|
1.
|
No authors listed:. Tamoxifen for early
breast cancer: an overview of the randomized trials. Early Breast
Cancer Trialists’ Collaborative Group. Lancet. 351:1451–1467.
1998.
|
|
2.
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Bartel DP and Chen CZ: Micromanagers of
gene expression: the potentially widespread influence of metazoan
microRNAs. Nat Rev Genet. 5:396–400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ventura A and Jacks T: MicroRNAs and
cancer: short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Miller TE, Ghoshal K, Ramaswamy B, et al:
MicroRNA-221/222 confers tamoxifen resistance in breast cancer by
targeting p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Xin F, Li M, Balch C, et al: Computational
analysis of microRNA profiles and their target genes suggests
significant involvement in breast cancer antiestrogen resistance.
Bioinformatics. 25:430–434. 2009. View Article : Google Scholar
|
|
11.
|
Clark GM, Osborne CK and McGuire WL:
Correlations between estrogen receptor, progesterone receptor, and
patient characteristics in human breast cancer. J Clin Oncol.
2:1102–1109. 1984.
|
|
12.
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999.
|
|
13.
|
No authors listed:. Systemic treatment of
early breast cancer by hormonal, cytotoxic, or immune therapy. 133
randomised trials involving 31,000 recurrences and 24,000 deaths
among 75,000 women. Early Breast Cancer Trialists’ Collaborative
Group. Lancet. 339:1–15. 1992.
|
|
14.
|
El-Ashry D, Miller DL, Kharbanda S,
Lippman ME and Kern FG: Constitutive Raf-1 kinase activity in
breast cancer cells induces both estrogen-independent growth and
apoptosis. Oncogene. 15:423–435. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Osborne CK: Aromatase inhibitors in
relation to other forms of endocrine therapy for breast cancer.
Endocr Relat Cancer. 6:271–276. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Howell A, Osborne CK, Morris C and
Wakeling AE: ICI 182,780 (Faslodex): development of a novel, ‘pure’
antiestrogen. Cancer. 89:817–825. 2000.
|
|
17.
|
Buzdar AU: Advances in endocrine
treatments for post-menopausal women with metastatic and early
breast cancer. Oncologist. 8:335–341. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Hutcheson IR, Knowlden JM, Madden TA, et
al: Oestrogen receptor-mediated modulation of the EGFR/MAPK pathway
in tamoxifen-resistant MCF-7 cells. Breast Cancer Res Treat.
81:81–93. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Osborne CK, Pippen J, Jones SE, et al:
Double-blind, randomized trial comparing the efficacy and
tolerability of fulvestrant versus anastrozole in postmenopausal
women with advanced breast cancer progressing on prior endocrine
therapy: results of a North American trial. J Clin Oncol.
20:3386–3395. 2002. View Article : Google Scholar
|
|
20.
|
Early Breast Cancer Trialists’
Collaborative Group (EBCTCG): Effects of chemotherapy and hormonal
therapy for early breast cancer on recurrence and 15-year survival:
an overview of the randomised trials. Lancet. 365:1687–1717.
2005.PubMed/NCBI
|
|
21.
|
Baum M, Buzdar AU, Cuzick J, et al: ATAC
Trialists’ Group Anastrozole alone or in combination with tamoxifen
versus tamoxifen alone for adjuvant treatment of postmenopausal
women with early breast cancer: first results of the ATAC
randomised trial. Lancet. 359:2131–2139. 2002.
|
|
22.
|
Mouridsen H, Gershanovich M, Sun Y, et al:
Phase III study of letrozole versus tamoxifen as first-line therapy
of advanced breast cancer in postmenopausal women: analysis of
survival and update of efficacy from the International Letrozole
Breast Cancer Group. J Clin Oncol. 21:2101–2109. 2003. View Article : Google Scholar
|
|
23.
|
Massarweh S and Schiff R: Unraveling the
mechanisms of endocrine resistance in breast cancer: new
therapeutic opportunities. Clin Cancer Res. 13:1950–1954. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Musgrove EA and Sutherland RL: Biological
determinants of endocrine resistance in breast cancer. Nat Rev
Cancer. 9:631–643. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Huber-Keener KJ, Liu XP, Wang Z, et al:
Differential gene expression in Tamoxifen-resistant breast cancer
cells revealed by a new analytical model of RNA-Seq data. PLoS One.
