Contents
Introduction
Brief overview of the mechanisms of endocrine
resistance in breast cancer
miRNA: Biogenesis and function
miRNAs and breast cancer
Role of miRNAs in the endocrine resistance of breast
cancer
Future prospects
Conclusion
Introduction
Although extensive research into the molecular
mechanisms involved in breast cancer has been performed, challenges
remain in the early diagnosis and management of patients with
breast cancer, such as unpredictable responses and the development
of resistance to adjuvant therapies (including endocrine therapy).
Current endocrine therapies for females with estrogen receptor
(ER)-positive breast cancer have led to substantial improvements in
outcomes (1). ER-targeted therapy
has improved the quality of life and survival of millions of
females with breast cancer worldwide in the past three decades, but
its success has been limited by de novo and acquired
resistance. Significant efforts have been undertaken to resolve the
molecular mechanisms that lead to endocrine resistance. miRNAs are
evolutionarily conserved short non-coding RNA genes that were first
identified over a decade ago (2–4).
According to various studies over the past decade, miRNAs are an
important and prevalent regulatory layer of gene expression that
acts at the post-transcriptional level (5,6), and
have also been implicated to be pivotal in common human diseases,
including cancer (7). miRNAs are
rapidly emerging as a novel class of biomarkers with a unique set
of biological and chemical properties that makes them extremely
attractive candidates for clinical implementation in cancer. Since
miRNA deregulation in breast cancer was first reported in 2005
(8), there have been numerous
studies on the aberrant expression of various miRNAs and their
roles in the mechanisms of endocrine resistance in breast cancer
(9,10). miRNAs may have a significant role in
the development of endocrine resistance, as well as affecting the
progression and proliferation of breast cancer cells. miRNA-based
analysis is an area of research that has rapidly accelerated since
it began. This is due to the significant effect of miRNA-mediated
gene regulation and the clear potential of these extremely small
molecules for future diagnostic, predictive and therapeutic
applications. miRNAs may significantly enhance the under-standing
of the mechanisms of endocrine resistance, and provide real-time
information concerning the development of such resistance.
This review addresses efforts to understand how the
miRNA profile is altered upon the development of resistance; the
critical regulatory role of miRNAs in conferring resistance to
commonly used endocrine agents; and how these differentially
expressed miRNAs may serve as prognostic and predictive markers and
novel therapeutic targets for overcoming endocrine resistance.
Brief overview of the mechanisms of
endocrine resistance in breast cancer
The term endocrine therapy is applied to breast
cancer treatments that target the ER by blocking receptor binding
with an antagonist or by depriving the tumor of estrogen. The ER,
which has nuclear (genomic) and non-nuclear (non-genomic)
functions, is the predominant driver of the majority of types of
breast cancer (11). With at least
70% of breast cancers exhibiting high ER expression, which is known
to contribute to tumor growth and progression (12), Beatson’s findings revolutionized the
management of breast cancer, leading to the discovery of the
selective ER modulator (SERM), tamoxifen. This agent has been the
mainstay endocrine therapy for breast cancer for the past 25 years
(13). Tamoxifen was shown to
improve survival in early breast cancer (1) as well as quality of life for patients
with advanced breast cancer (14).
Over the past decade, several novel endocrine agents against
ER-positive breast cancer, which either lower the estrogen ligand
for the ER (aromatase inhibitors) or degrade the ER (ICI 182,780
fulvestrant), have also demonstrated activity in various clinical
settings (15,16). These agents, which result in a more
effective inhibition of ER signaling, have been demonstrated to be
clinically effective and are now an indispensable part of the
present treatment strategies for breast cancer (17–22).
Regardless of these significant advances in the
treatment of patients with initially hormone-sensitive breast
cancer, resistance to all forms of endocrine therapy remains a
major clinical issue; a significant proportion of patients
experience recurrence caused by intrinsic or acquired resistance to
endocrine agents within 15 years (20). At present, resistance to endocrine
therapy is considered to be a progressive, step-wise phenomenon
induced by the selective pressure of hormonal agents. Breast cancer
cells are converted from an estrogen-dependent phenotype, which is
responsive to endocrine manipulation, to a non-responsive phenotype
and eventually to an estrogen-independent phenotype. The molecular
mechanism of endocrine resistance involves alterations to the ER
and its co-regulators, receptor tyrosine kinase (RTK) signaling,
cell cycle regulators, the cell survival pathway and apoptosis
(23,24). The mechanism of endocrine resistance
is further complicated by the results of a study that utilized a
next generation sequencing (NGS) approach and a novel
bioinformatics model to compare the transcriptomes of
tamoxifen-sensitive and -resistant breast cancer cells (25). The authors identified differential
expression of 1,215 mRNA and 513 small RNA transcripts clustered
around ERα function, cell cycle regulation,
transcription/translation and mitochondrial dysfunction. The extent
of the alterations observed at multiple levels of gene regulation
demonstrates the ability of the tamoxifen-resistant cells to
modulate global gene expression (25).
Although knowledge of the mechanism of endocrine
resistance is developing, a more detailed understanding of the
molecular mechanisms and regulatory pathways involved in breast
cancer cells is required to improve the design of novel anti-tumor
drugs and inform the selection of optimal therapeutic
strategies.
miRNA: Biogenesis and function
miRNAs are a contemporary class of tiny non-coding
endogenous RNA molecules that are only 18–25 nucleotides in length.
It has been proposed that the discovery of miRNAs as regulators of
gene expression represents a paradigm-changing event in biology and
medicine (26). miRNA was first
described in Caenorhabditis elegans in the laboratory
studies of Victor Ambros and Gary Ruvkun (27,28).
The authors identified lin-4, which was originally considered to be
a biological entity specific to the C. elegans nematode
(27,28). Subsequently, miRNA research has
rapidly expanded, with the realization that miRNAs are critical to
the development of multicellular organisms and the basic functions
of cells (29). At present, ∼1,100
miRNAs and 16,228,619 predicted miRNA target sites in 34,911
distinct 3′UTR from isoforms of 19,898 human genes have been
identified in the human genome (http://www.microrna.org/) (30). As they are fundamental to genetic
regulation, miRNAs are estimated to regulate approximately
one-third of all human transcripts (31).
It is well-recognized that miRNAs are involved in
the regulation of multiple cellular processes, including
proliferation, apoptosis, cell-cycle regulation and differentiation
(29). Aberrant expression and
function of miRNAs have been associated with numerous diseases and
disorders (29). Notably,
abnormalities in miRNA activity have been implicated in the
initiation and progression of cancer, as miRNA alterations are
ubiquitous among human cancers, where they may function as
oncogenes or tumor suppressor genes. miRNAs function as key gene
regulators capable of silencing gene expression at the
post-transcriptional level.
miRNAs and breast cancer
In the last 5 years, miRNA studies of breast cancer
have represented a valuable area of research, having resulted in
new knowledge on the molecular basis of the disease, tools for
molecular classification, new markers with diagnostic and
prognostic relevance, as well as the identification of novel breast
cancer-predisposing genes (32). It
has been suggested that, as well as altering estrogen-responsive
gene expression, miRNAs may simultaneously achieve a secondary
level of post-transcriptional regulation of pathways implicated in
breast cancer and resistance to endocrine therapy (33).
The association between miRNAs and breast cancer has
been elucidated by studies investigating the expression of miRNAs
in breast cancer cell lines and clinical samples. miRNA expression
studies in breast cancer have indicated the importance and
potential use of miRNAs as disease classifiers and prognostic
tools. Lu et al analyzed the systematic expression of 217
miRNAs from 334 samples (including breast cancer) and observed that
the miRNA profiles were informative, reflecting the developmental
lineage and differentiation state of the tumors, with a general
downregulation of miRNAs noted in the tumors compared with normal
tissues (34). Another study
identified 29 miRNAs that were differentially expressed in breast
cancer tissue compared with normal tissue, as well as a further set
of 15 miRNAs that were able to correctly discriminate between tumor
and normal tissue (8). Increasing
data has revealed that miRNAs modulate the network of signal
transduction pathways associated with drug resistance by
upregulating drug efflux transporters and anti-apoptotic proteins,
undergoing epithelial-mesenchymal transition (EMT) and forming
cancer stem cells [review (35)].
