Introduction
It is generally accepted that the accumulation of
mutations in critical genes takes major responsibility for the
evolution of cancer (1). This
abnormal biological phenomenon appears and develops under the
control of transcription factors and microRNAs (miRNAs/miRs). In
recent years, research on miRNA, small [21–24 nucleotides (nt)]
non-coding RNA molecules, has been more widely acknowledged
(2). miRNA affects the expression
of genes at a post-transcriptional level by binding to
complementary sequences on target mRNA, resulting in translational
repression or target degradation and gene silencing. miRNA
participates in various biological processes, including
proliferation, differentiation and apoptosis, independently or
together with transcription factors (TFs). Increasing evidence
demonstrates that miRNA is of great importance in the development
of malignant tumors (3,4). According to this research, miRNAs are
involved in numerous significant biological functions and signal
transduction pathways, and also in cervical tumorigenesis (5).
A TF is a protein that binds to specific DNA
sequences, thereby controlling the flow (or transcription) of
genetic information from DNA to mRNA. TFs regulate the expression
of genes at a transcriptional level so that they may exert an
effect on malignant cell transformation (6,7).
The miRNA and genes, including TFs, involved in
cervical cancer are the key elements of the analysis based on
computational methods in the present study.
Not all the elements in the complete collection of
genes and miRNA can be equally considered in the data mining
process. A certain number act abnormally in cervical cancer, which
is known as differential expression. Unusual changes similar to
this occurring for genes and miRNA may be one of the factors
resulting in cancer (8,9).
Genes and miRNA related to cervical cancer were
located, but not all of them were differentially-expressed.
Considering that the appearance of cancer is ectopic in the human
body, the differentially-expressed genes and miRNA can be regarded
as the potential factors that have negative carcinogenic effects.
Therefore, the present study was focused on the
differentially-expressed genes and miRNA, with the assistance of
the secondary related ones.
miRNA has multiple connections with various target
genes. These genes are indispensable materials in uncovering the
role of miRNA in cancers. Numerous resources, including
computational predicted methods and experimentally validated
databases, provide sufficient data to acquire the associations
between miRNA and the corresponding target gene.
Certain miRNAs are located inside genes that are
bound to miRNA as their host genes. Rodriguez et al
indicated that miRNAs are transcribed in parallel with their host
transcripts, and two types of transcription (exonic and intronic)
were identified that indicate that miRNAs may require slightly
different mechanisms of biogenesis (10). Baskerville et al indicated
that intronic miRNA and its host gene have a closer association
than that of exonic miRNA and its corresponding host genes
(11). Intronic miRNA and its host
genes are usually coordinately expressed in biological progression,
and they usually work together to conduct biological functions and
affect the alteration of signaling pathways (12). Studies have demonstrated that their
differential expression could contribute to the progression of
cancer (13,14). Therefore, we suggest that miRNAs can
work together with their host gene in the regulatory system.
In the present study, the underlying networks
composed of miRNA, target genes, TFs, host genes of miRNA and the
regulatory associations represented in human cervical cancer were
visualized. Various data was manually collected, including
experimentally validated associations between miRNAs and their
targets, experimentally validated associations between TFs and
miRNAs, and associations of miRNAs and their host genes; they were
reserved as fundamental resources to uncover regulatory mechanisms
of genes and miRNA in cervical cancer. The differentially-expressed
and secondary related genes and miRNAs were collected from
databases and the literature. Eventually the networks associated
with cervical cancer at three levels were built based on these
materials. The first network was the differentially-expressed
network constructed from differentially-expressed genes,
differentially-expressed miRNA and host genes of
differentially-expressed miRNA. The second network was the cervical
cancer-related network, which was constructed from related genes,
related miRNAs and host genes of cancer-related miRNAs. The third
network was the global network, which consisted of all the elements
extracted from the basic source data. The differentially-expressed
network was the most significant network, and it deserves more
attention due to the differentially-expressed characteristics of
its constituents. Comparisons were made to find similarities and
differences between the three networks, and separate significant
regulatory pathways were extracted that play key roles in cervical
cancer. The present study revealed certain important core signal
networks of TFs, miRNA, targets of miRNA and host genes of miRNA in
cervical cancer. The present study will contribute to the
understanding of the pathogenesis and the development of therapy
for cervical cancer.
