Introduction
Breast cancer is the most commonly occurring
malignancy in women. Worldwide, it is estimated that more than one
million women are diagnosed with breast cancer each year and
>410,000 succumb to the disease (1). Despite great improvements over the
past decades in conventional breast cancer treatment approaches,
including surgery, radiotherapy, chemotherapy and endocrine
therapy, breast cancer recurrence, metastasis and mortality rates
remain high. With the considerable progress in molecular cancer
biology, targeted therapy is emerging as the most promising
strategy in cancer therapeutics. In breast cancer, the use of
trastuzumab treatment in Her-2-positive patients has been
considered as a successful example of modern oncology (2). However, given the complexity of breast
cancer biology itself and the limited application range of each
targeted therapy, the exploration of new cancer targets with the
aim of developing novel therapeutic approaches remains a pressing
need in breast cancer research.
Synuclein-γ (SNCG) is a human gene localized at
10q23.20–23.3. SNCG cDNA is ∼5 kb in length and comprised of five
exons that translate into a protein of 127 amino acids (3,4). The
SNCG protein belongs to the family of synuclein proteins (5). Synucleins are small soluble proteins
expressed primarily in the neural tissue and certain tumors
(6). The three synucleins that
comprise the entire protein family (α-synuclein, β-synuclein and
SNCG) are characterized by a highly conserved α-helical
lipid-binding motif with similarity to the class-A2 lipid-binding
domains of the exchangeable apolipoproteins (7). Members of this family are considered
to have important pathogenic roles in various neurodegenerative
disorders, including Parkinson’s disease, dementia with Lewy bodies
and Alzheimer’s disease (8–10). Since SNCG was first observed to be
overexpressed in advanced breast carcinoma (11), but not in normal or benign breast
tissue, it was previously named breast cancer specific gene 1
(BCSG1). At present, accumulating evidence has shown that the
over-expression of this gene is also implicated in numerous other
cancer types, including ovarian, lung, liver, esophagus, colon and
prostate (12–15), and correlated with the later stages
of the diseases. In addition, SNCG was also suggested to be
associated with antimicrotubule drug resistance in breast cancer
treatment (16). Gene function
studies further indicated that SNCG is associated with tumor
progression through a series of mechanisms which include the
stimulation or promotion of cell proliferation, invasion and
metastasis (17–20). All this evidence indicated that SNCG
is a tumor marker and a knockdown of SNCG oncogene expression may
be an effective strategy in breast cancer treatment.
RNA interference (RNAi) is a specific gene silencing
phenomenon mediated by double-stranded RNA, which is an ancient and
highly conserved gene regulatory mechanism (21). Since its discovery, RNAi technology
has rapidly become an effective gene-silencing tool and has been
broadly applied in a number of biological research fields. The
double-stranded small RNAi molecule functions in the nanomolar
range and is far more effective than the antisense approach which
was popular 10–15 years ago. In cancer research, RNAi intervention
has been used for targeting numerous gene products involved in
carcinogenesis, including key molecules crucial for cancer cell
proliferation, tumor-host interactions and tumor resistance to
chemo- or radiotherapy. With the ever-evolving understanding of
molecular cancer biology, great opportunities for using RNAi
technology in cancer therapy are rapidly being created. The
selection of appropriate gene targets is also becoming a critical
parameter for the potential success of RNAi-based cancer
therapies.
In the present study, a vector-based RNAi strategy
was used to suppress the expression of SNCG in the breast cancer
MCF-7 cell line and its effects on the clonogenicity and invasion
of the transfected cells were evaluated. It was observed that the
specific downregulation of SNCG was achieved at the mRNA and
protein levels by this approach and the clonogenic and invasion
capabilities of the cancer cells were significantly diminished. We
concluded that SNCG suppression mediated by RNAi may become an
effective therapeutic approach for the treatment of breast
cancer.
Materials and methods
Cell culture
The human breast cancer MCF-7 cells were purchased
from Nanjing Kenou Biology Co. Ltd. (Nanjing, China) and grown in
RPMI-1640 culture medium containing 10% fetal bovine serum (FBS),
100 U/ml penicillin and 100 U/ml streptomycin in 75 cm2
flasks and incubated in a humidified incubator (37°C, 5%
CO2). The study was approved by the Institutional Review
Board of Zhengzhou University, Zhengzhou, China.
Construction of SNCG RNAi plasmid
expression vectors
Based on the full SNCG cDNA sequence, four pairs of
siRNA DNA chains were designed using Ambion and Takara siRNA design
programs to produce the following sequences: 5′-CCAA
GGAGAATGTTGTACAGA-3′, 5′-CAAGACCAAGGAGAAT GTTGT-3′,
5′-GCCAAGACCAAGGAGAATGTT-3′ and 5′-TGGTGAGCAGCGTCAACACTG-3′. The
negative control siRNA was designed as 5′-GTTCTCCGAACGTGTCAC GT-3′,
a sequence with no homology to human or mouse SNCG. All designed
sequences were then synthesized and constructed into pGPU6 vector
(Shanghai GenePharma Co. Ltd, Shanghai, China).