The involvement of miRNA-10b (36),
-335 (37), -373 and -520c
(38) has been identified in the
development of breast cancer metastases [review (39)].
Furthermore, present findings have indicated that
miRNAs are involved in the majority of stages of breast cancer
development, such as regulating self-renewal, the tumorigenicity of
breast cancer cells (40,41), apoptosis and angiogenesis [review
(42)].
Role of miRNAs in the endocrine resistance
of breast cancer
It is considered that miRNAs are associated with the
ER pathway and other pathways involved in endocrine resistance
(9,43). Although their involvement is
apparent, the study of the specific pathways they affect and the
mechanisms they aid in regulating has only recently begun. The
incorporation of miRNA regulation into the current models of the
molecular mechanism of endocrine resistance in breast cancer is
likely to be essential to achieve a complete understanding of
endocrine resistance.
miRNA expression is closely correlated
with endocrine resistance
miRNAs and the ER
The ER remains the best predictor for endocrine
therapy. However, it is reasonable to propose that miRNAs may
function as endocrine resistance stimulators and inhibitors, and be
involved in associated pathways by directly or indirectly
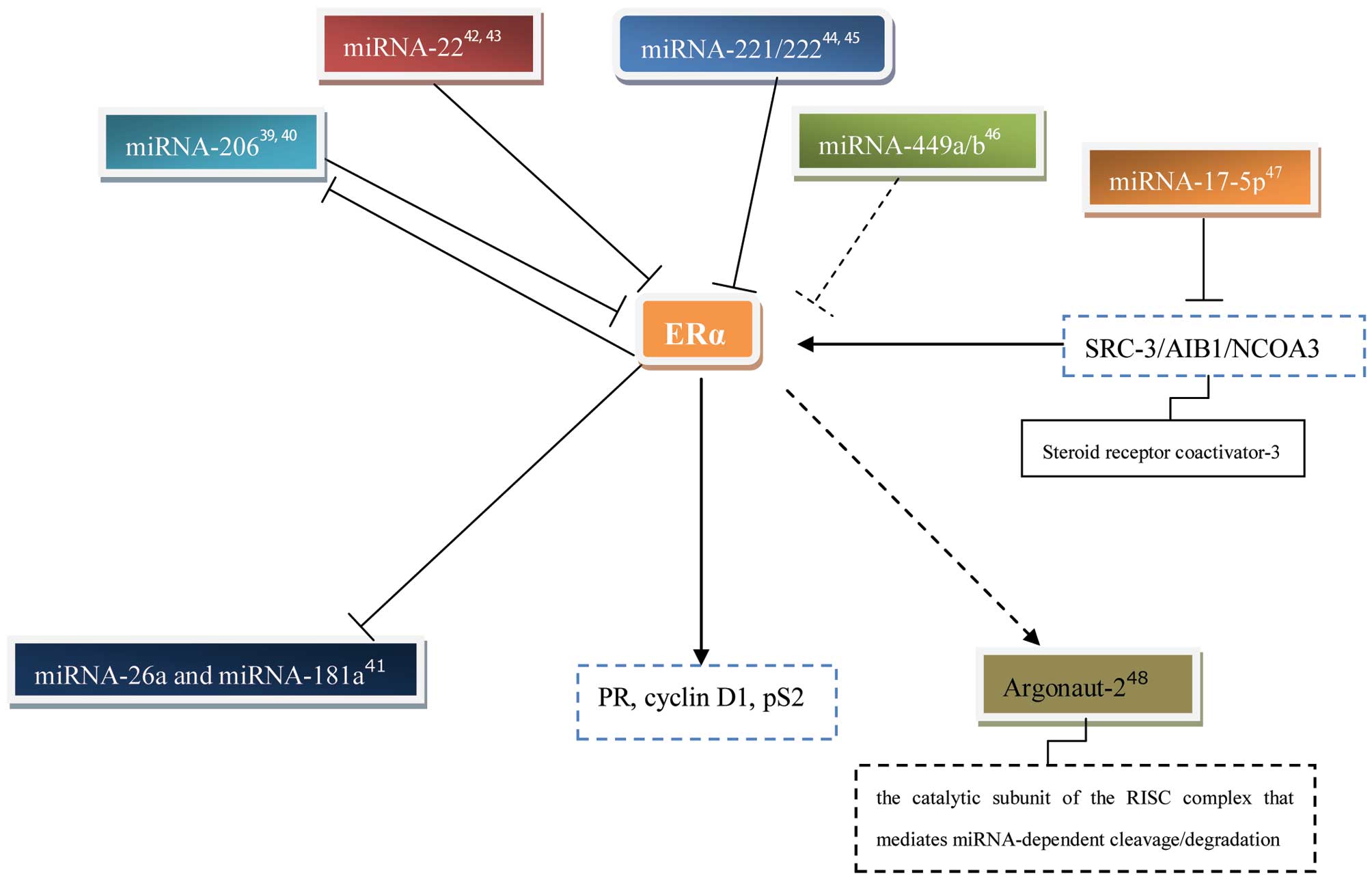
regulating genes. ERα mRNA has a long 3′-untranslated region
(3′UTR) of ∼4.3 kb, which has been reported to reduce mRNA
stability and comprise evolutionarily conserved miRNA target sites,
enabling it to be regulated by miRNAs.
The first indication of the post-transcriptional
regulation of ERα by miRNA in the context of breast cancer was
observed by Adams et al (43). Two miRNA-206-targeting sites were
identified in silico within the 3′UTR of human ERα mRNA, and
transfection of MCF-7 cells with pre-miRNA-206 specifically
decreased the ERα mRNA levels. Notably, the miRNA-206 levels were
higher in ERα-negative MB-MDA-231 cells compared with ERα-positive
MCF-7 cells. miRNA-206 expression was markedly inhibited by ERα
agonists, although not by ERβ agonists or progesterone. The authors
suggested that miRNA-206 may function in a mutually negative
feedback loop to temporally regulate ERα expression (43). Another study also demonstrated that
miRNA-206 is inversely correlated with ERα expression (44). Furthermore, transfecting MCF-7 cells
with an expression plasmid for pre-miRNA-206 reduced ERα mRNA
expression and the basal expression levels of the progesterone
receptor (PR), cyclin D1 and pS2 (well-established ERα-regulated
genes), and inhibited cell proliferation in a 17β-estradiol
(E2)-independent manner. The authors suggested that miRNA-206 may
be a novel candidate for endocrine therapy that specifically
targets ERα in breast cancer (44).
By contrast, an additional two studies did not identify
E2-independent miRNA-206 regulation of expression, which the
authors surmised may be consistent with the apparent lack of
miRNA-206 expression in MCF-7 cells (45,46).
Pandey and Picard suggested that miRNA-22 represses
ERα expression most markedly and directly through the 3′UTR,
resulting in a reduction in estrogen signaling (46). In addition, Xiong et al also
revealed that there was a significant inverse association between
the miRNA-22 levels and ERα protein expression in five breast
cancer cell lines and 23 clinical biopsies (47). miRNA-22 was frequently
down-regulated in the ERα-positive human breast cancer cell lines
and clinical samples. The authors suggested that miRNA-22 may be
pivotal in the pathogenesis of breast cancer by direct involvement
in the regulation of ERα (47).
miRNA-221/222 was also revealed to be involved in the suppression
of ERα protein expression, which is detailed in the following
section (48,49). In addition, Lau et al
examined the expression of miRNA-449a/b in 60 fresh-frozen breast
tumor samples, and observed that the expression of miRNA-449a/b was
inversely correlated with tumor grade and markedly associated with
the estrogen receptor status of the tumor. The authors suggested
that miRNA-449a/b may affect tamoxifen sensitivity in breast cancer
cells (50).
