncomine.org) as previously described (5,6), but
with minor modifications. The threshold was adjusted to P-value
<1E-4; fold-change, 2 and gene rank, top 1%. Seventeen different
arrays fulfilled these criteria (Table
I) and the top 200 coexpressed genes were extracted and
filtered to give one representative gene per study (removing
duplicates and partial expressed sequence tags). These filtered
gene lists were then compared to search for repeatedly coexpressed
genes over multiple studies. The frequency cut-off was 6 studies
(>30% of 17 studies). This generated a meta-analysis list for
Fam83D. The web-based Database for Annotation, Visualization and
Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov) was used to assess
enriched gene ontology terms within the gene lists produced by the
coexpression data analysis (7,8). The
results were corrected for multiple testing using the Benjamini and
Hochberg false discovery rate (FDR) correction.
 | Table I.Arrays used in coexpression
analysis. |
Table I.
Arrays used in coexpression
analysis.
| No. | Array name |
|---|
| 1 | Lingren
Bladder |
| 2 | Lee Brain |
| 3 | Bittner Breast |
| 4 | Richardson Breast
2 |
| 5 | Meyniel
Ovarian |
| 6 | Lu Breast |
| 7 | HAO Esophagus |
| 8 | Anglesio
Ovarian |
| 9 | Bittner
Multicancer |
| 10 | Janoueix-Lerosey
Brain |
| 11 | Lee Brain 2 |
| 12 | Skrzypczak
Colorectal 2 |
| 13 | Ma Breast 2 |
| 14 | Giordano Adrenal
2 |
| 15 | Yang Renal |
| 16 | Loi Breast 3 |
| 17 | Bittner
Thyroid |
Correlation between Fam83D and
cancer
The oncomine cancer microarray database was used to
study gene expression of Fam83D in various tumor types and in their
normal control tissues. Only the gene transcriptome data from the
same study, generated with the same methodology, were used. All
gene expression data were log-transformed, median-centered per
array, and standard deviation was normalized to one per array
(9). Student’s t-test was used for
differential expression analysis, and only studies with P-value
less than 1E-4 and fold-change greater than two were
considered.
Results
Fam83D is coexpressed with genes involved
in mitosis
Using the Oncomine cancer microarray database Fam83D
was searched for coexpressed genes. Fig. 1 indicates the methodological
workflow of the meta-analysis and the selected multi-array studies
for Fam83D. Following meta-analysis, 150 genes were found to be
coexpressed in six or more studies (Table II). DAVID was used to perform gene
ontology (GO) term enrichment analysis to obtain characteristics of
the set of significant genes from our meta-analyses. This analysis
provides a list of gene functions, which are overrepresented in a
gene set. Analysis of the 150 Fam83D-coexpressed genes with the
DAVID functional annotation tool (GOTERM BP FAT) resulted in 181 GO
categories (cut-off, P<0.05; count ≥2 and fold enrichment
>1.5) (data not shown). To produce a more comprehensive and
structured view of the annotation terms, a DAVID clustering
analysis under high-stringency conditions was performed, resulting
in 42 annotation clusters matching the statistical criteria
(P<0.0001, count ≥10 and fold enrichment >1.5) (Table III). Subsequently, the
aforementioned DAVID annotation tool was used for identification of
putative KEGG pathways associated with Fam83D-coexpressed genes.
Consequently, five pathways associated with the cell cycle, mitosis
and related signaling pathways were significantly enriched with
Fam83D-coexpressed genes (P<0.05 and fold enrichment >1.5)
(Table IV). In addition, DAVID was
used for predicting putative diseases that linked with
Fam83D-coexpressed genes using the Genetic Association Database.
The results revealed that breast and colorectal cancers were
significantly enriched with these genes (P<0.05 and fold
enrichment >1.5) (Table V).
 | Table II.Fam83D-coexpressed genes. |
Table II.