Transfection of plasmid DNA and selection
of stable cell lines
MCF-7 cells were seeded in a 24-well plate one day
prior to transfection and transfected with 1.6 μg SNCG RNAi
plasmid DNA and 4 μl transfection reagent Lipofectamine 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA) when the cells
reached ∼90% confluence. Together with the four SNCG specific RNAi
plasmids, transfection of a non-specific RNAi negative control
plasmid, an empty vector control plasmid and the Lipofectamine 2000
control were also performed. After 48 h, the transfected cells were
plated on to 6-well plates and selected with G418 at 800 mg/l.
Fresh culture media containing G418 was replaced every two or three
days and stable cell lines were established in ∼2 weeks. The RNAi
expression status in all stable lines was visually monitored under
microscope by GFP signal.
Immunocytochemistry assay
The SNCG expression level was detected using the SP
hypersensitivity kit (Beijing Biosynthesis Biotechnology Co., Ltd.,
Beijing, China) according to the manufacturer’s instructions. SNCG
primary antibody was purchased from Santa Cruz Biotechnology Inc.
(Santa Cruz, CA, USA) and PBS was applied as negative control.
Positive SNCG staining was shown as yellow to brown signals located
in the cytoplasm. Under a light microscope, 10 high power fields
were randomly selected and scored with the following criteria
(22): a) for the extent of
positive staining: 0 = positive cells <1%, 1 = 1–25%, 2 =
26–50%, 3 = 51–75% and 4 = 76–100%; b) for staining intensity: 0 =
no color, 1 = pale-yellow, 2 = brown to yellow, 3 = tan. The final
score was equal to a × b and was used for statistical analysis.
Reverse transcription polymerase chain
reaction (RT-PCR) assay
Cells (5×105) from each SNCG RNAi-stable
cell line and negative controls were used for total RNA extraction
using the TRIzol reagent (Invitrogen Life Technologies). The RNA
concentration and quality were determined with an ultraviolet
spectrophotometer. The reverse transcription reaction was performed
using the RT-PCR kit purchased from Invitrogen Life Technologies.
The primer sequences used for SNCG detection were as follows:
upstream primer, 5′-CAAGAAGGG CTTCTCCATCGCCAAGG-3′ and downstream
primer, 5′-CCTCTTTCTCTTTGGATGCCACACCC-3′. ACTB was used as
the internal control and the primer sequences were: upstream
primer, 5′-GATGACGATATCGCTGCGCT-3′ and downstream primer,
5′-CTAGGCACCAGGTAAGTGAC-3′. The PCR amplification reaction was as
follows: 94°C for 30 sec, 55°C for 30 sec, 72°C for 1 min (for 30
cycles) and a final extension at 72°C for 10 min. The PCR products
were analyzed by electrophoresis on a 1% agarose gel and were
visualized by ethidium bromide staining under ultraviolet light.
The expression level of SNCG was indicated by the photodensity of
the PCR band on the image and normalized with the corresponding
internal ACTB band intensity for each sample.
Clonogenic assay
Along with parallel MCF-7 cells, empty vector and
non-specific transfection cells, SNCG-RNAi-3-stable cells (with the
most potent SNCG knockdown efficiency) were grown in RPMI-1640
media (with 10% FBS) to 70% confluence, trypsinized and counted.
Cells (2×104) were seeded on 60-mm tissue culture dishes
(in triplicate) and incubated for 14 days. Fresh media were added
every three to five days. At day 14, the culture media were removed
and the cells were washed with PBS three times. Cells were fixed
with methanol for 15 min at room temperature, followed by
hemotoxylin staining for 2–3 min and washed with PBS. All colonies
with a cell count >50 were identified under a microscope. The
clonogenic rate was calculated as: average colony number / seeded
cell number × 100.
Boyden chamber cell invasiveness
assay
The cell invasion assay was performed using Boyden
chambers (Corning Co., Corning, NY, USA). Cells (5×105)
from each stable and control cell line were suspended in 100
μl serum-free medium and placed into the upper compartments
of the Boyden chambers. The lower compartments of the chambers were
filled with 200 μl serum-containing medium and the cells
were allowed to migrate for 24 h. Following the incubation, the
cells on the lower surface of the filter were fixed in cold ethanol
and stained with 0.5% crystal violet (CV) for 30 min. Five random
fields were then selected and counted at magnification ×200. Data
representing the average cell numbers of the five fields were
compared between all the experimental and control groups.
Statistical analysis
All results were expressed as the mean ± SD.