Maillot et al demonstrated that a set of 23
miRNAs (including miRNA-181a, -21, -181b, -26a, -200c, -26b, -27b
and -23b) were downregulated by E2 in various ER-positive breast
tumor cell lines (45). The RNA
precursors of the miRNAs were the primary transcriptionally
repressed targets of ER. In addition, transcriptome analysis
revealed that the E2-repression of miRNA-26a and -181a regulated
numerous genes associated with cell growth and proliferation,
including the progesterone receptor (PGR) gene (45).
miRNAs are also able to affect estrogen-regulated
gene expression by inhibiting the expression of the coactivator
SRC-3/AIB1/NCOA3. miRNA-17-5p was demonstrated to inhibit the
translation of SRC-3/AIB1/NCOA3 in a study by Hossain et al
(51). Transfection of CHO-K1 cells
with ERα and miRNA-17-5p inhibited E2-stimulated ERE-driven
luciferase reporter activity by 50%. This study also demonstrated
that the transfection of MCF-7 cells with miRNA-17-5p reduced
E2-induced proliferation and endogenous cyclin D1 transcription
(51). Furthermore, the expression
level of Argonaute-2 (Ago2), the catalytic subunit of the
RNA-induced silencing complex (RISC) that mediates miRNA-dependent
cleavage/degradation, is higher in ERα-negative/human epidermal
growth factor receptor 2 (HER2)-positive cells compared with
ERα-positive/HER2 negative human breast cancer cell lines and
tumors (52). E2 and the
ERα-agonist, propyl pyrazole triol (PPT), were able to increase the
expression of Ago2 protein in MCF-7 cells. In addition, the high
expression of Ago2 in ERα-negative cells was severely limited by
the inhibition of the epidermal growth factor receptor
(EGFR)/mitogen-activated protein kinase (MAPK) signaling pathway,
indicating that EGF acts through the MAPK pathway to increase Ago2
protein stability. However, the mechanism by which E2 and PPT
increase Ago2 protein levels, potentially through ERα, remains
poorly understood. Ago2 overexpression in MCF-7 cells increased the
ERα protein levels by three-fold, regardless of also increasing the
levels of miRNA-206 that suppress ERα (52). This study indicated that other
factors involved in miRNA biogenesis may also be involved in the
mutual regulation between ER and miRNAs.
miRNAs and other endocrine resistance
associated pathways
In addition to estrogen-mediated gene expression,
hormone-regulated miRNAs may provide an additional level of
post-transcriptional regulation for signaling pathways critically
involved in the endocrine resistance of breast cancer. The
expression levels of a number of miRNAs in ER-positive breast
cancer have been directly associated with responses to endocrine
therapy (9).
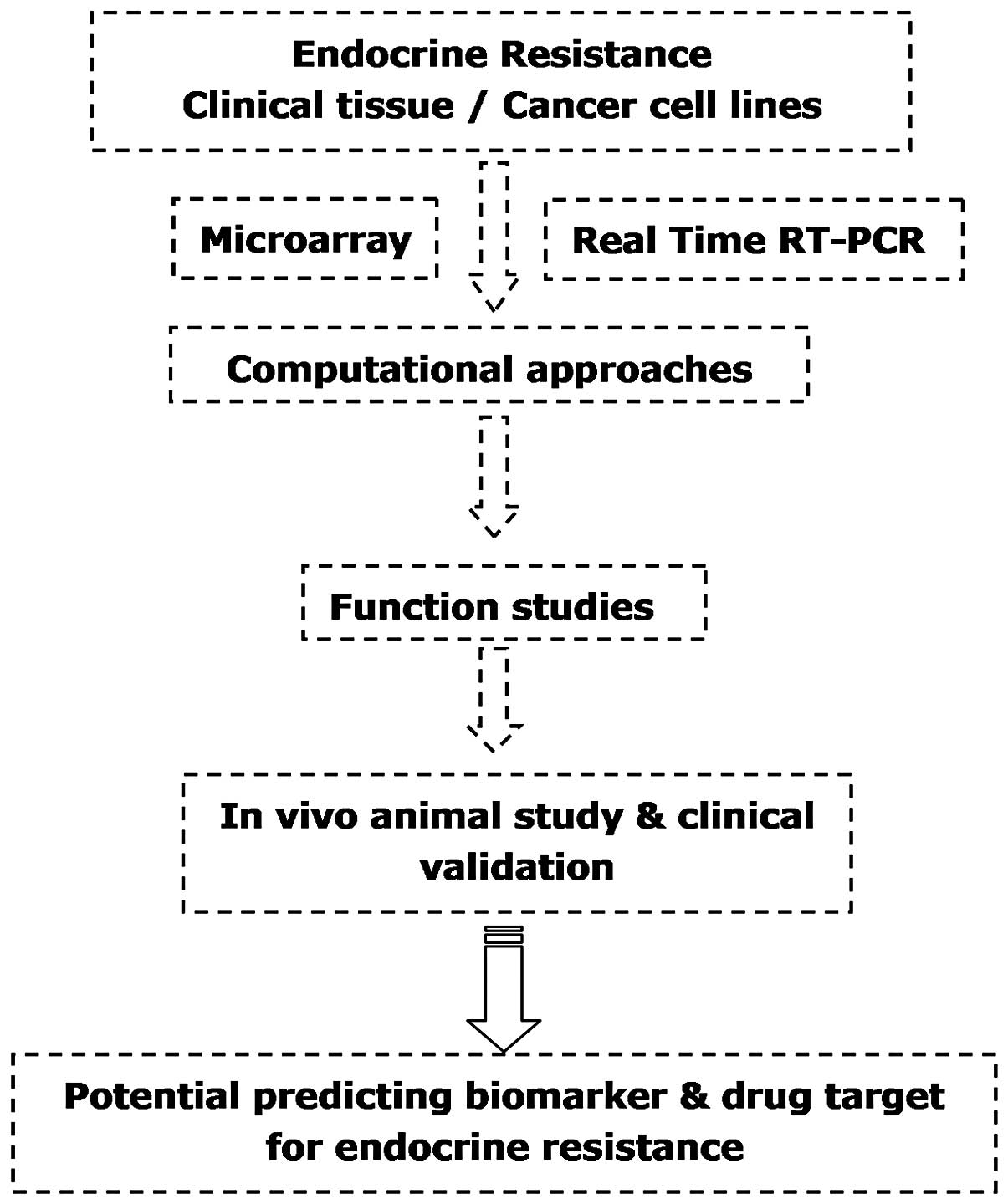
The model of research (Fig. 1) in the majority of studies
investigating the potential roles of aberrantly expressed miRNAs in
acquired endocrine resistance is as follows: i) microarray data are
utilized to compare endocrine-resistant and -sensitive breast
cancer cells and clinical samples, revealing the differential
expression of miRNAs; ii) computational approaches (shown in
Table I) are utilized to predict
the potential targets and signaling pathways of the aberrantly
expressed miRNAs in silico; iii) functional studies in
vitro are implemented to validate the predicted regulatory
roles of the miRNAs via ectopic expression and downregulation; and
iv) to support the significance of the studies performed in cell
lines, clinical data are analyzed and in vivo animal studies
are conducted to further confirm the association between miRNAs and
endocrine resistance.
 | Table I.Potential targets of miRNA and
pathway prediction tools. |
Table I.
