Introduction
Breast cancer is the most common malignancy in
females (1). Treatments for breast
cancer patients include surgery and chemotherapy, as well as
radiation, hormone and molecular-targeted therapies, and yet
metastasis and recurrence remain clinical challenges for a
substantial proportion of patients. Biomarkers for breast cancer
are urgently required for early diagnosis, patient stratification
and prognosis determination.
The Vav proteins are guanine nucleotide exchange
factors for GTPases of the Rho family. Vav proteins are involved in
cell signaling and tumorigenesis (2,3). The first
report of a Vav protein (now known as Vav1) was in 1989, as the
result of cell transformation experiments that determined it was a
human oncogene (4). Subsequent to
this discovery, two more Vav proteins, Vav2 and Vav3, have been
identified in mammals (5). Vav2 and
Vav3 are expressed in the majority of tissues, while Vav1 is mostly
expressed in cells of hematopoietic lineage (6).
The Vav3 oncogene is involved in various cellular
signaling processes, including cytoskeleton organization, calcium
influx, gene transcription, cell transformation, cell proliferation
and apoptosis (2). Vav3 has been
found to be overexpressed in human prostate cancer cells and has
been proposed to promote the tumorigenesis of prostate cancer
(7,8).
Vav3 enhances cell growth and proliferation by activating androgen
receptor-mediated signaling pathways (8). Breast cancer and prostate cancer are
hormone-dependent tumors, whose growth is mediated by their
respective hormone receptors. Vav3 is an upstream mediator of
Ras-related C3 botulinum toxin substrate 1, which enhances the
transcriptional activity of estrogen receptor α (ER-α) in breast
cancer cells (9). In addition, Vav3
is epigenetically regulated during the development of breast cancer
(10). Thus, it is intriguing to
postulate that the progression and maintenance of breast cancer
relies on the deregulation of the Vav3 oncogene.
In the present study, the expression of Vav3 in
breast cancer and benign breast lesion tissues was analyzed, and
the clinical and prognostic significance of Vav3 expression in
human breast cancer was evaluated.
Materials and methods
Cell culture
The human breast cancer cell lines, MCF7 and
MDA-MB-231, and tamoxifen-resistant (TAM-R) breast cancer cells
were kindly provided by Dr Ping Fan (University of Virginia Health
Sciences System, Charlottesville, VA, USA). In addition, non-tumor
human breast epithelial MCF10A cells were obtained from the
American Type Culture Collection (Rockville, MD, USA). MCF7,
MDA-MB-231 and MCF10A cells were cultured in Dulbecco's modified
Eagle's medium (Invitrogen, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (Gibco BRL, Gaithersburg, MD, USA) at 37°C
in a humidified atmosphere of 5% CO2. TAM-R cells
derived from MCF7 cells were continuously cultured in the above
medium containing 10−7 mol/l tamoxifen.
Western blot analysis
Total proteins from the MCF7, MDA-MB-231, TAM-R and
MCF10A cells were extracted on ice with cell lysis buffer (Cell
Signaling Technology, Inc., Danvers, MA, USA). Equal amounts of
protein were separated by 10% SDS-PAGE and transferred to a
polyvinylidene difluoride membrane. The membranes were blocked with
2% skimmed milk in phosphate-buffered saline (PBS) at room
temperature for 1 h and probed with the primary polyclonal goat
anti-human Vav3 (1:300; cat. no. ab21208; Abcam, Cambridge, MA,
USA) and monoclonal mouse anti-human glyceraldehyde-3-phosphate
dehydrogenase (1:1,000; cat. no. 60004-1, Proteintech Group,
Chicago, IL, USA) antibodies overnight at 4°C in PBS containing
0.1% Tween-20 (PBST) and 1% skimmed milk. The membranes were then
washed four times in PBST and incubated with monoclonal goat
anti-rabbit horseradish peroxidase-conjugated secondary antibody
(1:1,000; cat. no. STAR54, Bio-Rad Laboratories, Inc., Hercules,
CA, USA). Signals were developed with enhanced chemiluminescent
reagents (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Patients and follow-up
This study was approved by the Ethics Committee of
the First Affiliated Hospital of Nanjing Medical University
(Nanjing, Jiangsu, China). A cohort of 173 breast cancer patients
and 19 patients with benign breast disease (fibroadenosis or
fibroadenoma) were recruited for the study. Written informed
consent was obtained from all patients. The patients underwent
surgical treatment between the beginning of 2004 and the end of
2007. ER, progesterone receptor (PR) and human epidermal growth
factor receptor 2 (HER2) status were determined by
immunohistochemistry (Table I).