Statistical analyses were performed using SPSS 10.0 statistical
software to perform t-tests. All statistical analyses were
two-sided and comparisons were made in which P<0.05 was
considered to indicate statistically significant differences.
Results
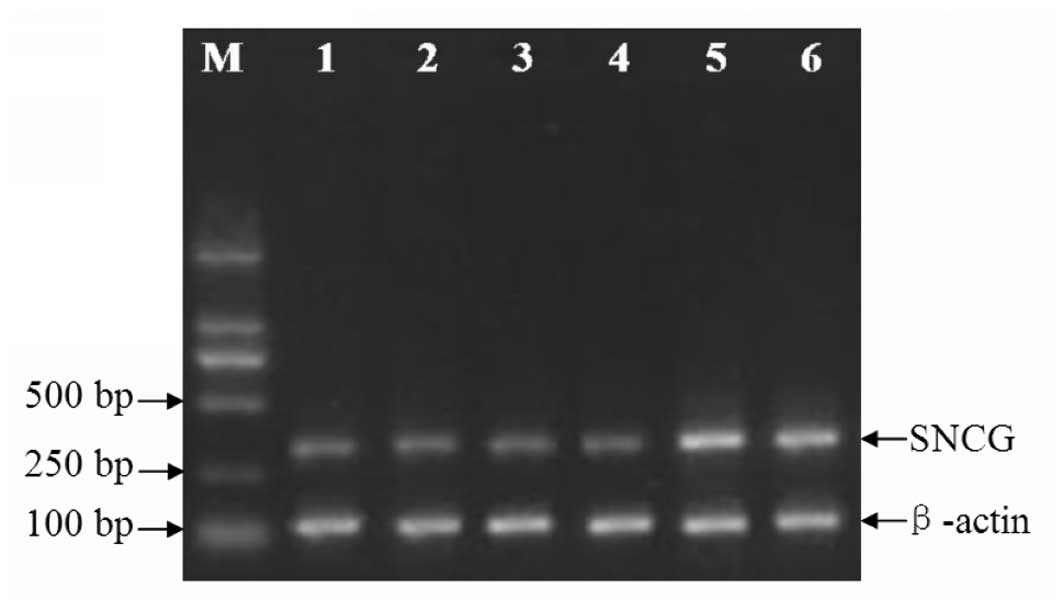
SNCG RNAi significantly inhibits SNCG
mRNA expression
Four SNCG RNAi expression plasmids (SNCG-siRNA-1,
-2, -3 and -4) and a non-specific plasmid were constructed in
pGPU-6 vectors. After the corresponding stable cell lines were
established, the SNCG mRNA expression levels were measured with
RT-PCR and compared among the groups. As demonstrated in Fig. 1 and Table I, SNCG mRNA expression was
significantly diminished in the four SNCG RNAi groups in comparison
with the parallel control (P<0.05), whereas the levels stayed
almost unchanged in the non-specific RNAi and empty vector groups
(P>0.05).
 | Table IExpression of SNCG mRNA in MCF-7 cells
(mean ± SD). |
Table I
Expression of SNCG mRNA in MCF-7 cells
(mean ± SD).
| Transfected
construct | SNCG mRNA
concentration |
|---|
| SNCG-siRNA-1 | 0.624±0.010a |
| SNCG-siRNA-2 | 0.626±0.013a |
| SNCG-siRNA-3 | 0.634±0.008a |
| SNCG-siRNA-4 | 0.631±0.010a |
| Non-specific siRNA
control | 0.976±0.076b |
| Empty vector
control | 0.983±0.052b |
| F | 147.42 |
| P-value | <0.001 |