Fam83D-coexpressed genes.
| 1 ANLN | 51 DLGAP5 | 101 MYBL2 |
| 2 APOBEC3B | 52 DSCC1 | 102 NCAPG |
| 3 ATAD2 | 53 DTL | 103 NCAPG2 |
| 4 AURKA | 54 E2F7 | 104 NCAPH |
| 5 AURKB | 55 E2F8 | 105 NDC80 |
| 6 BIRC5 | 56 ECT2 | 106 NEK2 |
| 7 BUB1 | 57 ERCC6L | 107 NUF2 |
| 8 BUB1B | 58 ESPL1 | 108 NUSAP1 |
| 9 C11orf82 | 59 EXO1 | 109 IP5 |
| 10 C15orf42 | 60 EZH2 | 110 PBK |
| 11 C16ORF75 | 61 FAM54A | 111 PHF19 |
| 12 CASC5 | 62 FAM64A | 112 PLK1 |
| 13 CCNA2 | 63 FANCI | 113 PLK4 |
| 14 CCNB1 | 64 FBXO5 | 114 POLE2 |
| 15 CCNB2 | 65 FEN1 | 115 PRC1 |
| 16 CDC20 | 66 FOXM1 | 116 PTTG1 |
| 17 CDC25A | 67 GGH | 117 RACGAP1 |
| 18 CDC25B | 68 GIN | 118 RAD51 |
| 19 CDC25C | 69 GINS2 S1 | 119 RAD54L |
| 20 CDC45 | 70 GINS4 | 120 RECQL4 |
| 21 CDC6 | 71 GMNN | 121 RFC3 |
| 22 CDC7 | 72 GPSM2 | 122 RFC4 |
| 23 CDCA2 | 73 GTSE1 | 123 RNASEH2A |
| 24 CDCA3 | 74 HELLS | 124 RRM2 |
| 25 CDCA5 | 75 HJURP | 125 SGOL2 |
| 26 CDCA7 | 76 HMMR | 126 SHCBP1 |
| 27 CDCA8 | 77 KIAA0101 | 127 SLC7A5 |
| 28 CDK1 | 78 KIF11 | 128 SMC4 |
| 29 CDKN3 | 79 KIF14 | 129 SPAG5 |
| 30 CDT1 | 80 KIF15 | 130 SPC24 |
| 31 CENPA | 81 KIF18B | 131 SPC25 |
| 32 CENPE | 82 KIF20A | 132 STIL |
| 33 CENPF | 83 KIF23 | 133 TACC3 |
| 34 CENPI | 84 KIF2C | 134 TFRC |
| 35 CENPJ | 85 KIF4A | 135 TIMELESS |
| 36 CENPK | 86 KIFC1 | 136 TK1 |
| 37 CENPM | 87 KPNA2 | 137 TOP2A |
| 38 CENPN | 88 LMNB1 | 138 TPX2 |
| 39 CENPW | 89 MAD2L1 | 139 TRIM59 |
| 40 CEP55 | 90 MASTL | 140 TRIP13 |
| 41 CHEK1 | 91 MCM10 | 141 TROAP |
| 42 CKAP2 | 92 MCM2 | 142 TTK |
| 43 CKAP2L | 93 MCM4 | 143 TYMS |
| 44 CKS1B | 94 MCM6 | 144 UBE2C |
| 45 CKS2 | 95 MCM7 | 145 UBE2S |
| 46 DBF4 | 96 MCM8 | 146 UBE2T |
| 47 DEPDC1 | 97 MELK | 147 UHRF1 |
| 48 DEPDC1B | 98 MKI67 | 148 WHSC1 |
| 49 DHFR | 99 MLF1IP | 149 ZNF367 |
| 50 DIAPH3 | 100 MYBL1 | 150 ZWINT |
 | Table III.Functional enrichment of
Fam83D-coexpressed genes. |
Table III.