Materials and methods
Dataset of experimentally validated
targeting associations between miRNAs and corresponding target
genes
The targeting associations between miRNAs and
corresponding genes were acquired based on the data provided by
Tarbase 5.0 and miRTarBase (15,16).
The list consisted of 6,749 entries describing the targeting
interactions of 426 miRNAs and 2,029 genes.
Dataset of experimentally validated
regulating associations between TFs and corresponding miRNAs
The data controlling signal flow from TFs to miRNA
was acquired from TransmiR, a manually constructed database of
regulating associations between TFs and miRNA (17); this included 862 entries of
regulating associations between 153 transcription factors and 220
miRNAs.
Dataset of miRNAs and host genes
The association mapping host genes and their
respective miRNAs were established based on the data from miRBase
and the National Centre for Biotechnology Information (NCBI)
(http://www.ncbi.nlm.nih.gov/) and
miRBase (18). The result contained
1,419 entries between 1,136 host genes and 1,209 miRNAs.
Differentially-expressed and related
miRNAs associated with cervical cancer
The differentially-expressed miRNAs in cervical
cancer were mainly extracted from mir2Disease, a manually curated
database collecting and reorganizing data on
differentially-expressed miRNA in various human diseases (19). Furthermore, supplementary miRNAs
were included by a literature search. In the same way, related
miRNAs were collected. In total, 11 differentially-expressed miRNAs
and 12 related miRNAs were acquired.
Differentially-expressed genes and
related genes associated with cervical cancer
The differentially-expressed genes in cervical
cancer were gathered from several sources, including Cancer
Genetics Web (http://www.cancerindex.org/geneweb/), the single
nucleotide polymorphism database of the NCBI (http://www.ncbi.nlm.nih.gov/snp/) and Kyoto
Encyclopedia of Genes and Genomes (20). Again, the literature search was
necessary as a complementary resource.
For genes with a secondary correlation to cervical
cancer, additional sources were used. The first was the GeneCards
database, from which 203 genes of high confidence levels were
collected (21). An algorithm
called P-match operating was adopted on the concept of pattern
matching and weight matrix approaches to identify TF binding sites
in DNA sequences (22). From the
University of California, Santa Cruz Genome Browser (UCSC), the
1,000-nt promoter region sequences of differentially-expressed
genes and target genes of differentially-expressed miRNA were
acquired, following an input of P-match (23). The vertebrate matrix and restricted
high-quality criterion were chosen, and the products were referred
to as part of 1,000-nt TFs in secondary related genes. Again, data
mining from the literature search was conducted to provide
complementary information. As always, the differentially-expressed
genes were regarded as part of the related genes. In total, 38
differentially expressing genes and 241 related genes were
collected.
Construction of networks at three
levels
In order to discuss the regulating mechanism between
the involved genes and miRNA in the undercover networks of cervical
cancer, a series of procedures were designed to investigate the
interactions extracted from the data acquired. The raw data became
a designed relational model and was used as the basic source for
further processing. According to previous processes, the models can
be described as follows: U1 = [(M,
GT)/M targets
GT]; U2 =
[(GTF, M)/GTF
targets M]; and U3 =
[(GH, M)/GH is
the host gene of M]. M refers to miRNAs,
GT represents the target gene of
corresponding miRNA, GTF is the
transcriptional factor that regulates the expression of miRNA and
GH is miRNA host genes. Respectively, they
display the ways in which miRNAs and genes interact with each
other. Based on the preinstalled structure, the data was organized
into factor pairs marked by interaction types. The search procedure
was conducted to extend the original connections to deeper meshwork
rather than direct interactions between isolated nodes. The
networks are represented at three levels, the
differentially-expressed, related and global networks, together
with the network with regard to 1,000-nt TFs. Through use of
prefabricated programs, the differentially-expressed and related
networks could be extracted at corresponding levels with elements
filtered as required. The differentially-expressed network lists
associations involving differentially-expressed genes and miRNA,
together with host genes. Similarly, the related network is of
related genes and miRNAs together with host genes.