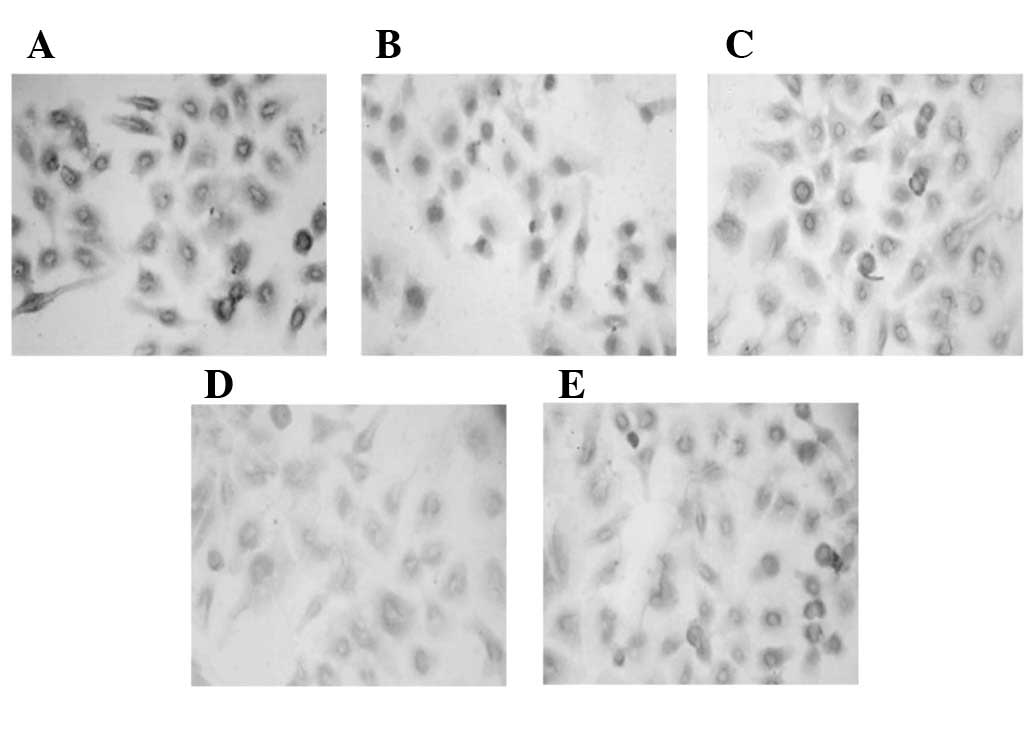
SNCG RNAi significantly inhibits SNCG
protein expression
The SNCG protein expression levels were assessed by
immunocytochemistry for all the stable cell lines, followed by
statistical analyses. In accordance with the results for SNCG
expression at the mRNA level, SNCG protein expression was also
significantly reduced in the four SNCG RNAi groups (P<0.05), in
comparison with the parallel controls (Fig. 2 and Table II).
 | Table IIExpression of SNCG protein in MCF-7
cells (mean ± SD). |
Table II
Expression of SNCG protein in MCF-7
cells (mean ± SD).
| Transfected
construct | SNCG protein
concentration |
|---|
| SNCG-siRNA-1 | 4.27±0.12a |
| SNCG-siRNA-2 | 4.19±0.22a |
| SNCG-siRNA-3 | 4.15±0.14a |
| SNCG-siRNA-4 | 4.17±0.13a |
| Non-specific siRNA
control | 7.92±0.22b |
| Empty vector
control | 8.02±0.13b |
| F | 481.06 |
| P-value | <0.001 |
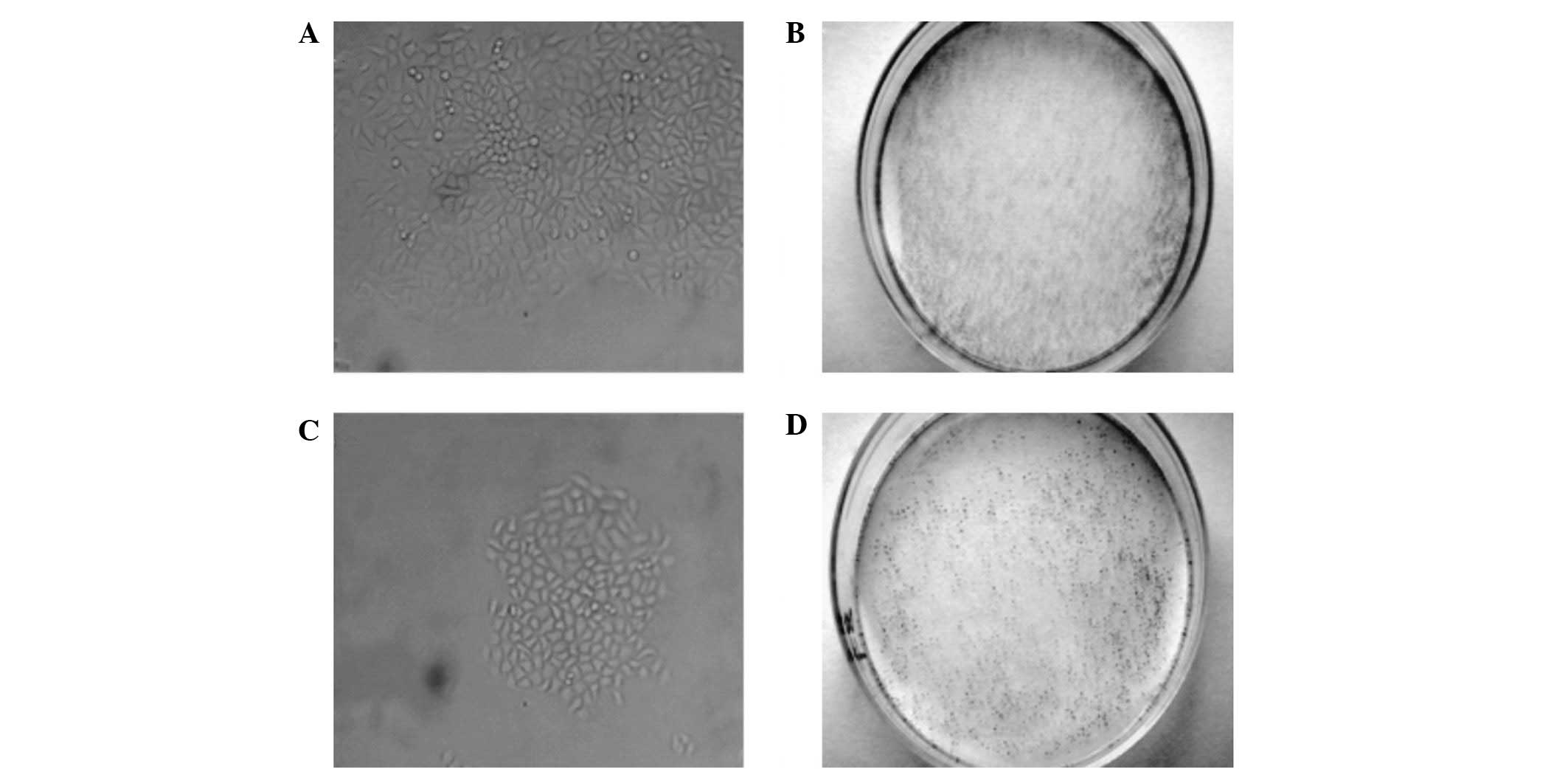
SNCG suppression mediated by RNAi
significantly diminishes the clonogenicity of transfected MCF-7
cells
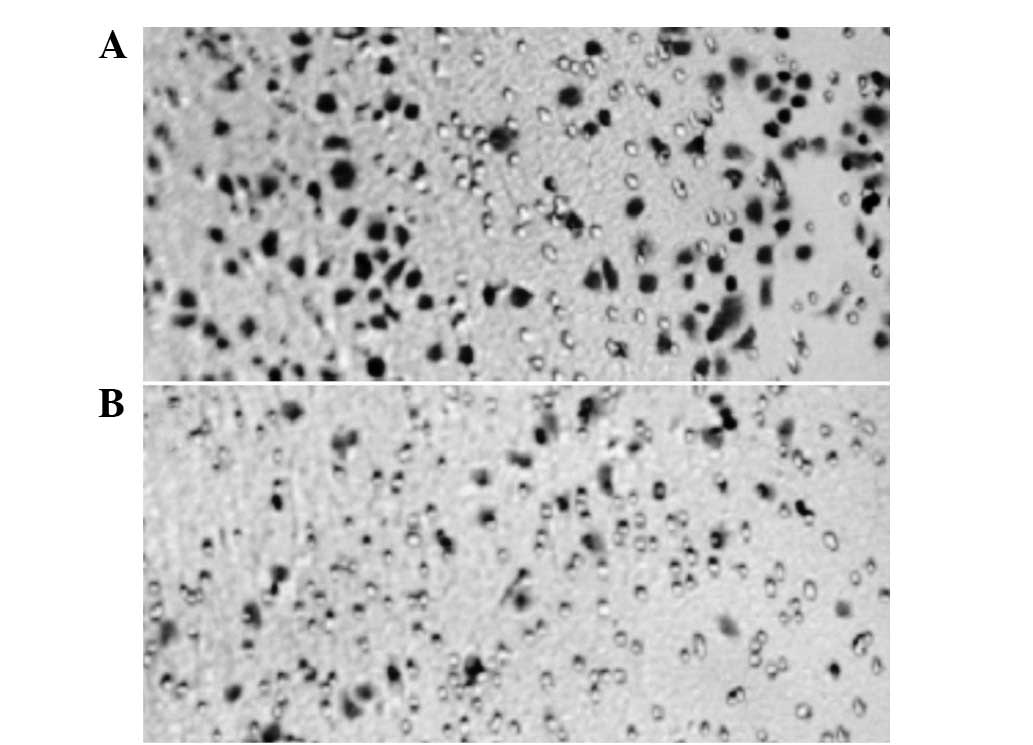
To investigate the effects of SNCG suppression
mediated by RNAi on the clonogenicity of MCF-7 cells, the
SNCG-siRNA-3-stable cell line (with the most potent SNCG knock-down
efficiency) was used for comparison with the parallel and other