 | Table I.Clinicopathological characteristics of
173 breast cancer patients. |
Table I.
Clinicopathological characteristics of
173 breast cancer patients.
| Characteristic | Number of
patients | % |
|---|
| Age at diagnosis,
years |
|
|
| ≤35 | 6 | 3.5 |
|
35–55 | 100 | 57.8 |
|
>55 | 67 | 38.7 |
| Tumor size, cm |
|
|
| ≤2 | 97 | 56.1 |
| 2–5 | 66 | 38.2 |
|
>5 | 10 | 5.7 |
| Lymph node stage |
|
|
| N0 | 94 | 54.3 |
| N1 | 45 | 26.0 |
| N2 | 20 | 11.6 |
| N3 | 14 | 8.1 |
| Histological
subtype |
|
|
| Invasive
ductal carcinoma | 151 | 87.3 |
| Invasive
lobular carcinoma | 12 | 7.0 |
| Ductal
carcinoma in situ | 4 | 2.3 |
|
Othera | 6 | 3.4 |
| Estrogen
receptor |
|
|
|
Positive | 109 | 63.0 |
|
Negative | 64 | 37.0 |
| Progesterone
receptor |
|
|
|
Positive | 119 | 68.8 |
|
Negative | 54 | 31.2 |
| HER2 |
|
|
|
Positive | 46 | 26.6 |
|
Negative | 127 | 73.4 |
| TNM staging |
|
|
| I | 62 | 35.8 |
| II | 75 | 43.4 |
| III | 35 | 20.2 |
| IV | 1 | 0.6 |
The patients were followed up every three months for
the first two years, every six months for the next three years and
once a year after five years. Chest computed tomography,
mammography or breast sonography (for patients ≤35 years old),
radionuclide bone scans, abdominal sonography, serum tumor marker
analysis and detailed physical examinations were routinely
performed. No patients were lost to follow-up.
Tissue microarray construction
A tissue microarray from the 192 selected patients
was constructed as previously described (11–15). In
brief, hematoxylin and eosin-stained sections of the primary tumors
were reviewed, and areas of tumors were marked on the slides.
Tissue microarrays were constructed by removing 1-mm cores from
selected paraffin-fixed tissue blocks and transferring them to a
recipient paraffin block using a Manual Tissue Arrayer (Beecher
Instruments, Silver Spring, MD, USA). Cores were spaced at
intervals of 1.5 mm. All samples were spotted in duplicate,
corresponding to the respective areas of the same original paraffin
block. Sections of 4-µm thick histological cuts were obtained from
the tissue microarray and fixed onto glass slides with adhesive
film.
Immunohistochemistry
After the 4-µm tissue microarray sections were
deparaffinized, endogenous peroxidase activity was blocked with
0.3% hydrogen peroxide for 10 min, followed by incubation with a
polyclonal primary antibody against Vav3 (1:50, Upstate
Biotechnology, Lake Placid, NY, USA) for 1 h. Subsequent to washing
in PBS three times, sections were incubated for 40 min with the
secondary antibody (BioGenex Laboratories, Inc., San Ramon, CA,
USA) at room temperature. After washing, the sections were
incubated with streptavidin-conjugated peroxidase (BioGenex).