7:e413332012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View Article : Google Scholar
|
|
27.
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
28.
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans.
Cell. 75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar
|
|
32.
|
O’Day E and Lal A: MicroRNAs and their
target gene networks in breast cancer. Breast Cancer Res.
12:2012010.
|
|
33.
|
Masri S, Liu Z, Phung S, Wang E, Yuan YC
and Chen SA: The role of microRNA-128a in regulating TGFbeta
signaling in letrozole-resistant breast cancer cells. Breast Cancer
Res Treat. 124:89–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Tian W, Chen J, He H and Deng Y: MicroRNAs
and drug resistance of breast cancer: basic evidence and clinical
applications. Clin Transl Oncol. 15:335–342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Tavazoie SF, Alarcón C, Oskarsson T, et
al: Endogenous human microRNAs that suppress breast cancer
metastasis. Nature. 451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Huang QH, Gumireddy K, Schrier M, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Negrini M and Calin GA: Breast cancer
metastasis: a microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Yu F, Yao H, Zhu P, et al: Iet-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Lyng MB, Lænkholm AV, Søkilde R, Gravgaard
KH, Litman T and Ditzel HJ: Global microRNA expression profiling of
high-risk ER+ breast cancers from patients receiving adjuvant
tamoxifen mono-therapy: a DBCG study. PLoS One.
7:e361702012.PubMed/NCBI
|
|
42.
|
Cortés-Sempere M and Ibáñez de Cáceres I:
microRNAs as novel epigenetic biomarkers for human cancer. Clin
Transl Oncol. 13:357–362. 2011.PubMed/NCBI
|
|
43.
|
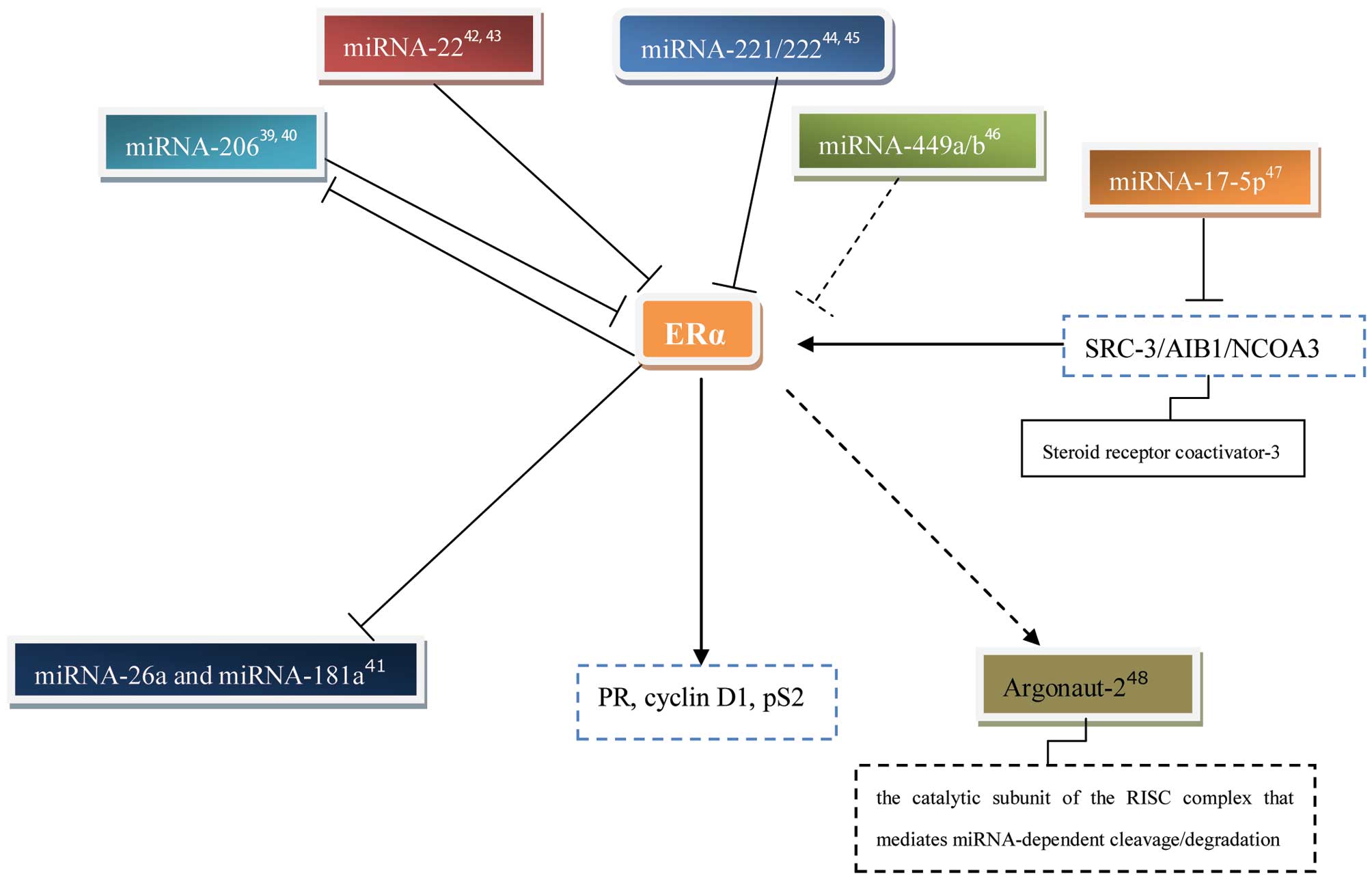
Adams BD, Furneaux H and White BA: The
micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen
receptor-alpha (ERalpha) and represses ERalpha messenger RNA and
protein expression in breast cancer cell lines. Mol Endocrinol.
21:1132–1147. 2007. View Article : Google Scholar
|
|
44.
|
Kondo N, Toyama T, Sugiura H, Fujii Y and
Yamashita H: miR-206 expression is down-regulated in estrogen
receptor alpha-positive human breast cancer. Cancer Res.
68:5004–5008. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Maillot G, Lacroix-Triki M, Pierredon S,
et al: Widespread estrogen-dependent repression of microRNAs
involved in breast tumor cell growth. Cancer Res. 69:8332–8340.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Pandey DP and Picard D: miR-22 inhibits
estrogen signaling by directly targeting the estrogen receptor