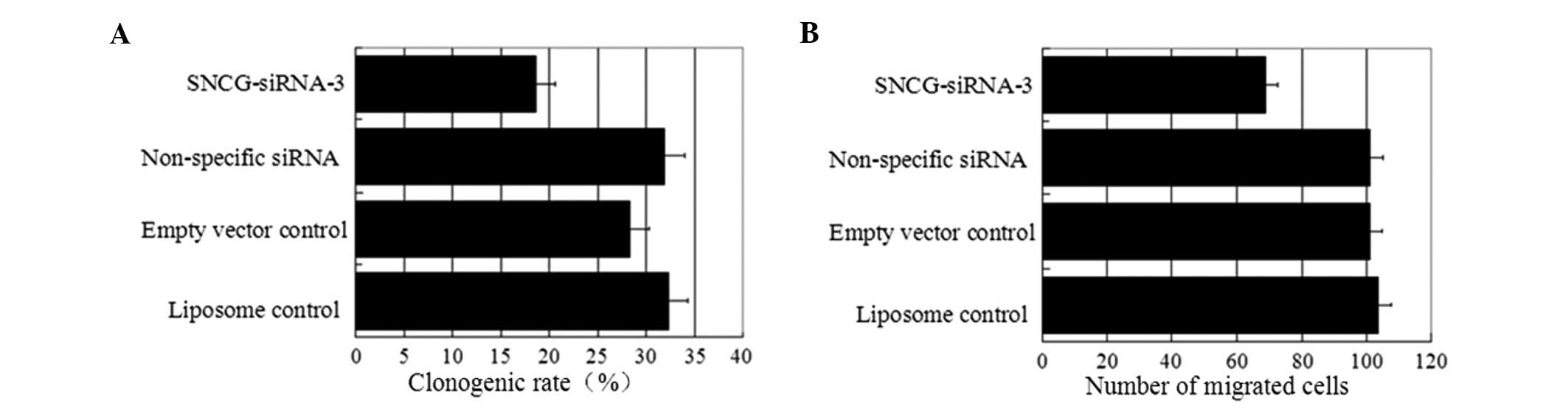
transfection controls. As demonstrated in Figs. 3 and 5A, the sizes and number of colonies formed
in the SNCG-siRNA-3 line were observed to be smaller and lower than
in the parallel control, respectively. The clonogenic rate was
significantly reduced (P<0.05) in comparison with the controls
(Table III), whereas no significant
difference was observed in the non-specific and empty vector
controls compared with the parallel control.
 | Table IIIEffects of SNCG-siRNA suppression on
clonogenicity in MCF-7 cells. |
Table III
Effects of SNCG-siRNA suppression on
clonogenicity in MCF-7 cells.
| Group | Clonogenic rate
(%) |
|---|
| Liposome
control | 32.33±2.52 |
| Empty vector
control | 28.33±3.51 |
| Non-specific siRNA
control | 32.00±4.36 |
| SNCG-siRNA-3 | 18.67±1.53a |
| F | 12.189 |
| P-value | 0.002 |
SNCG suppression mediated by RNAi
significantly inhibits the invasiveness of transfected MCF-7
cells
The effects of SNCG suppression mediated by RNAi on
cell invasiveness were assessed by Boyden chamber assays. As shown
in Figs. 4 and 5B, the number of migrated cells was
observed to be lower in the SNCG-siRNA-3 line compared with the
parallel control. The differences in invasiveness between
SNCG-siRNA-3 and all other controls were significant, as
demonstrated in Table IV, whereas
