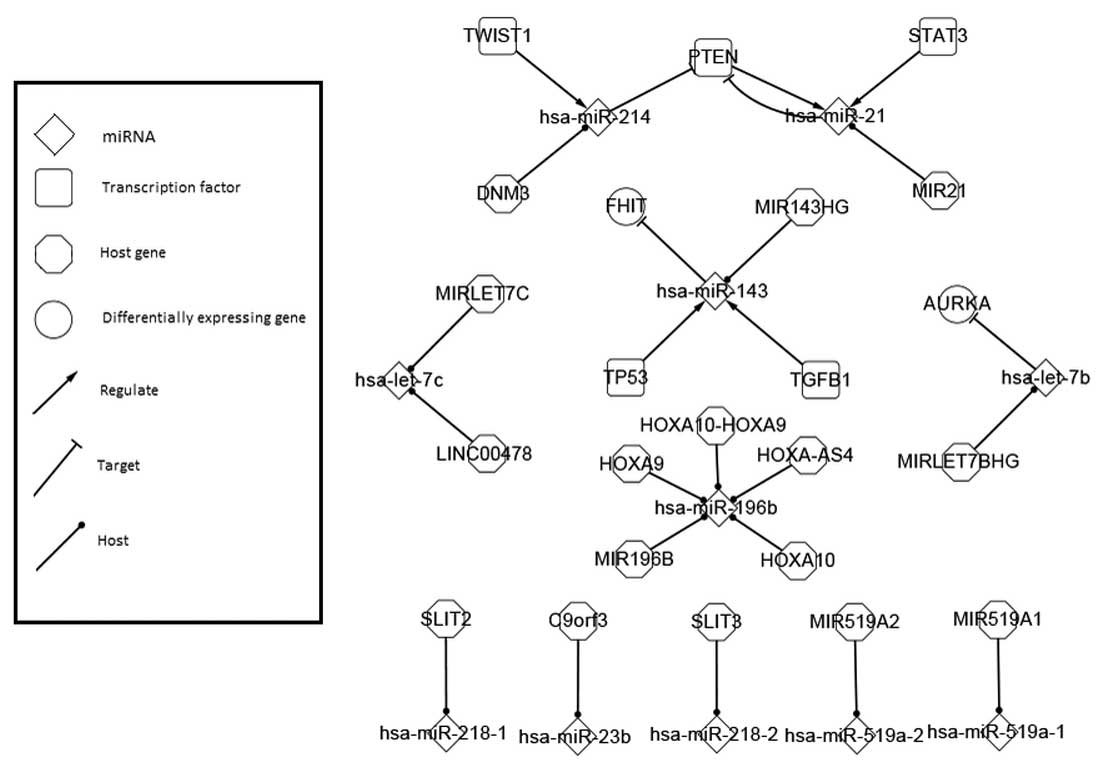
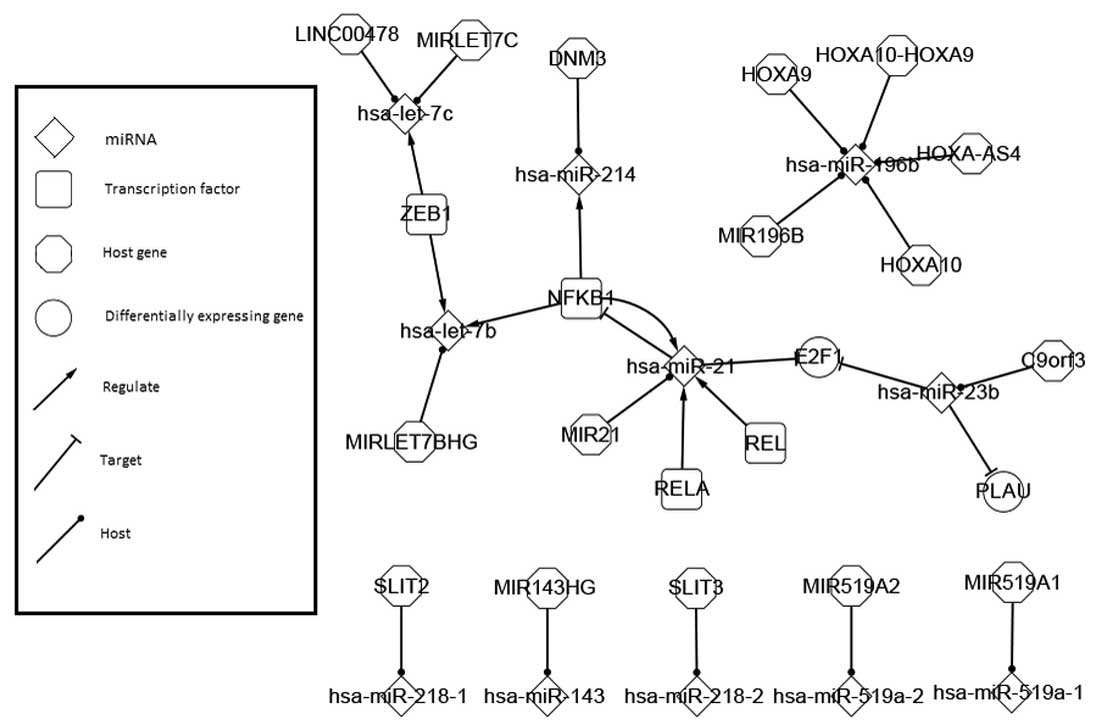
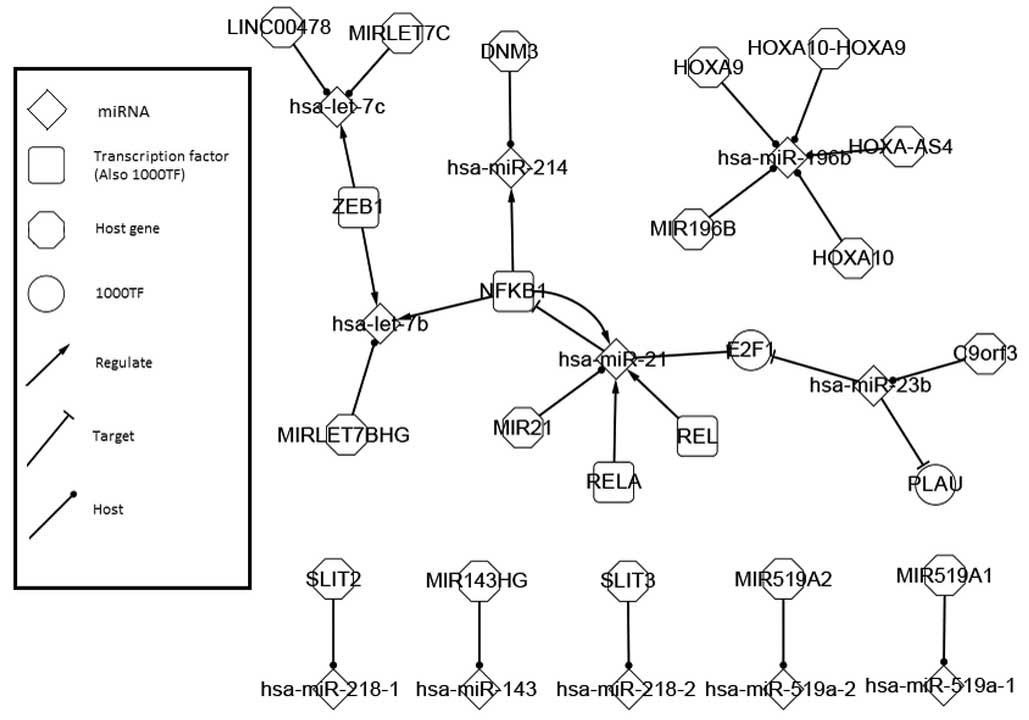
|
1
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, et al: Mutations of the BRAF gene in human
cancer. Nature. 417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hamada S, Satoh K, Miura S, Hirota M,
Kanno A, et al: miR-197 induces epithelial-mesenchymal transition
in pancreatic cancer cells by targeting p120 catenin. J Cell
Physiol. 228:1255–1263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nohata N, Hanazawa T, Kinoshita T, Okamoto
Y and Seki N: MicroRNAs function as tumor suppressors or oncogenes:
Aberrant expression of microRNAs in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 40:143–149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma D, Zhang YY, Guo YL, Li ZJ and Geng L:
Profiling of microRNA-mRNA reveals roles of microRNAs in cervical
cancer. Chin Med J (Engl). 125:4270–4276. 2012.PubMed/NCBI
|
|
6
|
Latchman DS: Transcription factors: An
overview. Int J Exp Pathol. 74:417–422. 1993.PubMed/NCBI
|
|
7
|
Libermann TA and Zerbini LF: Targeting
transcription factors for cancer gene therapy. Curr Gene Ther.
6:17–33. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chaudhry P, Srinivasan R and Patel FD:
Differential expression of Fas family members and Bcl-2 family
members in benign versus malignant epithelial ovarian cancer (EOC)
in North Indian population. Mol Cell Biochem. 368:119–126. 2012.
View Article : Google Scholar
|
|
9
|
Abdel-Rahman WM, Ruosaari S, Knuutila S
and Peltomäki P: Differential roles of EPS8 in carcinogenesis: loss
of protein expression in a subset of colorectal carcinoma and
adenoma. World J Gastroenterol. 18:3896–3903. 2012. View Article : Google Scholar
|
|
10
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar
|
|
12
|
Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia
W, et al: Intronic miR-301 feedback regulates its host gene, ska2,
in A549 cells by targeting MEOX2 to affect ERK/CREB pathways.