alpha mRNA. Mol Cell Biol. 29:3783–3790. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Xiong J, Yu D, Wei N, et al: An estrogen
receptor alpha suppressor, microRNA-22, is downregulated in
estrogen receptor alpha-positive human breast cancer cell lines and
clinical samples. FEBS J. 277:1684–1694. 2010. View Article : Google Scholar
|
|
48.
|
Zhao JJ, Lin J, Yang H, et al:
MicroRNA-221/222 negatively regulates estrogen receptor alpha and
is associated with tamoxifen resistance in breast cancer. J Biol
Chem. 283:31079–31086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Manavalan TT, Teng Y, Appana SN, et al:
Differential expression of microRNA expression in
tamoxifen-sensitive MCF-7 versus tamoxifen-resistant LY2 human
breast cancer cells. Cancer Lett. 313:26–43. 2011. View Article : Google Scholar
|
|
50.
|
Lau L, Chan K and Khoo U: Identification
of microRNAs associated with tamoxifen resistance in breast cancer.
In: Proceedings of the 101st Annual Meeting of the American
Association for Cancer Research (AACR); (abstract 2040),. 2010
|
|
51.
|
Hossain A, Kuo MT and Saunders GF:
Mir-17-5p regulates breast cancer cell proliferation by inhibiting
translation of AIB1 mRNA. Mol Cell Biol. 26:8191–8201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Adams BD, Claffey KP and White BA:
Argonaute-2 expression is regulated by epidermal growth factor
receptor and mitogen-activated protein kinase signaling and
correlates with a transformed phenotype in breast cancer cells.
Endocrinology. 150:14–23. 2009. View Article : Google Scholar
|
|
53.
|
Rao X, Di Leva G, Li M, et al:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Fan M, Yan PS, Hartman-Frey C, et al:
Diverse gene expression and DNA methylation profiles correlate with
differential adaptation of breast cancer cells to the antiestrogens
tamoxifen and fulvestrant. Cancer Res. 66:11954–11966. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Cittelly DM, Das PM, Spoelstra NS, et al:
Downregulation of miR-342 is associated with tamoxifen resistant
breast tumors. Mol Cancer. 9:3172010. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Lu Y, Roy S, Nuovo G, et al:
Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of
tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein
and modulating mitogenic signal. J Biol Chem. 286:42292–42302.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Lønning PE: Lack of complete
cross-resistance between different aromatase inhibitors; a real
finding in search for an explanation? Eur J Cancer. 45:527–535.