no significant differences were observed among the controls.
 | Table IVEffects of SNCG-siRNA suppression in
MCF-7 cells invasiveness assessed by Boyden chamber assays. |
Table IV
Effects of SNCG-siRNA suppression in
MCF-7 cells invasiveness assessed by Boyden chamber assays.
| Group | Number of migrated
cells |
|---|
| Liposome
control | 103.53±1.33 |
| Empty vector
control | 100.93±1.81 |
| Non-specific siRNA
control | 101.20±1.71 |
| SNCG-siRNA-3 | 68.80±10.43a |
| F | 572.108 |
| P-value | <0.001 |
Discussion
SNCG has been shown to be an unfavorable prognostic
marker in a variety of cancer types (11–15).
In breast cancer, the expression of SNCG was observed to be closely
correlated with the disease stage, lymph node involvement,
metastasis, tumor size and Her-2 status, but not with estrogen
receptor (ER) and progesterone (PR) expression status (23). Overexpression of SNCG in breast
cancer cells leads to the significant stimulation of cell
proliferation (19), increased
invasiveness and profound augmentation of metastasis in nude mice
(24). Moreover, SNCG was also
reported to be correlated with ERα overexpression (20), antimicrotubule drug resistance
(25) and an accelerated rate of
chromosomal instability (26). All
these findings suggest that SNCG may be a potential therapeutic
target in breast cancer treatment.