Potential targets of miRNA and
pathway prediction tools.
| Tool | Method for
prediction and ranking | Website |
|---|
| Target Scan | Stringent seed
pairing, conservation, UTR context | http://www.targetscan.org/ |
| PicTar | Stringent seed
pairing, free energy, conservation, probability being target to set
of miRNAs | http://pictar.mdc-berlin.de/ |
| miRanda | Moderately
stringent seed pairing, free energy, conservation | http://www.microrna.org/ |
| miR Base | Target predictions
using miRanda algorithm with varied parameters | http://www.mirbase.org/ |
| PITA | Seed pairing, site
accessibility, total interaction energy, site number | http://genie.weizmann.ac.il/pubs/mir07/ |
Miller et al investigated the role of miRNAs
in acquiring resistance to tamoxifen by comparing the miRNA
profiles of tamoxifen-resistant with tamoxifen-sensitive MCF-7
breast cancer cell lines by microarray analysis (9). The results showed the significant
upregulation of eight miRNAs and marked downregulation of seven
miRNAs in tamoxifen-resistant MCF-7 breast cancer cells compared
with tamoxifen-sensitive cells. Furthermore, the expression levels
of miRNA-221 and -222 were also significantly elevated in
HER2/neu-positive primary human breast cancer tissues, which are
known to be resistant to endocrine therapy, compared with
HER2/neu-negative tissue samples. The ectopic expression of
miRNA-221/222 rendered the parental MCF-7 cells resistant to
tamoxifen. The protein level of the cyclin-dependent kinase
inhibitor 1B (p27Kip1) cell cycle inhibitor, a known target of
miRNA-221/222, was reduced by 50% in tamoxifen-resistant cells and
28–50% in tamoxifen-resistant cells that overexpressed
miRNA-221/222 (9). This was the
first study to demonstrate a correlation between miRNA-221/222
expression and HER2/neu overexpression in primary breast tumors
that are generally resistant to tamoxifen therapy.
Almost simultaneously, another study revealed the
distinct expression of a panel of miRNAs in a comparison between
ERα-positive and -negative breast cancer cell lines and primary
tumors, using a custom miRNA microarray platform containing 515
miRNAs (48). Similar to the
previously mentioned study, the miRNA-221/222 levels were also
observed to be higher in ERα-negative breast cancer cell lines and
human breast tumors compared with those that were ERα-positive.
Furthermore, two miRNA-221 and -222 seed elements were identified
in the 3′UTR of ERα. Ectopic expression of miRNA-221 and -222 in
ERα-positive MCF-7 and T47D cells suppressed the expression of ERα
protein, but not mRNA. Notably, this rendered the cells resistant
to tamoxifen. By contrast, knockdown of miRNA-221 and -222 in
ERα-negative MDA-MB-468 cells partially restored the ERα protein
expression and increased tamoxifen-induced apoptosis. These
findings indicated that miRNA-221 and -222 are pivotal in the
regulation of ERα expression, and may serve as potential targets
for restoring ERα expression and responding to endocrine therapy in
a subset of breast cancers (48).
Another study revealed a role of miRNA-221/222 in the underlying
mechanism of acquired fulvestrant resistance in MCF7 cells
(53). The overexpression of
miRNA-221/222 in ERα-positive cell lines confers
hormone-independent growth and fulvestrant resistance through
offsetting the effects of E2 depletion or fulvestrant-induced cell
death. Furthermore, potential miRNA-221/222 targets were identified
by global gene expression profiles, revealing that miRNA-221/222
overexpression resulted in the deregulation of multiple oncogenic
signaling pathways associated with drug resistance (53). β-catenin is activated by
miRNA-221/222, while transforming growth factor β (TGF-β)-mediated
growth inhibition is repressed. These effects are critical for
estrogen-independent growth and fulvestrant resistance (53). Lau et al studied the
expression profiles of >600 miRNAs in a pair of
tamoxifen-sensitive ZR75 and -resistant AK47 breast cancer cell
lines (50). A total of 65 miRNAs
were identified as being significantly downregulated in the
tamoxifen-resistant cell line, while 44 miRNAs were revealed to be
upregulated. Consistent with the findings discussed previously, the
profiling results also indicated that miRNA-221 and -222 were
overexpressed in the tamoxifen-resistant breast cancer cells
(50).
Manavalan et al used microarray to identify
97 differentially expressed miRNAs that were regulated by
4-hydroxytamoxifen in a comparison between MCF-7
endocrine-sensitive and -resistant breast cancer cells (49). The microarray expression data showed
that the expression of miR-10a, -22, -29a, -125b, -181a and-222 was
lower in EtOH-treated MCF-7 cells compared with that in LY2 cells.
In contrast, the expression of miR-21, -93 and -200a,b, and c was
lower in EtOH-treated LY2 cells compared with in MCF-7 cells. Of
these miRNAs, only miR-21 and miR-181a were regulated by E2 in
MCF-7 cells. The authors also observed that miRNA-221/222 was
overexpressed in TAM-resistant breast cancer cell lines,
suppressing ERα expression. Bioinformatic analysis to evaluate the
biological significance of miRNAs identified 36 potential gene
targets among those regulated by 4-hydroxytamoxifen in MCF-7 cells.
The targets of the miRNAs were predicted in silico using
identification software. Functional and network analyses of the
changes in differential miRNA gene expression were performed using
Ingenuity Pathways Analysis (IPA) 8.8 (Ingenuity®
Systems, Redwood City, CA, USA; http://www.ingenuity.com). Agreement in the direction
of anticipated regulation was detected for 12 putative targets.
Pyruvate decarboxylase regulator (PDCD4), B-cell
CLL/lymphoma 2 (BCL2), cytochrome P450 1B1 (CYP1B1)
and v-erb-b2 erythroblastic leukemia viral oncogene homolog 3
(ERBB3) were revealed to be differentially expressed between
the two cell lines. However, the role of the interaction between
the target genes and miRNAs for endocrine resistance (Fig. 2) were not clearly confirmed. The
miRNAs with opposite expression patterns between the two cell lines
may have been involved in the endocrine resistance (49).
To investigate the role of miRNAs in resistance to
fulvestrant, a study used experimental microarray data to compare
the global miRNA expression patterns between fulvestrant-resistant
MCF7-FR cells and their drug-sensitive parental ER-positive MCF7
cells (10). The authors identified
14 downregulated miRNAs (let-7i, miRNA-181a, -191, -199b, -204,
-211, -212, -216, -328, -373, -424, -204 and -191) in MCF7-FR
cells, and then used TargetScan and the Parallel Implicit
Time-Integration Algorithm (PITA) to predict potential target
genes. Consequently, pathway analyses predicted that these miRNAs
would regulate well-described cancer-associated signaling pathways,
such as the TGF-β, Wnt, MAPK and mammalian target of rapamycin
(mTOR) pathways (10). It has been
reported that fulvestrant-resistant breast cancer cells were able
to utilize Wnt/β-catenin and EGFR/ErbB2 signaling pathways to
establish estrogen-independence and autocrine-regulated
proliferation (54). These results
suggest a significant role for miRNA-regulated gene expression in
the onset of breast cancer anti-estrogen resistance. An enhanced
understanding of this phenomenon may lead to improved therapies for
this often fatal condition.
miRNA-342 was observed to be significantly
downregulated in breast tumor samples and cell models of tamoxifen
resistance. Restoring miRNA-342 expression in the resistant MCF-7
cell lines resensitized these cells to tamoxifen-induced apoptosis.
In addition to a role in apoptosis, IPA revealed that miRNA-342 may
affect multiple stages of the cell cycle through cyclin B1
suppression, numerous breast cancer 1 (BRCA1) activities, p53 cell
cycle checkpoint function and phosphatase and tensin homolog (PTEN)
tumor suppressor activity. Overall, these results suggest that
miRNA-342 modulates the expression of genes involved in
tamoxifen-mediated tumor cell apoptosis and cell cycle progression
(55).
Lu et al observed the significant
overexpression of miRNA-181b, -221 and -222 in tamoxifen-resistant
MCF-7 cells. In addition, anti-miRNA-222 or -181b, in combination
with tamoxifen, suppressed the growth of tamoxifen-resistant
xenografts in mice through tissue inhibitor of metalloproteinase 3
(TIMP3), which is a common target of miRNA-221, -222 and -181b.
Downregulation of TIMP3 enabled the growth of tamoxifen-resistant
cells by alleviating its inhibitory effect on a disintegrin and
metallopeptidase domain 10 (ADAM10) and ADAM17, which are essential
for tamoxifen-resistant cell growth. Furthermore, the authors
indicated that TIMP3 and the miRNAs may modulate EGF-induced MAPK
and AKT phosphorylation. Based on these findings, the authors
proposed that miRNA-221, -222 and -181b facilitate growth factor
signaling in tamoxifen-resistant breast cancer by down-regulating
TIMP3, and corresponding anti-miRNAs may be utilized to render
these tumors responsive to tamoxifen (56).