Results and Discussion
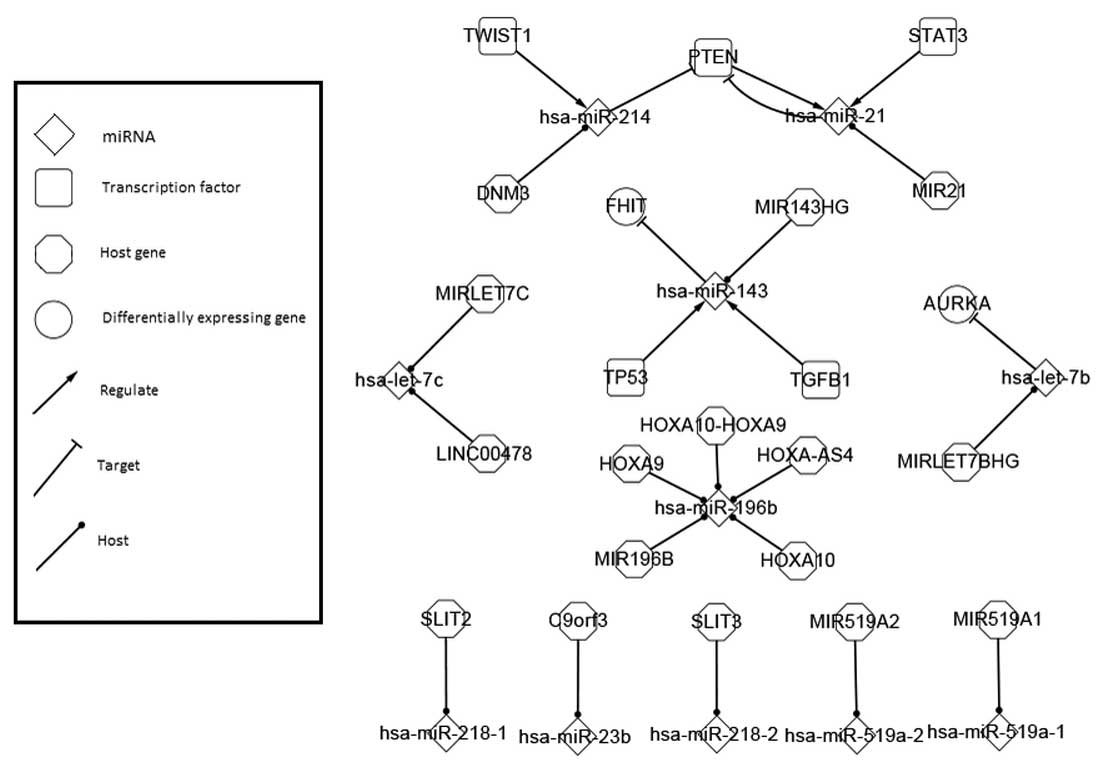
Differentially-expressed network in
cervical cancer
The differentially-expressed network is primarily
discussed in the present study due to its possession of
differentially-expressed biological factors that have been
experimentally validated in cervical cancer. The visual
representation is shown in Fig.
1.
Seven genes and 10 miRNAs appearing in Fig. 1 were experimentally validated as
differentially-expressed in cervical cancer. The sub-network
centering on PTEN was the principal component in the network.
TWIST1, hsa-miR-214, PTEN and hsa-miR-21 bind together as an
ordered control chain. hsa-miR-21 targeted its regulator PTEN,
which formed a self-adaption feedback loop, bringing a balance
mechanism and involving STAT3 and MIR21 in the system as
participants, similar to DNM3. Consequently, two host genes, two
transcription factors and one miRNA affected the expression of PTEN
and hsa-miR-21. At the same time, these two core factors controlled
each other. The circling between hsa-miR-21 and PTEN made them
dominant and dominating factors simultaneously, and turned the path
to a bidirectional net crux.