Functional enrichment of
Fam83D-coexpressed genes.
| Term | Count | % | P-value | Fold | FDR |
|---|
| GO:0007049 - Cell
cycle | 88 | 59.1 | 1.90E-74 | 11.2 | 1.31E-71 |
| GO:0000279 - M
phase | 65 | 43.6 | 9.23E-68 | 19.5 | 3.19E-65 |
| GO:0022403 - Cell
cycle phase | 69 | 46.3 | 3.78E-67 | 16.5 | 8.71E-65 |
| GO:0022402 - Cell
cycle process | 73 | 49 | 2.29E-63 | 12.8 | 3.96E-61 |
| GO:0000278 -
Mitotic cell cycle | 62 | 41.6 | 1.39E-59 | 16.5 | 1.92E-57 |
| GO:0007067 -
Mitosis | 53 | 35.6 | 7.11E-59 | 23.8 | 8.19E-57 |
| GO:0000280 -
Nuclear division | 53 | 35.6 | 7.11E-59 | 23.8 | 8.19E-57 |
| GO:0000087 - M
phase of mitotic cell cycle | 53 | 35.6 | 2.01E-58 | 23.4 | 1.99E-56 |
| GO:0048285 -
Organelle fission | 53 | 35.6 | 7.15E-58 | 22.9 | 6.18E-56 |
| GO:0051301 - Cell
division | 53 | 35.6 | 1.10E-51 | 17.7 | 8.47E-50 |
| GO:0006260 - DNA
replication | 31 | 20.8 | 8.29E-28 | 16.1 | 5.73E-26 |
| GO:0007059 -
Chromosome segregation | 22 | 14.8 | 1.82E-24 | 26.8 | 1.14E-22 |
| GO:0006259 - DNA
metabolic process | 40 | 26.8 | 3.13E-24 | 7.81 | 1.80E-22 |
| GO:0051726 -
Regulation of cell cycle | 33 | 22.1 | 7.82E-23 | 9.84 | 4.16E-21 |
| GO:0007017 -
Microtubule-based process | 29 | 19.5 | 1.31E-21 | 11.3 | 6.46E-20 |
| GO:0007051 -
Spindle organization | 15 | 10.1 | 6.83E-18 | 32.9 | 3.15E-16 |
| GO:0000070 -
Mitotic sister chromatid segregation | 14 | 9.4 | 1.12E-17 | 38.4 | 4.82E-16 |
| GO:0000819 - Sister
chromatid segregation | 14 | 9.4 | 1.71E-17 | 37.4 | 6.93E-16 |
| GO:0007346 -
Regulation of mitotic cell cycle | 21 | 14.1 | 3.98E-17 | 13.6 | 1.53E-15 |
| GO:0010564 -
Regulation of cell cycle process | 19 | 12.8 | 5.90E-17 | 16.5 | 4.00E-15 |
| GO:0000226 -
Microtubule cytoskeleton organization | 20 | 13.4 | 3.60E-16 | 13.4 | 1.15E-14 |
| GO:0000075 - Cell
cycle checkpoint | 15 | 10.1 | 3.02E-13 | 16.3 | 9.93E-12 |
| GO:0051276 -
Chromosome organization | 27 | 18.1 | 1.98E-12 | 5.5 | 6.22E-11 |
| GO:0007126 -
Meiosis | 13 | 8.72 | 2.54E-10 | 13.1 | 7.63E-09 |
| GO:0051327 - M
phase of meiotic cell cycle | 13 | 8.72 | 2.54E-10 | 13.1 | 7.63E-09 |
| GO:0051321 -
Meiotic cell cycle | 13 | 8.72 | 3.23E-10 | 12.8 | 9.29E-09 |
| GO:0007093 -
Mitotic cell cycle checkpoint | 10 | 6.71 | 3.39E-10 | 23 | 9.37E-09 |
| GO:0007010 -
Cytoskeleton organization | 23 | 15.4 | 3.87E-10 | 5.21 | 1.03E-08 |
| GO:0051329 -
Interphase of mitotic cell cycle | 13 | 8.72 | 4.58E-10 | 12.5 | 1.17E-08 |
| GO:0051325 -
Interphase | 13 | 8.72 | 6.43E-10 | 12.1 | 1.59E-08 |
| GO:0006974 -
Response to DNA damage stimulus | 21 | 14.1 | 9.27E-10 | 5.56 | 2.21E-08 |
| GO:0007088 -
Regulation of mitosis | 10 | 6.71 | 4.08E-09 | 17.6 | 9.40E-08 |
| GO:0051783 -