The absence of clinical cross-resistance among the
three aromatase inhibitors (AIs) and tamoxifen indicates that the
mechanisms of resistance to these endocrine therapy agents are not
identical (57). Masri et al
identified 49 hormone-responsive miRNAs in hormone-refractory cell
lines using microarray analysis. A number of hormone-responsive
miRNAs were inversely expressed between the androgen independent
(AI), long-term estrogen-deprived (LTEDaro) and tamoxifen-resistant
cell lines. Subsequently, the authors focused on miRNA-128a, which
was hormone-responsive and differentially overexpressed in
letrozole-resistant cell lines. Furthermore, TGF-β receptor 1
(TGFβR1) and SMAD2 were confirmed to be targets of miRNA-128a. The
inhibition of endogenous miRNA-128a resulted in resensitization of
the letrozole-resistant lines to the growth inhibitory effects of
TGF-β (33).
HER2Δ16 (a clinically important oncogenic isoform of
HER2) expression promotes endocrine resistance in HER2/ERα-positive
breast tumors via the suppression of miRNA-15a/16, which targets
BCL-2 (58). The authors suggested
that their study provided a template for unique therapeutic
interventions, which combine tamoxifen with the modulation of
miRNAs (58). Furthermore, this
study provided unique insights into the molecular complexity of
endocrine-resistant Her2- and ERα-positive breast cancer. It is
notable that this study demonstrated a new approach to
investigating the role of miRNAs in endocrine resistance, which
utilized bioinformatics tools to select endocrine
resistance-associated protein-targeting miRNAs.
The 14-3-3 family member and conserved protein,
14-3-3 ζ, is upregulated by tamoxifen, and this increased
expression is correlated with early disease recurrence (59). A study revealed that the tamoxifen
upregulation of 14-3-3 ζ results from its ability to rapidly
downregulate miRNA-451 that specifically targets 14-3-3 ζ (6). Increasing the level of miRNA-451, by
overexpression, downregulated 14-3-3 ζ. This, in turn, suppressed
cell proliferation and colony formation, markedly reduced the
activation of HER2, EGFR and MAPK signaling, increased apoptosis
and, notably, restored the growth-inhibitory effectiveness of SERMs
in endocrine-resistant cells (60).
In addition to the role of miRNAs in acquired
endocrine resistance, a study investigated their critical roles in
estrogen-independent growth, which results in intrinsic tamoxifen
resistance (61). An in vivo
selection system was used against an miRNA library. ER-positive
MCF-7 cells were initially infected with the pooled library and
subsequently, the cells were orthotopically injected into female
nude mice depleted of estrogen. Further studies highlighted the
enrichment of miRNA-101, which alone is able to promote
estrogen-independent growth and lead to tamoxifen resistance
without targeting the ER. Finally, the direct suppression of
membrane associated guanylate kinase (Magi-2) by miRNA-101 was
demonstrated to provide a molecular link for miRNA-101-mediated
activation of Akt through the suppression of PTEN activity. Given
these findings, the authors suggested that miRNA-101 may serve as a
biomarker for the subpopulation of ER-positive patients with breast
cancer that are intrinsically resistant to tamoxifen (61).
miRNAs may predict responses to
endocrine therapy
The ability of miRNAs to be used as predictive
biomarkers has been demonstrated in numerous studies (62,63).
miRNAs have also been shown to exhibit potential for predicting
responses to endocrine therapy in breast cancer.
miRNA profiling was used by Rothé et al as a
complementary tool to assess whether miRNA expression was able to
predict the clinical outcome of patients with breast cancer who
were treated with tamoxifen alone (64). The authors evaluated the expression
of miRNA-210 in a cohort of 89 ER-positive patients with breast
cancer. A high level of miRNA-210 expression was observed to be
associated with a higher risk of recurrence compared with lower
levels of miRNA-210. Therefore, miRNA-210 was shown to be
associated with a poor clinical outcome in tamoxifen treatment
(64).
A study investigated the association between five
putative candidate miRNAs and outcome in ERα-positive patients with
breast cancer who received tamoxifen therapy for the advanced
disease (65). The authors revealed
an association between high expression levels of miRNA-30a-3p, -30c
and -182 and clinical benefit/longer progression-free survival
(PFS). Subsequently, the authors searched the published predicted
target databases and none of these miRNAs were identified to have
ER mRNA as a target, suggesting that the effects of the studied
miRNAs on responses to tamoxifen were indirectly associated with
the modulation of ER levels. Furthermore, the authors utilized
global testing pathway analysis together with gene expression data
to clarify the underlying mechanisms by which these miRNAs affected
the outcome of tamoxifen treatment. miRNA-30c was shown to be
positively associated with ERα and negatively associated with EGFR.
Furthermore, miRNA-30c and -30a-3p were negatively correlated with
the ras-related C3 botulinum toxin substrate 1 (RAC1) signaling
pathway, which is driven by platelet-derived growth factor receptor
α (PDGFR-α). Pathway analysis also showed that miRNA-30a-3p was
positively correlated with BCL2-mediated cellular
survival/ceramide-induced apoptosis, which corresponded with a
favorable role for increased BCL2 expression in breast cancer
treated with endocrine therapy. However, following data mining, the
putative targets of these miRNAs did not demonstrate a specific
mechanism and were not associated with the aforementioned pathways.
Consequently, the authors proposed that the putative targets were
not informative, and that further research was required to
elucidate the exact mechanisms underlying the association between
the identified miRNAs and the outcome of tamoxifen treatment
(65).
Increasing levels of miRNA-26a were observed to be
significantly (P<0.005) associated with clinical benefit and
prolonged time to progression (TTP) in 235 patients with
ER-positive tumors, who were administered tamoxifen as the
first-line therapy for the treatment of metastatic disease
(66). This indicated that
miRNA-26a may serve as an optimal marker associated with the
outcome of tamoxifen therapy. The authors performed an exploratory
pathway analysis with a global testing approach (GTA) to identify
cyclin and cell cycle regulation genes; cyclin E1 (CCNE1) and
cyclin-dependent kinase 2 (CDC2) were the only genes that
contributed significantly and overlapped between miRNA-26a and
enhancer of zeste homolog 2 (EZH2). This study may aid clinicians
in the identification of patients resistant to tamoxifen (66). This finding also provides rationale
for the application of altered expression levels of specific miRNAs
as a predictive marker for endocrine-resistant breast cancer.
However, another study using global miRNA analysis
was performed on 152 ER-positive primary tumors from high-risk
patients with breast cancer, who had received adjuvant tamoxifen as
monotherapy and half of which had developed distant recurrence.
Based on the large sample size, there was no single miRNA profile
that was predictive of the outcome following adjuvant tamoxifen
treatment in a broad cohort of ER-positive patients with breast
cancer (41).
Limitations of the currently available
studies
Regardless of the notable achievements in the
previous studies concerned with the role of miRNAs in endocrine
resistance (summarized in Table
II), the implementation of miRNAs for clinical use remains at
an early stage. Furthermore, the utilization of this information is
limited by the following drawbacks of the available data.
 | Table II.Summary of miRNAs involved in the
endocrine resistance of breast cancer. |
Table II.