According to the organized data, hsa-miR-21 was
found to be differentially-expressed in ≥16 carcinomas and PTEN was
differentially-expressed in 13. miR-21 is regarded as one of the
oncomiRs contributing to tumor cell growth (24,25).
With the famous tumor suppressor, PTEN, as a bridge, hsa-miR-214
was involved in the regulatory balance between has-miR-21 and PTEN
(26). miR-214 has been
demonstrated to inhibit cell growth, migration and invasion
(27). These three significant
biochemical cancer-related factors are expressed abnormally in a
cervical tumor organism and bind together closely as a distinctive
sub-system (28–30).
In another partial network centered on hsa-miR-143,
TP53 and TGFB1 jointly has been shown to regulate hsa-miR-143, with
a resulting signal flow to FHIT. As a generally acknowledged tumor
suppressor standing out in various cancer investigations, TP53 may
be a starting point for further discussion (31).
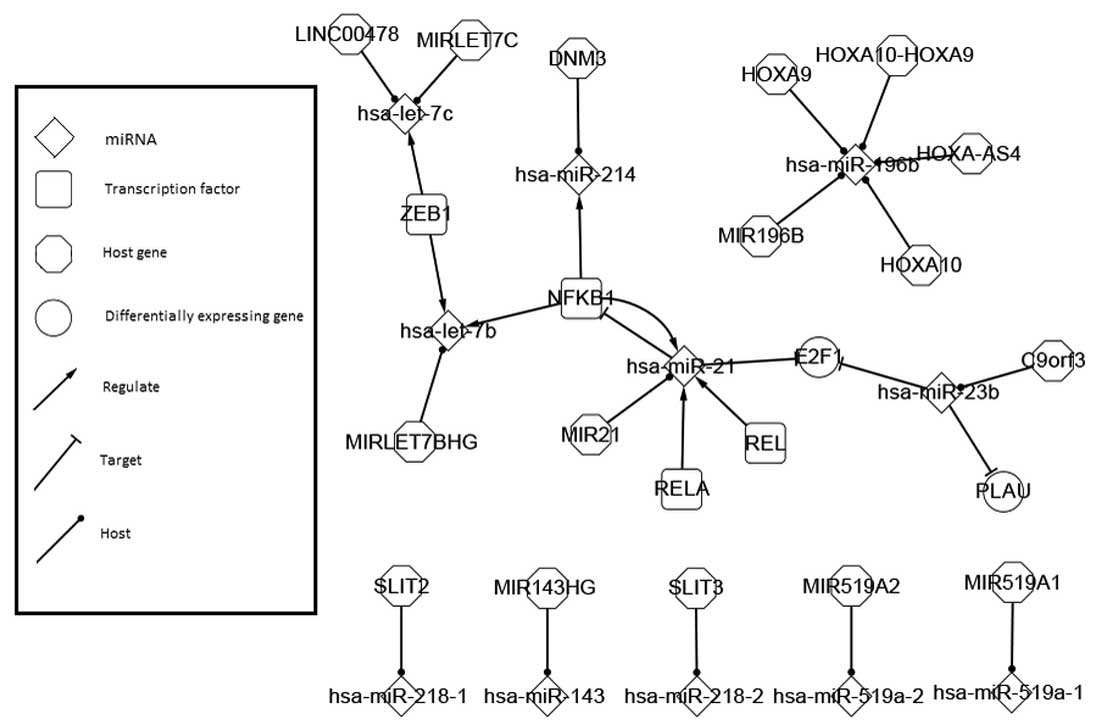
Cervical cancer-related network
The cervical cancer-related network expanded the
differentially-expressed network by absorbing more genes and miRNAs
whose association with cervical cancer are not as close as those of
the differentially-expressed ones. The network is demonstrated in
Fig. 2.