Several strategies using growth factor inhibitor and
antagonistic peptides for targeting aberrant SNCG expression have
been investigated (27,28). Although they have been reported as
having a level of effectiveness, the clinical application of these
approaches remains far away. Given that RNAi is such a
well-developed technology and effective delivery systems for RNAi
are under intensive investigation, we proposed that it would be
rational to study an SNCG targeting strategy mediated by RNAi
instead of other systems. In the present study, a series of SNCG
siRNA sequences were designed and validated, constructed into siRNA
expression vectors and introduced into MCF-7 cells. The present
results show that all these small molecules were able to
specifically and significantly suppress the SNCG expression at the
mRNA and protein levels. At the maximum response, 35.81 and 48.25%
reductions in SNCG mRNA and protein expression, respectively, were
observed in stable SNCG RNAi lines. By contrast, SNCG expression
was almost unchanged in the control groups.
On the basis of the significant suppressive effects
on SNCG expression, the effects of these SNCG RNAi molecules on the
clonogenicity and invasiveness of breast cancer cells were further
investigated. In comparison with the parallel MCF-7 control, as
much as 50.82% inhibition in clonogenicity and 44.33% in
invasiveness was observed in SNCG RNAi-transfected MCF-7 cells,
indicating that successful SNCG downregulation was able to
attenuate breast cancer malignancy and may possibly be a novel
approach for cancer treatment.
The mechanisms underlying SNCG’s involvement in
breast cancer pathogenesis have been studied in previous studies.
In addition to its involvement in the checkpoint complex (26), SNCG has also been shown to stimulate
the ligand-dependent transcriptional activity of ER in breast
cancer cells (20). It has also
been shown that SNCG stimulates ER signaling by acting as a
chaperone through a multi-protein chaperone complex with Hsp70 and
Hsp90 (19). Given the essential
role of estrogen in modulating the cellular growth and
differentiation of the mammary gland, any factors affecting this
signaling pathway should be considered to be critical in breast
cancer pathogenesis. Moreover, the overexpression of SNCG was
demonstrated to correlate with elevated levels of matrix
metalloproteases (MMPs) (20,24,29), a
well recognized mechanism associated with the characteristic
invasive and metastatic properties of cancer cells (29).
Taken together, the present findings that SNCG
suppression mediated by RNAi may significantly diminish breast
cancer cell malignancy in terms of clonogenicity and invasiveness,
suggested that SNCG may be exploited as a therapeutic target
(30–33) and have the potential to become a
novel approach in breast cancer treatment. However, it should be
considered that although the RNAi strategy is effective and
promising for gene-specific targeting and new RNAi delivery methods
are under intensive investigation, efficient RNAi delivery methods
are still lacking. Additionally, the phenomena of off-target RNAi
is also commonly encountered in practice and any RNAi-based
medicine must be carefully evaluated prior to its use in
clinics.
Acknowledgements
The present study was supported by the
Specified Key Research Grant of Henan Province (No. 92102310224)
and Zhengzhou Committee of Science and Technology (No.
10LJRC173).
References
|
1
|
Coughlin SS and Ekwueme DU: Breast cancer
as a global health concern. Cancer Epidemiol. 33:315–318. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garnock-Jones KP, Keating GM and Scott LJ:
Trastuzumab: A review of its use as adjuvant treatment in human
epidermal growth factor receptor 2 (HER2)-positive early breast
cancer. Drugs. 70:215–239. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lavedan C, Leroy E, Dehejia A, et al:
Identification, localization and characterization of the human
gamma-synuclein gene. Hum Genet. 103:106–112. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Surgucheva I, Weisman AD, Goldberg JL, et
al: γ-Synuclein as a marker of retinal ganglion cells. Mol Vis.
14:1540–1548. 2008.
|
|
5
|
George JM: The synucleins. Genome Biol 3:
REVIEWS3002. 2002
|
|
6
|
Ahmad M, Attoub S, Singh MN, et al:
Gamma-synuclein and the progression of cancer. FASEB. 21:3419–3430.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shen PH, Fan QX, Li YW, et al: SNCG shRNA
suppressed breast cancer cell xenograft formation and growth in
nude mice. Chinese Med J (Engl). 124:1524–1528. 2011.PubMed/NCBI
|
|
8
|
Lavedan C: The synuclein family. Genome
Res. 8:871–880. 1998.
|
|
9
|
Von Bohlen und Halbach O: Synucleins and
their relationship to Parkinson’s disease. Cell Tissue Res.
318:163–174. 2004.
|
|
10
|
Iwai A: Properties of NACP/alpha-synuclein
and its role in Alzheimer’s disease. Biochim Biophys Acta.
1502:95–109. 2000.PubMed/NCBI
|
|
11
|
Ji H, Liu Y, Jia T, et al: Identification
of a breast cancer-specific gene, BCSG1, by direct differential
cDNA sequencing. Cancer Res. 57:759–764. 1997.PubMed/NCBI
|
|
12
|
Bruening W, Giasson BI, Klein-Szanto AJ,
et al: Synucleins are expressed in the majority of breast and
ovarian carcinomas and in preneoplastic lesions of the ovary.