2009.PubMed/NCBI
|
|
58.
|
Cittelly DM, Das PM, Salvo VA, Fonseca JP,
Burow ME and Jones FE: Oncogenic HER2Δ16 suppresses miR-15a/16 and
deregulates BCL-2 to promote endocrine resistance of breast tumors.
Carcinogenesis. 31:2049–2057. 2010.
|
|
59.
|
Frasor J, Chang EC, Komm B, et al: Gene
expression preferentially regulated by tamoxifen in breast cancer
cells and correlations with clinical outcome. Cancer Res.
66:7334–7340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Bergamaschi A and Katzenellenbogen BS:
Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes
breast cancer cell survival and endocrine resistance. Oncogene.
31:39–47. 2012.PubMed/NCBI
|
|
61.
|
Sachdeva M, Wu H, Ru P, Hwang L, Trieu V
and Mo Y: MicroRNA-101 promotes estrogen independent growth and
confers tamoxifen resistance in ER positive breast cancer cells.
In: Proceedings of the 101st Annual Meeting of the American
Association for Cancer Research (AACR); (abstract 2104),. 2010
|
|
62.
|
Ji J, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Murakami Y, Tanaka M, Toyoda H, et al:
Hepatic microRNA expression is associated with the response to
interferon treatment of chronic hepatitis C. BMC Med Genomics.
3:482010. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Rothé F, Ignatiadis M, Chaboteaux C, et
al: Global microRNA expression profiling identifies MiR-210
associated with tumor proliferation, invasion and poor clinical
outcome in breast cancer. PLoS One. 6:e209802011.PubMed/NCBI
|
|
65.
|
Rodríguez-González FG, Sieuwerts AM, Smid
M, et al: MicroRNA-30c expression level is an independent predictor
of clinical benefit of endocrine therapy in advanced estrogen
receptor positive breast cancer. Breast Cancer Res Treat.
127:43–51. 2011.
|
|
66.
|
Jansen MP, Reijm EA, Sieuwerts AM, et al:
High miR-26a and low CDC2 levels associate with decreased EZH2
expression and with favorable outcome on tamoxifen in metastatic
breast cancer. Breast Cancer Res Treat. 133:937–947. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
67.
|
Mueller DW and Bosserhoff AK: Role of
miRNAs in the progression of malignant melanoma. Br J Cancer.
101:551–556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Xi Y, Nakajima G, Gavin E, et al:
Systematic analysis of microRNA expression of RNA extracted from
fresh frozen and formalin-fixed paraffin-embedded samples. RNA.
13:1668–1674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Lawrie CH, Gal S, Dunlop HM, et al:
Detection of elevated levels of tumour-associated microRNAs in
serum of patients with diffuse large B-cell lymphoma. Br J
Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Gilad S, Meiri E, Yogev Y, et al: Serum
microRNAs are promising novel biomarkers. PLoS One. 3:e31482008.
View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Zhao H, Shen J, Medico L, Wang D,
Ambrosone CB and Liu S: A pilot study of circulating miRNAs as
potential biomarkers of early stage breast cancer. PLoS One.
5:e137352010. View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Hutvágner G, Simard MJ, Mello CC and
Zamore PD: Sequence-specific inhibition of small RNA function. PLoS
Biol. 2:E982004.
|
|
76.
|
Meister G, Landthaler M, Dorsett Y and
Tuschl T: Sequence-specific inhibition of microRNA- and
siRNA-induced RNA silencing. RNA. 10:544–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Ørom UA, Kauppinen S and Lund AH:
LNA-modified oligonucleotides mediate specific inhibition of
microRNA function. Gene. 372:137–141. 2006.PubMed/NCBI
|
|
78.
|
Krützfeldt J, Rajewsky N, Braich R, et al:
Silencing of microRNAs in vivo with ‘antagomirs’. Nature.
438:685–689. 2005.
|