Summary of miRNAs involved in the
endocrine resistance of breast cancer.
| miRNAs | miRNA
expression | Putative
target | Drug | Sample source
| Reference |
|---|
| Cell line | Patient | Xenograft |
|---|
| -221, -222 | ↑ | p27Kip1 | Tamoxifen | MCF-7 | Yes | No | (9) |
| ↑ | ERα | Tamoxifen |
MCF-7/T47D
MDA-MB-468 | Yes | No | (48) |
| ↑ | β-catenin,
TGF-β | Fulvestrant | MCF-7 | No | No | (53) |
| ↑ | - | Tamoxifen | AK47 | No | No | (50) |
| ↑ | ERα, PDCD4,
BCL2, CYP1B1, ERBB3 | Tamoxifen | MCF-7 | No | No | (49) |
|
let-7i,-181a,-191-199b,
-373*,-204, -211, -212, -216, -328,-424, -204, -191,
let-7i | ↓ | TGF-β, Wnt MAPK,
mTOR | Fulvestrant | MCF-7 | No | No | (10) |
| -342 | ↓ | cyclin B1, BRCA1
p53, PTEN | Tamoxifen | MCF-7 | No | No | (55) |
| -222,-181b | ↑ | TIMP3, MAPK, AKT,
ADAM10, ADAM17 | Tamoxifen | MCF-7 | Yes | Yes | (56) |
| -128a | ↑ | TGFβR1, SMAD2 | Letrozole | MCF-7 | No | No | (33) |
| -101 | ↑ | Magi-2, Akt,
PTEN | Tamoxifen | MCF-7 | No | Yes | (61) |
| -210 | ↑ | - | Tamoxifen | - | Yes | No | (64) |
| -15a/16 | ↓ | BCL2 | Tamoxifen | MCF-7 | No | Yes | (58) |
| -30c | ↑ | HER, RAC1 | Tamoxifen | - | Yes | No | (65) |
| -26a | ↑ | CCNE1, CDC2,
EZH2 | Tamoxifen | - | Yes | No | (66) |
| -451 | ↓ | 14-3-3 ζ, HER2,
EGFR, MAPK | Tamoxifen | MCF-7 | No | No | (60) |
The majority of the current findings are limited to
cell line and animal models, although a small number of studies
have been based on clinical specimens with a limited sample size.
Moreover, the accuracy of the miRNA signatures has not been
adequately evaluated, leading to difficulties in determining
whether aberrant miRNA expression in breast cancer tissues is able
to reliably differentiate endocrine-sensitive patients from
endocrine-resistant patients. Additional studies are required to
determine the roles of miRNAs in a clinical setting, to generate
conclusive results. It is essential that the potential of the miRNA
expression profiles is carefully validated prior to being adopted
for clinical applications.
Furthermore, the differences in the expression
levels of miRNAs have been observed in different
endocrine-resistant cell lines, and should be further investigated
to determine whether the differences are only associated with the
characteristics of the cell types or caused by more complex
mechanisms. Technical and analytical variations may contribute to
inconsistencies between different miRNA profiling studies, and
standardization and confirmation with further analytical approaches
are critical for the validation of these findings.
In addition, the majority of the targets and
signaling pathways in which miRNAs are involved have been predicted
by computational approaches and require further investigation in
functional studies. Mueller and Bosserhoff suggested that the lack
of understanding with regard to the fundamental rules for the
pairing of miRNAs to their target sequences leads to deficiencies
in current miRNA target prediction algorithms. Identifying and
verifying further miRNA-target gene interactions may improve the
reliability of the present algorithms (67).
Moreover, the majority of studies have been based on
microarray analysis, which detects significant expression through
certain criteria. Potential miRNAs with essential functions that do
not change significantly enough to meet the criteria may be
ignored. Furthermore, it is unlikely that all differential
expression patterns are equal. Among a number of up- or
downregulated miRNAs, some are passengers and others are drivers,
and this is a critical difference.
Additionally, it is beneficial to examine the
sensitivity and specificity of miRNA expression profiles in
clinical situations. Future studies should use adequate sample
sizes to discover a large number of potential endocrine-resistant
breast cancer-associated miRNAs. The specificity and sensitivity of
any single miRNA may be limited; establishing a panel of
symmetrical and validated miRNAs expression profiles may produce
the best result.
Further, the main targets and regulators of miRNAs
must be identified for the role of miRNAs in complex multistep
endocrine resistance to be understood. As miRNA biogenesis is a
multistage process, defects in miRNA processing have also been
shown to enhance tumorigenesis (68). It may be hypothesized that the
altered biogenesis of miRNAs is also a significant factor that
contributes to the development of endocrine resistance in breast
cancer. To the best of the authors’ knowledge, such research is not
yet available. This is likely to provide an invaluable opportunity
for studying the role of miRNAs in endocrine resistance.
Future prospects
Although miRNA research is in the early stage, there
is rationale for speculating that miRNAs are likely to have a
significant impact on improving the management of patients with
breast cancer who exhibit endocrine resistance. With the
accumulation of studies concerning the clinical significance of
miRNAs, these small molecules are predicted to have potential for
the development of novel predictive and therapeutic approaches.
Prediction of the development of
endocrine resistance
At present, endocrine resistance in breast cancer is
predicted by utilizing the ‘wait and see’ approach. In this
situation, endocrine resistance, which is measured
radiographically, and appreciable changes in tumor recurrence and
metastases may only be detected in the long-term, following the
initiation of therapy, resulting in unacceptable and devastating
outcomes. With miRNAs, responses and resistance to therapy are
likely to be detected at an early stage during the course of
treatment. As the technology evolves and is able to detect miRNAs
with sufficient sensitivity and specificity, we suggest that it
will allow the prediction of resistance to endocrine therapy.
Additional studies are required to determine whether
the potential responses to specific endocrine treatment for
patients with breast cancer may be efficiently classified by the
patients’ miRNA expression profiles. If these are demonstrated to
be sensitive enough to detect the early presence of endocrine
resistance and absent or minimal resistance in endocrine-sensitive
individuals in future clinical trials, miRNAs are likely to be
promising biomarkers for predicting responses and resistance as
patients with breast cancer receive endocrine therapy.
The advent of advanced technologies has allowed
clinical tissues from patients of known responsiveness or
resistance to endocrine therapy to be used as a means of obtaining
an enhanced understanding of the potential mechanisms of endocrine
resistance. miRNAs, which are stable molecules that have been shown
to be well-preserved in formalin-fixed paraffin-embedded tissues,
as well as fresh snap-frozen specimens (69), have enormous potential to serve as
ideal biomarkers. However, the difficulty in obtaining the tumor
tissue when the resistance has developed, as opposed to prior to
therapy, retards this approach (24). Additionally, biopsies of patients
with metastatic disease in the lung, bone or liver are complicated
to perform, and are associated with high morbidity rates. However,
such tissue is crucial for the molecular profiling of resistant
tumors, in order to understand the underlying pathways.
miRNAs in circulation are readily accessible and may
be sampled relatively noninvasively. The presence of miRNAs in
serum was first described in patients with diffuse large B-cell
lymphoma (70). Subsequently,
several studies have identified similar results regarding the
presence of miRNAs in circulation and their potential to serve as
novel biomarkers for diseases and physiological states, including
malignancy, diabetes mellitus and pregnancy (71–73).
For breast cancer, the first example of genome-wide miRNA
expression profiling in the circulation was by Zhao et al,
who utilized microarray-based expression profiling to identify
circulating miRNAs that were differentially expressed in 20
patients with breast cancer and 20 controls. The authors identified
26 miRNAs with at least two-fold differential expression between
the cases and controls, indicating potential for the development of
a signature of circulating miRNAs that may function as a diagnostic
biomarker of breast cancer (74).
miRNAs in circulation enable rapid and repeated
monitoring of miRNA expression profile changes through the course
of endocrine treatments, which is likely to provide insights into
the molecular evolution of the tumor. This, in turn, is likely to
enhance the personalized treatment of endocrine therapy for
patients with breast cancer.
Therapeutic potential
As the deregulation of miRNAs is involved in the
development of endocrine resistance in breast cancer, there is
potential for targeting miRNAs to serve as therapeutic guides for
personalized medicine. miRNAs are promising in the field of drug
development. in that they are likely to provide directions for
overcoming endocrine resistance. With regard to the complex
mechanism of endocrine resistance, the ability of miRNAs to
regulate several target genes is particularly important. Thus, by
targeting one miRNA, it is possible to target multiple pathways
involved in the formation of endocrine resistance.
Strategies for regulating miRNA expression in
endocrine resistant patients include the normalization of
deregulated miRNAs and targeting specific miRNAs that may allow for
the prevention or resensitization of resistance to hormonal
therapies. Antisense oligonucleotides have been shown to block the
function of miRNAs in numerous studies (75–77).