Biochem Biophys Res Commun. 396:978–982. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Poliseno L, Salmena L, Riccardi L, Fornari
A, Song MS, Hobbs RM, et al: Identification of the miR-106b~25
microRNA cluster as a proto-oncogenic PTEN-targeting intron that
cooperates with its host gene MCM7 in transformation. Sci Signal.
3:ra292010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Das Ghosh D, Bhattacharjee B, Sen S, Premi
L, Mukhopadhyay I, et al: Some novel insights on HPV16 related
cervical cancer pathogenesis based on analyses of LCR methylation,
viral load, E7 and E2/E4 expressions. PLoS One.
7:e446782012.PubMed/NCBI
|
|
15
|
Sethupathy P, Corda B and Hatzigeorgiou
AG: TarBase: A comprehensive database of experimentally supported
animal microRNA targets. RNA. 12:192–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, et al: miRTarBase: a database curates experimentally
validated microRNA-target interactions. Nucleic Acids Res.
39:D163–D169. 2011. View Article : Google Scholar
|
|
17
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: a
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kozomara A and Griffiths-Jones S: miRBase
integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, et al: miR2Disease: a manually curated database for
microRNA deregulation in human disease. Nucleic Acids Res.
37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Safran M, Dalah I, Alexander J, et al:
GeneCards Version 3: the human gene integrator. Database (Oxford).
5:20102010.
|
|
22
|
Chekmenev DS, Haid C and Kel AE: P-Match:
transcription factor binding site search by combining patterns and
weight matrices. Nucleic Acids Res. 33:W432–W437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, et al: The UCSC genome browser database:
update 2011. Nucleic Acids Res. 39:D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu TX, Lim EJ, Itskovich S, Besse JA,
Plassard AJ, et al: Targeted ablation of miR-21 decreases murine
eosinophil progenitor cell growth. PLoS One. 8:e593972013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Muppala S, Mudduluru G, Leupold JH, Buergy
D, Sleeman JP and Allgayer H: CD24 induces expression of the
oncomir miR-21 via Src, and CD24 and Src are both
post-transcriptionally downregulated by the tumor suppressor
miR-34a. PLoS One. 8:e595632013. View Article : Google Scholar
|
|
26
|
Bubien V, Bonnet F, Brouste V, Hoppe S,
Barouk-Simonet E, et al; French Cowden Disease Network. High
cumulative risks of cancer in patients with PTEN hamartoma tumour
syndrome. J Med Genet. 50:255–263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang F, Liu M, Li X and Tang H: MiR-214
reduces cell survival and enhances cisplatin-induced cytotoxicity
via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett.
587:488–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hou LL, Gao C, Chen L, Hu GQ and Xie SQ:
Essential role of autophagy in fucoxanthin-induced cytotoxicity to
human epithelial cervical cancer HeLa cells. Acta Pharmacologica
Sinica. 34:1403–1410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lui WO, Pourmand N, Patterson BK and Fire
A: Patterns of known and novel small RNAs in human cervical cancer.
Cancer Res. 67:6031–6043. 2007. View Article : Google Scholar
|
|
30
|
Yang ZZ, Chen S, Luan X, Li YX, Liu M, Li
X, Liu T and Tang H: MicroRNA-214 is aberrantly expressed in
cervical cancers and inhibits the growth of HeLa cells. IUBMB Life.
61:1075–1082. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matlashewski G, Lamb P, Pim D, Peacock J,
Crawford L and Benchimol S: Isolation and characterization of a
human p53 cDNA clone: expression of the human p53 gene. EMBO J.
3:3257–3262. 1984.PubMed/NCBI
|
|
32
|
Fukazawa T, Maeda Y, Matsuoka J, Yamatsuji
T, Shigemitsu K, et al: Inhibition of Myc effectively targets KRAS
mutation-positive lung cancer expressing high levels of Myc.
Anticancer Res. 30:4193–4200. 2010.
|
|
33
|
Wu X, Ajani JA, Gu J, Chang DW, Tan W, et
al: MicroRNA expression signatures during malignant progression
from Barrett’s esophagus to esophageal adenocarcinoma. Cancer Prev
Res (Phila). 6:196–205. 2013.
|
|
34
|
Yamazaki H, Chijiwa T, Inoue Y, Abe Y,
Suemizu H, et al: Overexpression of the miR-34 family suppresses
invasive growth of malignant melanoma with the wild-type p53 gene.
Exp Ther Med. 3:793–796. 2012.PubMed/NCBI
|