Cancer. 88:2154–2163. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao W, Liu H, Liu W, et al: Abnormal
activation of the synuclein-gamma gene in hepatocellular carcinomas
by epigenetic alteration. Int J Oncol. 28:1081–1088.
2006.PubMed/NCBI
|
|
14
|
Zhou CQ, Liu S, Xue LY, et al:
Down-regulation of gamma-synuclein in human esophageal squamous
cell carcinoma. World J Gastroenterol. 9:1900–1903. 2003.PubMed/NCBI
|
|
15
|
Liu H, Liu W, Wu Y, et al: Loss of
epigenetic control of synuclein-gamma gene as a molecular indicator
of metastasis in a wide range of human cancers. Cancer Res.
65:7635–7643. 2005.PubMed/NCBI
|
|
16
|
Singh VK and Jia Z: Targeting
synuclein-gamma to counteract drug resistance in cancer. Expert
Opin Ther Targets. 12:59–68. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Spence MJ, Zhang LY, et al:
Transcriptional suppression of synuclein gamma (SNCG) expression in
human breast cancer cells by the growth inhibitory cytokine
oncostatin M. Breast Cancer Res Treat. 62:99–107. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lu A, Zhang F, Gupta A and Liu L: Blockade
of AP1 trans-activation abrogates the abnormal expression of breast
cancer-specific gene 1 in breast cancer cells. J Biol Chem.
277:31364–31372. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Y, Liu Y, Goldberg ID and Shi YE:
Gamma synuclein, a novel heat-shock protein-associated chaperone,
stimulates ligand-dependent estrogen receptor alpha signaling and
mammary tumorigenesis. Cancer Res. 64:4539–4546. 2004. View Article : Google Scholar
|
|
20
|
Jiang Y, Liu Y, Lu A, et al: Stimulation
of estrogen receptor signaling by gamma synuclein. Cancer Res.
63:3899–3903. 2003.PubMed/NCBI
|
|
21
|
Ashihara E, Kawata E and Maekawa T: Future
prospect of RNA interference for cancer therapies. Curr Drug
Targets. 11:345–360. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krajewska M, Krajewski S, Epstein JI, et
al: Immunohistochemical analysis of bcl-2, bax, bcl-X, and mcl-1
expression in prostate cancers. Am J Pathol. 148:1567–1576.
1996.PubMed/NCBI
|
|
23
|
Guo J, Shou C, Meng L, et al: Neuronal
protein synuclein gamma predicts poor clinical outcome in breast
cancer. Int J Cancer. 121:1296–1305. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jia T, Liu YE, Liu J and Shi YE:
Stimulation of breast cancer invasion and metastasis by synuclein
gamma. Cancer Res. 59:742–747. 1999.PubMed/NCBI
|
|
25
|
Gupta A, Inaba S, Wong OK, et al: Breast
cancer-specific gene 1 interacts with the mitotic checkpoint kinase
BubR1. Oncogene. 22:7593–7599. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Inaba S, Li C, Shi YE, et al: Synuclein
gamma inhibits the mitotic checkpoint function and promotes
chromosomal instability of breast cancer cells. Breast Cancer Res
Treat. 94:25–35. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Singh VK, Zhou Y, Marsh JA, et al:
Synuclein-gamma targeting peptide inhibitor that enhances
sensitivity of breast cancer cells to antimicrotubule drugs. Cancer
Res. 67:626–633. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Y, Inaba S and Liu J: Inhibition of
synuclein-γ expression increases the sensitivity of breast cancer
cells to paclitaxel treatment. Int J Oncol. 29:289–295. 2006.
|
|
29
|
Surgucheva IG, Sivak JM, Fini ME, et al:
Effect of gamma-synuclein overexpression on matrix
metalloproteinases in retinoblastoma Y79 cells. Arch Biochem
Biophys. 410:167–176. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu H, Liu W, Wu Y, et al: Loss of
epigenetic control of synuclein-gamma gene as a molecular indicator
of metastasis in a wide range of human cancers. Cancer Res.
65:7635–7643. 2005.PubMed/NCBI
|
|
31
|
Martin TA, Gomez K, Watkins G, et al:
Expression of breast cancer specific gene-1 (BCSG-1/γ-synuclein) is
associated with tumor grade but not with clinical outcome of
patients with breast cancer. Oncol Rep. 16:207–212. 2006.PubMed/NCBI
|
|
32
|
Wu K, Weng Z, Tao Q, et al: Stage specific
expression of breast cancer specific gene γ-synuclein. Cancer
Epidemiol Biomark Prev. 12:920–925. 2003.
|
|
33
|
Dang RL, Shen FH, Xia QA, et al:
Expression of γ-synuclein protein in ovarian cancer. Mod Prev Med.
36:3551–3553. 2009.
|