The related network was of a much larger scale and
of higher complexity than the differentially-expressed network.
Certain factors were prominent, including the miRNAs of hsa-miR-21,
hsa-miR-23b, hsa-miR-34, hsa-miR-143, hsa-let-7c and hsa-let-7c,
and the TFs of PTEN, TP63, MYC, TP53 and KRAS.
Two more self-adaption feedbacks were identified
upon hsa-let-7c, hsa-miR-34 and MYC. MYC acted as the TF of
hsa-let-7b and hsa-let-7c, which target KRAS. This is accordant
with the outcome that KRAS mutation-positive lung cancer,
displaying high levels of MYC, could be treated by inhibiting MYC
transactivation function (32).
Also, MYC overexpression may disturb the tumor suppressor
capability of hsa-let-7c (33).
Meanwhile, another tumor suppressor, hsa-miR-34, holds a local
balance adjustment system with MYC as well (34).
In a similar way, the related network could be used
to identify more possible core factors, sub-networks and
motifs.
Global network of cervical cancer
The global network contained all the factors and
associations involved with miRNA-gene targeting associations,
TF-miRNA regulating associations and host gene-miRNA hosting
associations. The network accommodated the experimentally validated
signaling transmissions and potential routes for further
investigations.
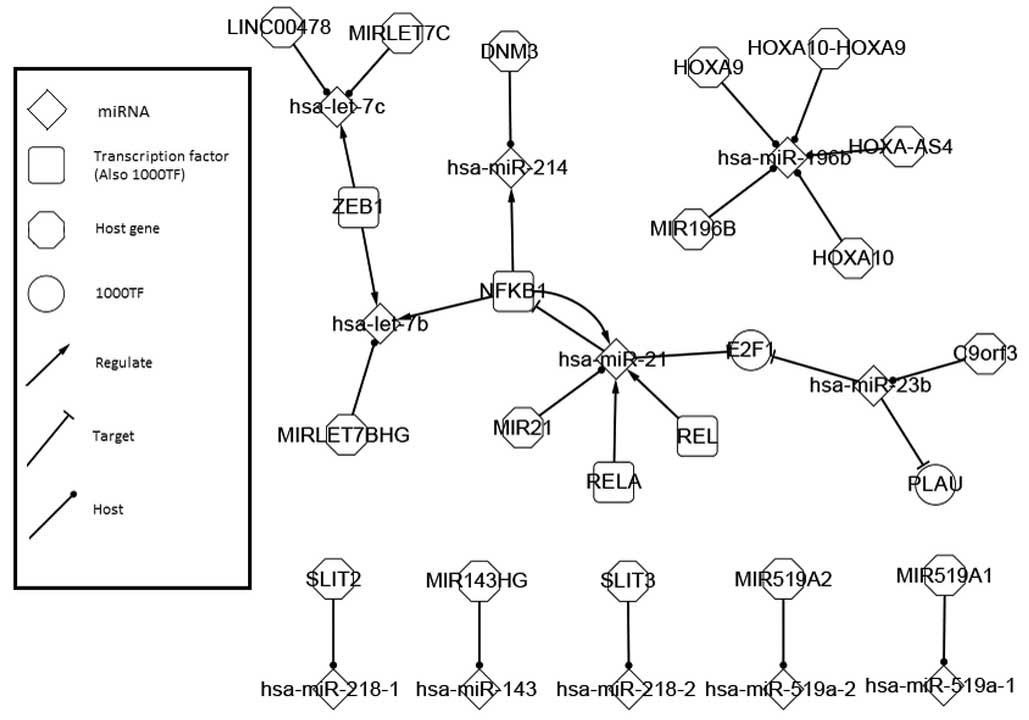
Comparison and analysis on features of
the 1,000-nt TF network
The interactions around the TFs that were acquired
previously and the base sequences of 1,000-nt were evaluated; they
were integrated with host genes and differentially-expressed miRNAs
to bring 1,000-nt TFs into the network system of cervical cancer
for overall clarification. The network is shown in Fig. 3.