Regulation of nuclear division | 10 | 6.71 | 4.08E-09 | 17.6 | 9.40E-08 |
| GO:0006261 -
DNA-dependent DNA replication | 10 | 6.71 | 5.64E-09 | 17 | 1.26E-07 |
| GO:0008283 - Cell
proliferation | 21 | 14.1 | 1.34E-08 | 4.76 | 2.89E-07 |
| GO:0048015 -
Phosphoinositide-mediated signaling | 11 | 7.38 | 1.75E-08 | 12.3 | 3.67E-07 |
| GO:0006323 - DNA
packaging | 11 | 7.38 | 2.71E-07 | 9.28 | 5.50E-06 |
| GO:0051640 -
Organelle localization | 10 | 6.71 | 3.45E-07 | 10.7 | 6.81E-06 |
| GO:0033554 -
Cellular response to stress | 21 | 14.1 | 9.19E-07 | 3.66 | 1.76E-05 |
| GO:0006281 - DNA
repair | 15 | 10.1 | 1.01E-06 | 5.22 | 1.88E-05 |
| GO:0007018 -
Microtubule-based movement | 10 | 6.71 | 1.98E-06 | 8.74 | 3.61E-05 |
| GO:0033043 -
Regulation of organelle organization | 11 | 7.38 | 6.71E-05 | 5.01 | 0.001188 |
 | Table IV.Pathway-based enrichment of
Fam83D-coexpressed genes. |
Table IV.
Pathway-based enrichment of
Fam83D-coexpressed genes.
| Term | Count | % | P-value | Fold | FDR |
|---|
| hsa04110: Cell
cycle | 24 | 16.1 | 1.16E-25 | 20.3 | 3.24E-24 |
| hsa03030: DNA
replication | 9 | 6.04 | 7.12E-10 | 26.5 | 9.97E-09 |
| hsa04114: Oocyte
meiosis | 12 | 8.05 | 2.66E-09 | 11.6 | 2.48E-08 |
| hsa04914:
Progesterone-mediated oocyte maturation | 10 | 6.71 | 5.97E-08 | 12.3 | 4.18E-07 |
| hsa04115: p53
signaling pathway | 6 | 4.03 | 3.66E-04 | 9.35 | 0.002048 |
 | Table V.Disease-based enrichment of
Fam83D-coexpressed genes. |
Table V.
Disease-based enrichment of
Fam83D-coexpressed genes.
| Term | Count | % | P-value | Fold | FDR |
|---|
| Breast cancer | 13 | 8.7 | 1.91E-06 | 4.9 | 1.39E-04 |
| Colorectal
cancer | 6 | 4.0 | 0.029838 | 3.2 | 0.669009 |
Fam83D is differentially expressed in
various cancers
We investigated the expression of Fam83D in cancer
using publicly available gene expression data from Oncomine
(Table VI). Fam83D has been found
to be upregulated in various tumors including in breast cancer
compared to normal breast (10); in
colorectal cancer compared to normal colon or rectum in three
independent studies (11–13); in gastric cancer compared to gastric
mucosa in two independent studies (14,15);
in hepatocellular carcinoma compared to normal liver in two
independent studies (16,17); in lung cancer compared to normal
lung in two independent studies (18,19)
and in vulva intraepithelial neoplasia compared to normal vulva
(20). Conversely, downregulation
of Fam83D was found in glioblastoma compared to neural stem cells
(21); in esophageal cancer
compared to normal esophagus (22)
and in leukemia compared to peripheral blood mononuclear cells
(23).
 | Table VI.Differential expression of Fam83D in
cancer types compared to their normal counterparts, using the
Oncomine cancer microarray database. |
Table VI.