Notably, the possibility of the delivery of antisense
oligonucleotides in vivo was demonstrated by Krützfeldt
et al, who showed that the intravenous administration of
antisense oligonucleotides against miRNA-16, -122, -192 and -194
resulted in a significant reduction in the expression of the
corresponding miRNA levels in various tissues (78). These results suggest that silencing
specific overexpressed miRNAs involved in endocrine resistance
in vivo may be a novel therapeutic strategy (available as
antisense oligonucleotides against miRNAs, locked nucleic acids,
anti-miRNAs or antagomirs). This process is theoretically similar
for the downregulated miRNAs, and the restoration of these miRNAs
may be another important therapeutic approach (available as miRNA
mimics).
At present, clinical trials that utilize the miRNA
approach to overcome endocrine resistance are not yet available
(www.clinicaltrials.gov). However, before miRNAs
become part of molecular therapies, further studies are required to
determine their roles in patients, as the majority of the current
findings are indirect, in vitro or both. With continuous
research on the biological characteristics of miRNAs and the
delivery systems, we propose that such strategies are likely to
become available in the near future.
Conclusion
Differentially expressed miRNAs of
endocrine-resistant patients with breast cancer may serve as
potential biomarkers for endocrine-resistant tumors. Furthermore,
the determination of the target genes/pathways of deregulated
miRNAs further enhances the understanding of the mechanism of
endocrine resistance and facilitates the development of novel
targeted therapeutic agents. In addition to altered expression, the
miRNAs themselves may be predictive of drug resistance development
and utilized as biomarkers in the personalized clinical management
of breast cancer.
Although significant progress has been made in
clarifying the functional contribution of miRNAs to the
modification of endocrine resistance in breast cancer, we are still
in the infancy of our understanding. The limitations of the current
studies are likely to be overcome by future continuous research via
strategic selection and utilization of an already robust and
expanding array of analytical tools. Further well-designed
preclinical and clinical studies in this field are required. These
scientific endeavors should lead to significant developments in the
future management of endocrine resistance in patients with breast
cancer. We propose that dynamic monitoring of circulating miRNAs
through the course of endocrine therapy may facilitate the
process.
Acknowledgements
This study was supported by the
Program for Innovative Research Team in Zhejiang Province (grant
no. 2010R50046), and by grants from the National Natural Science
Foundation of China (grant nos. 81000996/H1610 and 81071880). The
authors would like to apologize to those whose studies we were
unable to cite due to space constraints.
References
|
1.
|
No authors listed:. Tamoxifen for early
breast cancer: an overview of the randomized trials. Early Breast
Cancer Trialists’ Collaborative Group. Lancet. 351:1451–1467.
1998.
|
|
2.
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Bartel DP and Chen CZ: Micromanagers of
gene expression: the potentially widespread influence of metazoan
microRNAs. Nat Rev Genet. 5:396–400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ventura A and Jacks T: MicroRNAs and
cancer: short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Miller TE, Ghoshal K, Ramaswamy B, et al:
MicroRNA-221/222 confers tamoxifen resistance in breast cancer by
targeting p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Xin F, Li M, Balch C, et al: Computational
analysis of microRNA profiles and their target genes suggests
significant involvement in breast cancer antiestrogen resistance.
Bioinformatics. 25:430–434. 2009. View Article : Google Scholar
|
|
11.
|
Clark GM, Osborne CK and McGuire WL:
Correlations between estrogen receptor, progesterone receptor, and
patient characteristics in human breast cancer. J Clin Oncol.
2:1102–1109. 1984.
|
|
12.
|
Harvey JM, Clark GM, Osborne CK and Allred
DC: Estrogen receptor status by immunohistochemistry is superior to
the ligand binding assay for predicting response to adjuvant
endocrine therapy in breast cancer. J Clin Oncol. 17:1474–1481.
1999.
|
|
13.
|
No authors listed:. Systemic treatment of
early breast cancer by hormonal, cytotoxic, or immune therapy. 133
randomised trials involving 31,000 recurrences and 24,000 deaths
among 75,000 women. Early Breast Cancer Trialists’ Collaborative
Group. Lancet. 339:1–15. 1992.
|
|
14.
|
El-Ashry D, Miller DL, Kharbanda S,
Lippman ME and Kern FG: Constitutive Raf-1 kinase activity in
breast cancer cells induces both estrogen-independent growth and
apoptosis. Oncogene. 15:423–435. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Osborne CK: Aromatase inhibitors in
relation to other forms of endocrine therapy for breast cancer.
Endocr Relat Cancer. 6:271–276. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Howell A, Osborne CK, Morris C and
Wakeling AE: ICI 182,780 (Faslodex): development of a novel, ‘pure’
antiestrogen. Cancer. 89:817–825. 2000.
|
|
17.
|
Buzdar AU: Advances in endocrine
treatments for post-menopausal women with metastatic and early
breast cancer. Oncologist. 8:335–341. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Hutcheson IR, Knowlden JM, Madden TA, et
al: Oestrogen receptor-mediated modulation of the EGFR/MAPK pathway
in tamoxifen-resistant MCF-7 cells. Breast Cancer Res Treat.
81:81–93. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Osborne CK, Pippen J, Jones SE, et al:
Double-blind, randomized trial comparing the efficacy and
tolerability of fulvestrant versus anastrozole in postmenopausal
women with advanced breast cancer progressing on prior endocrine
therapy: results of a North American trial. J Clin Oncol.
20:3386–3395. 2002. View Article : Google Scholar
|
|
20.
|
Early Breast Cancer Trialists’
Collaborative Group (EBCTCG): Effects of chemotherapy and hormonal
therapy for early breast cancer on recurrence and 15-year survival:
an overview of the randomised trials. Lancet. 365:1687–1717.
2005.PubMed/NCBI
|
|
21.
|
Baum M, Buzdar AU, Cuzick J, et al: ATAC
Trialists’ Group Anastrozole alone or in combination with tamoxifen
versus tamoxifen alone for adjuvant treatment of postmenopausal
women with early breast cancer: first results of the ATAC
randomised trial. Lancet. 359:2131–2139. 2002.
|
|
22.
|
Mouridsen H, Gershanovich M, Sun Y, et al:
Phase III study of letrozole versus tamoxifen as first-line therapy
of advanced breast cancer in postmenopausal women: analysis of
survival and update of efficacy from the International Letrozole
Breast Cancer Group. J Clin Oncol. 21:2101–2109. 2003. View Article : Google Scholar
|
|
23.
|
Massarweh S and Schiff R: Unraveling the
mechanisms of endocrine resistance in breast cancer: new
therapeutic opportunities. Clin Cancer Res. 13:1950–1954. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Musgrove EA and Sutherland RL: Biological
determinants of endocrine resistance in breast cancer. Nat Rev
Cancer. 9:631–643. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Huber-Keener KJ, Liu XP, Wang Z, et al:
Differential gene expression in Tamoxifen-resistant breast cancer
cells revealed by a new analytical model of RNA-Seq data. PLoS One.
7:e413332012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View Article : Google Scholar
|
|
27.
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
28.
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans.
Cell. 75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar
|
|
32.
|
O’Day E and Lal A: MicroRNAs and their
target gene networks in breast cancer. Breast Cancer Res.
12:2012010.
|
|
33.
|
Masri S, Liu Z, Phung S, Wang E, Yuan YC
and Chen SA: The role of microRNA-128a in regulating TGFbeta
signaling in letrozole-resistant breast cancer cells. Breast Cancer
Res Treat. 124:89–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Tian W, Chen J, He H and Deng Y: MicroRNAs
and drug resistance of breast cancer: basic evidence and clinical
applications. Clin Transl Oncol. 15:335–342. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Tavazoie SF, Alarcón C, Oskarsson T, et
al: Endogenous human microRNAs that suppress breast cancer
metastasis. Nature. 451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Huang QH, Gumireddy K, Schrier M, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Biol. 10:202–210. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Negrini M and Calin GA: Breast cancer
metastasis: a microRNA story. Breast Cancer Res. 10:2032008.