As always, self-adaption feedback regulation was
assessed first; this existed between NFKB1 and hsa-miR-21 in this
circumstance. NFKB1 regulates hsa-miR-21, hsa-miR-214 and
hsa-let-7b. As discussed in the differentially-expressed network,
hsa-miR-21 and hsa-miR-214 are core biological factors in the
cervical cancer network and therefore, the reoccurrence here
strengthens the value of these particular miRNAs. With the help of
hsa-miR-21 and hsa-miR-214, NFKB1 takes part in the sub-system
where PTEN and has-miR-21 act as cores.
The present study defined an organized data system
and matched methods that can be computer-applied and transplanted
easily for the discussion of other cancer types.
Numerous genes and miRNAs with corresponding
specific biological features in cervical cancer were organized into
the form of associations representing their interactions with each
other as regulators or as those being regulated. The three levels
of networks demonstrated direct and indirect associations among
transcription factors, miRNAs, target genes and host genes in
different degrees of correlation with cervical cancer.
The networks provide core effective systems in
cervical cancer, particularly that of hsa-miR-214, PTEN and
hsa-miR-21. The methods used in the present study could be utilized
in full for further investigations on cancer-related networks.
References
|
1
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, et al: Mutations of the BRAF gene in human
cancer. Nature. 417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hamada S, Satoh K, Miura S, Hirota M,
Kanno A, et al: miR-197 induces epithelial-mesenchymal transition
in pancreatic cancer cells by targeting p120 catenin. J Cell
Physiol. 228:1255–1263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nohata N, Hanazawa T, Kinoshita T, Okamoto
Y and Seki N: MicroRNAs function as tumor suppressors or oncogenes:
Aberrant expression of microRNAs in head and neck squamous cell
carcinoma. Auris Nasus Larynx. 40:143–149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma D, Zhang YY, Guo YL, Li ZJ and Geng L:
Profiling of microRNA-mRNA reveals roles of microRNAs in cervical
cancer. Chin Med J (Engl). 125:4270–4276. 2012.PubMed/NCBI
|
|
6
|
Latchman DS: Transcription factors: An
overview. Int J Exp Pathol. 74:417–422. 1993.PubMed/NCBI
|
|
7
|
Libermann TA and Zerbini LF: Targeting
transcription factors for cancer gene therapy. Curr Gene Ther.
6:17–33. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chaudhry P, Srinivasan R and Patel FD:
Differential expression of Fas family members and Bcl-2 family
members in benign versus malignant epithelial ovarian cancer (EOC)
in North Indian population. Mol Cell Biochem. 368:119–126. 2012.
View Article : Google Scholar
|
|
9
|
Abdel-Rahman WM, Ruosaari S, Knuutila S
and Peltomäki P: Differential roles of EPS8 in carcinogenesis: loss
of protein expression in a subset of colorectal carcinoma and
adenoma. World J Gastroenterol. 18:3896–3903. 2012. View Article : Google Scholar
|
|
10
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar
|
|
12
|
Cao G, Huang B, Liu Z, Zhang J, Xu H, Xia
W, et al: Intronic miR-301 feedback regulates its host gene, ska2,
in A549 cells by targeting MEOX2 to affect ERK/CREB pathways.
Biochem Biophys Res Commun. 396:978–982. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Poliseno L, Salmena L, Riccardi L, Fornari
A, Song MS, Hobbs RM, et al: Identification of the miR-106b~25
microRNA cluster as a proto-oncogenic PTEN-targeting intron that
cooperates with its host gene MCM7 in transformation. Sci Signal.
3:ra292010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Das Ghosh D, Bhattacharjee B, Sen S, Premi
L, Mukhopadhyay I, et al: Some novel insights on HPV16 related
cervical cancer pathogenesis based on analyses of LCR methylation,
viral load, E7 and E2/E4 expressions. PLoS One.