Differential expression of Fam83D in
cancer types compared to their normal counterparts, using the
Oncomine cancer microarray database.
| Type of cancer | Overexpressed | Underexpressed | Ref. |
|---|
| Breast | + | | (10) |
| Cervical | + | | (20) |
| Colorectal | + | | (11–13) |
| Esophageal | | + | (22) |
| Gastric | + | | (14,15) |
| Glioblastoma | | + | (21) |
| Hepatocellular | + | | (16,17) |
| Leukemia | | + | (23) |
| Lung | + | | (18,19) |
Discussion
The main function of the cell cycle is to accurately
duplicate the entire genome and segregate a copy of each chromosome
precisely into two daughter cells. Maintenance of a correct
chromosome number is essential for the survival of an organism.
Errors in the cell division may lead to loss or gain of chromosomes
and consequently to aneuploidy. In mitotically dividing cells,
aneuploidy is a hallmark of cancer and many cancer cells are
characterized by high rates of chromosomal instability (CIN). CIN
leads to the persistent generation of new chromosomal variations,
to tumor progression and to the development of more aggressive
phenotypes (24). Centrosomes have
important roles in equal segregation of chromosomes through the
establishment of bipolar spindle formation during mitosis. Many
studies have reported that centrosome-located proteins are involved
in the regulation of centrosome organization (25,26).
Moreover, it has been demonstrated that deregulation of the
centrosome organization machinery is a clear source of centrosome
amplification (27). There is a
growing line of evidence to suggest that most solid tumors and many
hematopoietic malignancies contain cells with centrosome
abnormalities (28–30). For example, the centrosomal mitotic
kinases Aurora-A, Plk-1, Plk-4 and Nek2 are all Fam83D-coexpressed
genes (Table II), involved in
multiple mitotic events. These range from centrosome maturation to
centrosome separation, spindle formation and cytokinesis, and their
deregulation has been linked to centrosome abnormalities and
consequently carcinogenesis (31–35).
Therefore, all centrosome and bipolar spindle-associated proteins
are considered as putative cancer-related molecules. Santamaria
et al have demonstrated that Fam83D localizes to the mitotic
spindle, and Fam83D-depleted cells form shorter spindles and fail
to organize a correct metaphase plate (4). In this study, we showed that Fam83D is
coexpressed with many centrosome-located and mitosis-related genes,
which are involved in normal cell cycle progression as well as in
carcinogenesis. Notably, the majority of the coexpressed genes were
key molecules for entry into mitosis, mitotic progression and
cytokinesis. All these processes are related to centrosome
organization and important to the faithful segregation of
chromosomes. Therefore, we suggested that Fam83D may be involved in
equal segregation of chromosomes during mitosis. In concordance
with this hypothesis, our results also revealed that Fam83D is
differentially expressed in some cancers that are directly linked
to centrosome abnormalities, such as bladder (36), breast (37), lung (38), colorectal (30) or hepatocellular (39) carcinomas and leukemia (40).
In conclusion, we performed a meta-analysis for
Fam83D using in silico approaches. Our results revealed that
this molecule may be important for centrosome organization, mitotic
processes and also in carcinogenesis. In silico studies
support wet-lab approaches to finding new diagnostic, therapeutic
and prognostic factors by using various tools, software and
large-scale databases. However, the results of in silico
studies generally need confirmation by lab experiments. Therefore,
further investigation of the results presented in this study by
experimental approaches may increase our understanding of
centrosome organization, mitosis and carcinogenesis.
References
|
1.
|
P DeloukasLH MatthewsJ AshurstThe DNA
sequence and comparative analysis of human chromosome
20Nature414865871200110.1038/414865a11780052
|
|
2.
|
RD FinnJ MistryJ TateThe Pfam protein
families databaseNucleic Acids
Res38D211D222201010.1093/nar/gkp98519920124
|
|
3.
|
G SauerR KornerA HanischA RiesEA NiggHH
SilljeProteome analysis of the human mitotic spindleMol Cell
Proteomics43543200510.1074/mcp.M400158-MCP20015561729
|
|
4.