View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Yu F, Yao H, Zhu P, et al: Iet-7 regulates
self renewal and tumorigenicity of breast cancer cells. Cell.
131:1109–1123. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Lyng MB, Lænkholm AV, Søkilde R, Gravgaard
KH, Litman T and Ditzel HJ: Global microRNA expression profiling of
high-risk ER+ breast cancers from patients receiving adjuvant
tamoxifen mono-therapy: a DBCG study. PLoS One.
7:e361702012.PubMed/NCBI
|
|
42.
|
Cortés-Sempere M and Ibáñez de Cáceres I:
microRNAs as novel epigenetic biomarkers for human cancer. Clin
Transl Oncol. 13:357–362. 2011.PubMed/NCBI
|
|
43.
|
Adams BD, Furneaux H and White BA: The
micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen
receptor-alpha (ERalpha) and represses ERalpha messenger RNA and
protein expression in breast cancer cell lines. Mol Endocrinol.
21:1132–1147. 2007. View Article : Google Scholar
|
|
44.
|
Kondo N, Toyama T, Sugiura H, Fujii Y and
Yamashita H: miR-206 expression is down-regulated in estrogen
receptor alpha-positive human breast cancer. Cancer Res.
68:5004–5008. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Maillot G, Lacroix-Triki M, Pierredon S,
et al: Widespread estrogen-dependent repression of microRNAs
involved in breast tumor cell growth. Cancer Res. 69:8332–8340.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Pandey DP and Picard D: miR-22 inhibits
estrogen signaling by directly targeting the estrogen receptor
alpha mRNA. Mol Cell Biol. 29:3783–3790. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Xiong J, Yu D, Wei N, et al: An estrogen
receptor alpha suppressor, microRNA-22, is downregulated in
estrogen receptor alpha-positive human breast cancer cell lines and
clinical samples. FEBS J. 277:1684–1694. 2010. View Article : Google Scholar
|
|
48.
|
Zhao JJ, Lin J, Yang H, et al:
MicroRNA-221/222 negatively regulates estrogen receptor alpha and
is associated with tamoxifen resistance in breast cancer. J Biol
Chem. 283:31079–31086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Manavalan TT, Teng Y, Appana SN, et al:
Differential expression of microRNA expression in
tamoxifen-sensitive MCF-7 versus tamoxifen-resistant LY2 human
breast cancer cells. Cancer Lett. 313:26–43. 2011. View Article : Google Scholar
|
|
50.
|
Lau L, Chan K and Khoo U: Identification
of microRNAs associated with tamoxifen resistance in breast cancer.
In: Proceedings of the 101st Annual Meeting of the American
Association for Cancer Research (AACR); (abstract 2040),. 2010
|
|
51.
|
Hossain A, Kuo MT and Saunders GF:
Mir-17-5p regulates breast cancer cell proliferation by inhibiting
translation of AIB1 mRNA. Mol Cell Biol. 26:8191–8201. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Adams BD, Claffey KP and White BA:
Argonaute-2 expression is regulated by epidermal growth factor
receptor and mitogen-activated protein kinase signaling and
correlates with a transformed phenotype in breast cancer cells.
Endocrinology. 150:14–23. 2009. View Article : Google Scholar
|
|
53.
|
Rao X, Di Leva G, Li M, et al:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Fan M, Yan PS, Hartman-Frey C, et al:
Diverse gene expression and DNA methylation profiles correlate with
differential adaptation of breast cancer cells to the antiestrogens
tamoxifen and fulvestrant. Cancer Res. 66:11954–11966. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Cittelly DM, Das PM, Spoelstra NS, et al:
Downregulation of miR-342 is associated with tamoxifen resistant
breast tumors. Mol Cancer. 9:3172010. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Lu Y, Roy S, Nuovo G, et al:
Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of
tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein
and modulating mitogenic signal. J Biol Chem. 286:42292–42302.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Lønning PE: Lack of complete
cross-resistance between different aromatase inhibitors; a real
finding in search for an explanation? Eur J Cancer. 45:527–535.
2009.PubMed/NCBI
|
|
58.
|
Cittelly DM, Das PM, Salvo VA, Fonseca JP,
Burow ME and Jones FE: Oncogenic HER2Δ16 suppresses miR-15a/16 and
deregulates BCL-2 to promote endocrine resistance of breast tumors.
Carcinogenesis. 31:2049–2057. 2010.
|
|
59.
|
Frasor J, Chang EC, Komm B, et al: Gene
expression preferentially regulated by tamoxifen in breast cancer
cells and correlations with clinical outcome. Cancer Res.
66:7334–7340. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Bergamaschi A and Katzenellenbogen BS:
Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes
breast cancer cell survival and endocrine resistance. Oncogene.
31:39–47. 2012.PubMed/NCBI
|
|
61.
|
Sachdeva M, Wu H, Ru P, Hwang L, Trieu V
and Mo Y: MicroRNA-101 promotes estrogen independent growth and
confers tamoxifen resistance in ER positive breast cancer cells.
In: Proceedings of the 101st Annual Meeting of the American
Association for Cancer Research (AACR); (abstract 2104),. 2010
|
|
62.
|
Ji J, Shi J, Budhu A, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Murakami Y, Tanaka M, Toyoda H, et al:
Hepatic microRNA expression is associated with the response to
interferon treatment of chronic hepatitis C. BMC Med Genomics.
3:482010. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Rothé F, Ignatiadis M, Chaboteaux C, et
al: Global microRNA expression profiling identifies MiR-210
associated with tumor proliferation, invasion and poor clinical
outcome in breast cancer. PLoS One. 6:e209802011.PubMed/NCBI
|
|
65.
|
Rodríguez-González FG, Sieuwerts AM, Smid
M, et al: MicroRNA-30c expression level is an independent predictor
of clinical benefit of endocrine therapy in advanced estrogen
receptor positive breast cancer. Breast Cancer Res Treat.
127:43–51. 2011.
|
|
66.
|
Jansen MP, Reijm EA, Sieuwerts AM, et al:
High miR-26a and low CDC2 levels associate with decreased EZH2
expression and with favorable outcome on tamoxifen in metastatic
breast cancer. Breast Cancer Res Treat. 133:937–947. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
67.
|
Mueller DW and Bosserhoff AK: Role of
miRNAs in the progression of malignant melanoma. Br J Cancer.
101:551–556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Xi Y, Nakajima G, Gavin E, et al:
Systematic analysis of microRNA expression of RNA extracted from
fresh frozen and formalin-fixed paraffin-embedded samples. RNA.
13:1668–1674. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Lawrie CH, Gal S, Dunlop HM, et al:
Detection of elevated levels of tumour-associated microRNAs in
serum of patients with diffuse large B-cell lymphoma. Br J
Haematol. 141:672–675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Chen X, Ba Y, Ma L, et al:
Characterization of microRNAs in serum: a novel class of biomarkers
for diagnosis of cancer and other diseases. Cell Res. 18:997–1006.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Gilad S, Meiri E, Yogev Y, et al: Serum
microRNAs are promising novel biomarkers. PLoS One. 3:e31482008.
View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Zhao H, Shen J, Medico L, Wang D,
Ambrosone CB and Liu S: A pilot study of circulating miRNAs as
potential biomarkers of early stage breast cancer. PLoS One.
5:e137352010. View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Hutvágner G, Simard MJ, Mello CC and
Zamore PD: Sequence-specific inhibition of small RNA function. PLoS
Biol. 2:E982004.
|
|
76.
|
Meister G, Landthaler M, Dorsett Y and
Tuschl T: Sequence-specific inhibition of microRNA- and
siRNA-induced RNA silencing. RNA. 10:544–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Ørom UA, Kauppinen S and Lund AH:
LNA-modified oligonucleotides mediate specific inhibition of
microRNA function. Gene. 372:137–141. 2006.PubMed/NCBI
|
|
78.
|
Krützfeldt J, Rajewsky N, Braich R, et al:
Silencing of microRNAs in vivo with ‘antagomirs’. Nature.
438:685–689. 2005.
|