7:e446782012.PubMed/NCBI
|
|
15
|
Sethupathy P, Corda B and Hatzigeorgiou
AG: TarBase: A comprehensive database of experimentally supported
animal microRNA targets. RNA. 12:192–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu SD, Lin FM, Wu WY, Liang C, Huang WC,
Chan WL, et al: miRTarBase: a database curates experimentally
validated microRNA-target interactions. Nucleic Acids Res.
39:D163–D169. 2011. View Article : Google Scholar
|
|
17
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: a
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kozomara A and Griffiths-Jones S: miRBase
integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, et al: miR2Disease: a manually curated database for
microRNA deregulation in human disease. Nucleic Acids Res.
37:D98–D104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Safran M, Dalah I, Alexander J, et al:
GeneCards Version 3: the human gene integrator. Database (Oxford).
5:20102010.
|
|
22
|
Chekmenev DS, Haid C and Kel AE: P-Match:
transcription factor binding site search by combining patterns and
weight matrices. Nucleic Acids Res. 33:W432–W437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, et al: The UCSC genome browser database:
update 2011. Nucleic Acids Res. 39:D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu TX, Lim EJ, Itskovich S, Besse JA,
Plassard AJ, et al: Targeted ablation of miR-21 decreases murine
eosinophil progenitor cell growth. PLoS One. 8:e593972013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Muppala S, Mudduluru G, Leupold JH, Buergy
D, Sleeman JP and Allgayer H: CD24 induces expression of the
oncomir miR-21 via Src, and CD24 and Src are both
post-transcriptionally downregulated by the tumor suppressor
miR-34a. PLoS One. 8:e595632013. View Article : Google Scholar
|
|
26
|
Bubien V, Bonnet F, Brouste V, Hoppe S,
Barouk-Simonet E, et al; French Cowden Disease Network. High
cumulative risks of cancer in patients with PTEN hamartoma tumour
syndrome. J Med Genet. 50:255–263. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang F, Liu M, Li X and Tang H: MiR-214
reduces cell survival and enhances cisplatin-induced cytotoxicity
via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett.
587:488–495. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hou LL, Gao C, Chen L, Hu GQ and Xie SQ:
Essential role of autophagy in fucoxanthin-induced cytotoxicity to
human epithelial cervical cancer HeLa cells. Acta Pharmacologica
Sinica. 34:1403–1410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lui WO, Pourmand N, Patterson BK and Fire
A: Patterns of known and novel small RNAs in human cervical cancer.
Cancer Res. 67:6031–6043. 2007. View Article : Google Scholar
|
|
30
|
Yang ZZ, Chen S, Luan X, Li YX, Liu M, Li
X, Liu T and Tang H: MicroRNA-214 is aberrantly expressed in
cervical cancers and inhibits the growth of HeLa cells. IUBMB Life.
61:1075–1082. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matlashewski G, Lamb P, Pim D, Peacock J,
Crawford L and Benchimol S: Isolation and characterization of a
human p53 cDNA clone: expression of the human p53 gene. EMBO J.
3:3257–3262. 1984.PubMed/NCBI
|
|
32
|
Fukazawa T, Maeda Y, Matsuoka J, Yamatsuji
T, Shigemitsu K, et al: Inhibition of Myc effectively targets KRAS
mutation-positive lung cancer expressing high levels of Myc.
Anticancer Res. 30:4193–4200. 2010.
|
|
33
|
Wu X, Ajani JA, Gu J, Chang DW, Tan W, et
al: MicroRNA expression signatures during malignant progression
from Barrett’s esophagus to esophageal adenocarcinoma. Cancer Prev
Res (Phila). 6:196–205. 2013.
|
|
34
|
Yamazaki H, Chijiwa T, Inoue Y, Abe Y,
Suemizu H, et al: Overexpression of the miR-34 family suppresses
invasive growth of malignant melanoma with the wild-type p53 gene.
Exp Ther Med. 3:793–796. 2012.PubMed/NCBI
|