|
A SantamariaS NagelHH SilljeEA NiggThe
spindle protein CHICA mediates localization of the chromokinesin
Kid to the mitotic spindleCurr
Biol18723729200810.1016/j.cub.2008.04.04118485706
|
|
5.
|
BJ WilsonMeta-analysis of SUMO1BMC Res
Notes160200810.1186/1756-0500-1-60
|
|
6.
|
BJ WilsonV GiguereMeta-analysis of human
cancer microarrays reveals GATA3 is integral to the estrogen
receptor alpha pathwayMol
Cancer749200810.1186/1476-4598-7-4918533032
|
|
7.
|
W Huang daBT ShermanRA LempickiSystematic
and integrative analysis of large gene lists using DAVID
bioinformatics resourcesNat Protoc44457200919131956
|
|
8.
|
W Huang daBT ShermanRA
LempickiBioinformatics enrichment tools: paths toward the
comprehensive functional analysis of large gene listsNucleic Acids
Res37113200919033363
|
|
9.
|
DR RhodesJ YuK ShankerONCOMINE: a cancer
microarray database and integrated data-mining
platformNeoplasia616200410.1016/S1476-5586(04)80047-215068665
|
|
10.
|
AL RichardsonZC WangA De NicoloX
chromosomal abnormalities in basal-like human breast cancerCancer
Cell9121132200610.1016/j.ccr.2006.01.01316473279
|
|
11.
|
Y HongT DowneyKW EuPK KohPY CheahA
‘metastasis-prone’ signature for early-stage mismatch-repair
proficient sporadic colorectal cancer patients and its implications
for possible therapeuticsClin Exp Metastasis2783902010
|
|
12.
|
J Sabates-BellverLG Van der FlierM de
PaloTranscriptome profile of human colorectal adenomasMol Cancer
Res512631275200710.1158/1541-7786.MCR-07-0267
|
|
13.
|
M SkrzypczakK GorycaT RubelModeling
oncogenic signaling in colon tumors by multidirectional analyses of
microarray data directed for maximization of analytical
reliabilityPLoS
One5e13091201010.1371/journal.pone.001309120957034
|
|
14.
|
X ChenSY LeungST YuenVariation in gene
expression patterns in human gastric cancersMol Biol
Cell1432083215200310.1091/mbc.E02-12-083312925757
|
|
15.
|
M D’ErricoE de RinaldisMF BlasiGenome-wide
expression profile of sporadic gastric cancers with microsatellite
instabilityEur J Cancer45461469200919081245
|
|
16.
|
X ChenST CheungS SoGene expression
patterns in human liver cancersMol Biol
Cell1319291939200210.1091/mbc.02-02-0023.12058060
|
|
17.
|
E WurmbachYB ChenG KhitrovGenome-wide
molecular profiles of HCV-induced dysplasia and hepatocellular
carcinomaHepatology45938947200710.1002/hep.2162217393520
|
|
18.
|
ME GarberOG TroyanskayaK SchluensDiversity
of gene expression in adenocarcinoma of the lungProc Natl Acad Sci
USA981378413789200110.1073/pnas.24150079811707590
|
|
19.
|
J HouJ AertsB den HamerGene
expression-based classification of non-small cell lung carcinomas
and survival predictionPLoS
One5e10312201010.1371/journal.pone.001031220421987
|
|
20.
|
LA SantegoetsM SetersTJ HelmerhorstHPV
related VIN: highly proliferative and diminished responsiveness to
extracellular signalsInt J
Cancer121759766200710.1002/ijc.2276917471573
|
|
21.
|
J LeeS KotliarovaY KotliarovTumor stem
cells derived from glioblastomas cultured in bFGF and EGF more
closely mirror the phenotype and genotype of primary tumors than do
serum-cultured cell linesCancer
Cell9391403200610.1016/j.ccr.2006.03.03016697959
|
|
22.
|
SM KimYY ParkES ParkPrognostic biomarkers
for esophageal adenocarcinoma identified by analysis of tumor
transcriptomePLoS
One5e15074201010.1371/journal.pone.001507421152079
|
|
23.
|
T HaferlachA KohlmannL WieczorekClinical
utility of microarray-based gene expression profiling in the
diagnosis and subclassification of leukemia: report from the
International Microarray Innovations in Leukemia Study GroupJ Clin
Oncol2825292537201010.1200/JCO.2009.23.4732
|
|
24.
|
LA LoebA mutator phenotype in cancerCancer
Res6132303239200111309271
|
|
25.
|
O CizmeciogluM ArnoldR BahtzCep152 acts as
a scaffold for recruitment of Plk4 and CPAP to the centrosomeJ Cell
Biol191731739201010.1083/jcb.20100710721059844
|
|
26.
|
O CizmeciogluS WarnkeM ArnoldS DuensingI
HoffmannPlk-2 regulated centriole duplication is dependent on its
localization to the centrioles and a functional polo-box domainCell
Cycle735483555200810.4161/cc.7.22.707119001868
|
|
27.
|
D ZyssF GergelyCentrosome function in
cancer: guilty or innocent?Trends Cell
Biol19334346200910.1016/j.tcb.2009.04.00119570677
|
|
28.
|
BR BrinkleyManaging the centrosome numbers
game: from chaos to stability in cancer cell divisionTrends Cell
Biol111821200110.1016/S0962-8924(00)01872-911146294
|
|
29.
|
PE CarrollM OkudaHF HornCentrosome
hyperamplification in human cancer: chromosome instability induced
by p53 mutation and/or Mdm2
overexpressionOncogene1819351944199910.1038/sj.onc.120251510208415
|
|
30.
|
GA PihanA PurohitJ WallaceCentrosome
defects and genetic instability in malignant tumorsCancer
Res583974398519989731511
|
|
31.
|
R HabedanckYD StierhofCJ WilkinsonEA
NiggThe Polo kinase Plk-4 functions in centriole duplicationNat
Cell Biol711401146200510.1038/ncb132016244668
|
|
32.
|
DG HaywardAM FryNek-2 kinase in chromosome
instability and cancerCancer
Lett237155166200610.1016/j.canlet.2005.06.01716084011
|
|
33.
|
LY LuJL WoodL YeAurora-A is essential for
early embryonic development and tumor suppressionJ Biol
Chem2833178531790200810.1074/jbc.M80588020018801727
|
|
34.
|
XQ WangYQ ZhuKS LuiQ CaiP LuRT
PoonAberrant Polo-like kinase 1-Cdc25A pathway in metastatic
hepatocellular carcinomaClin Cancer
Res1468136820200810.1158/1078-0432.CCR-08-0626
|
|
35.
|
LY LuJL WoodK Minter-DykhousePolo-like
kinase 1 is essential for early embryonic development and tumor
suppressionMol Cell
Biol2868706876200810.1128/MCB.00392-0818794363
|
|
36.
|
Y YamamotoH MatsuyamaT FuruyaCentrosome
hyperamplification predicts progression and tumor recurrence in
bladder cancerClin Cancer
Res1064496455200410.1158/1078-0432.CCR-04-077315475431
|
|
37.
|
WL LingleWH LutzJN IngleNJ MaihleJL
SalisburyCentrosome hypertrophy in human breast tumors:
implications for genomic stability and cell polarityProc Natl Acad
Sci USA9529502955199810.1073/pnas.95.6.29509501196
|
|
38.
|
CK JungJH JungKY LeeCentrosome
abnormalities in non-small cell lung cancer: correlations with DNA
aneuploidy and expression of cell cycle regulatory proteinsPathol
Res Pract203839847200710.1016/j.prp.2007.08.00417913384
|
|
39.
|
T NakajimaM MoriguchiY MitsumotoCentrosome
aberration accompanied with p53 mutation can induce genetic
instability in hepatocellular carcinomaMod
Pathol17722727200410.1038/modpathol.3800115
|
|
40.
|
M GiehlA FabariusO FrankCentrosome
aberrations in chronic myeloid leukemia correlate with stage of
disease and chromosomal
instabilityLeukemia1911921197200510.1038/sj.leu.240377915858613
|