The intensity and extent of cytoplasm-positive
labeling for Vav3 in the tissue arrays were assessed
semi-quantitatively and scored as follows: 0, no staining; 1+, weak
and focal staining in <30% of the tissues; 2+, moderate
intensity staining in 30–50% of the tissues; or 3+, strong and
diffuse staining in >50% of the tissues. A score of 0 was
defined as negative for Vav3 labeling.
For hormone receptors, the staining intensity was
scored as follows: Negative, -; low, 1+; moderate, 2+; or strong,
3+. Invasive tumor cell nuclear staining ≥10% was considered
hormone receptor-positive, while <10% was considered negative.
The criteria for positive HER2 was 3+ uniform cell membrane
staining in >30% of tumor cells. Negative HER2 was 0, or 1+ cell
surface protein expression in any percentage of staining. HER2
scored as 2+ and 3+ in <30% of tumor cells by
immunohistochemistry was further confirmed by fluorescence in
situ hybridization.
Statistical analyses
Statistical analyses were performed using STATA 10.0
software (StataCorp LP, College Station, TX, USA). The Student's
t-test was used to determine the differences in Vav3 expression.
Differences in proportions were evaluated with the χ2 or
Fisher's exact tests. The Kaplan-Meier method was used to calculate
the non-parametric survival plots, and the difference was
determined by the log-rank test. Disease-free survival (DFS) was
calculated as the time from the date of diagnosis to the occurrence
of locoregional or distant metastasis. The overall survival (OS)
period was calculated from the date of diagnosis to mortality or
the date of last follow-up. The Cox regression model was used to
evaluate the prognostic significance of Vav3. P<0.05 was
considered to indicate a statistically significant difference.
Results
Vav3 oncogene is overexpressed in
human breast cancers
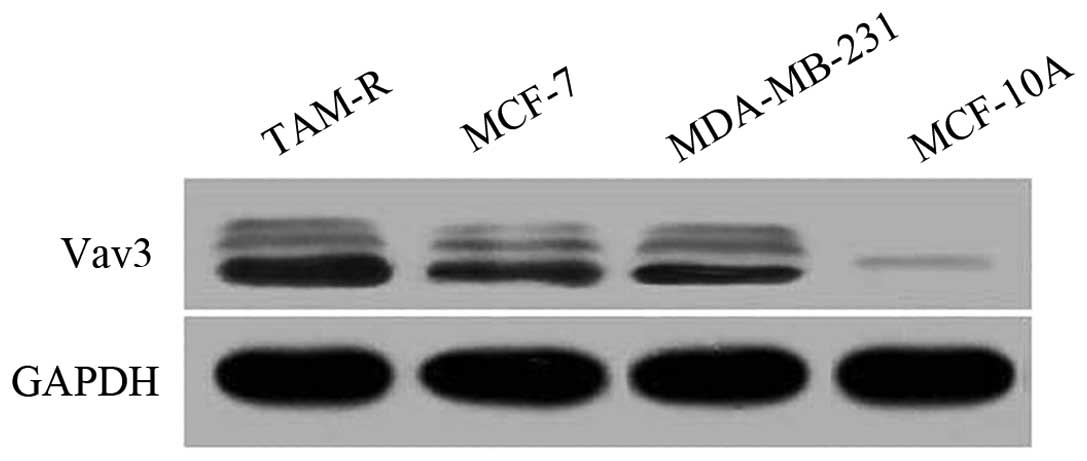
To determine the expression status of Vav3 in breast
cancers, the Vav3 protein levels in breast cancer cell lines were
first checked using western blot analysis. Compared with the breast
epithelial MCF10A cells, the cells of the breast cancer MCF7 and
MDA-MB-231 cell lines, and the TAM-R cells revealed an apparently
higher expression level of Vav3 (Fig.
1).
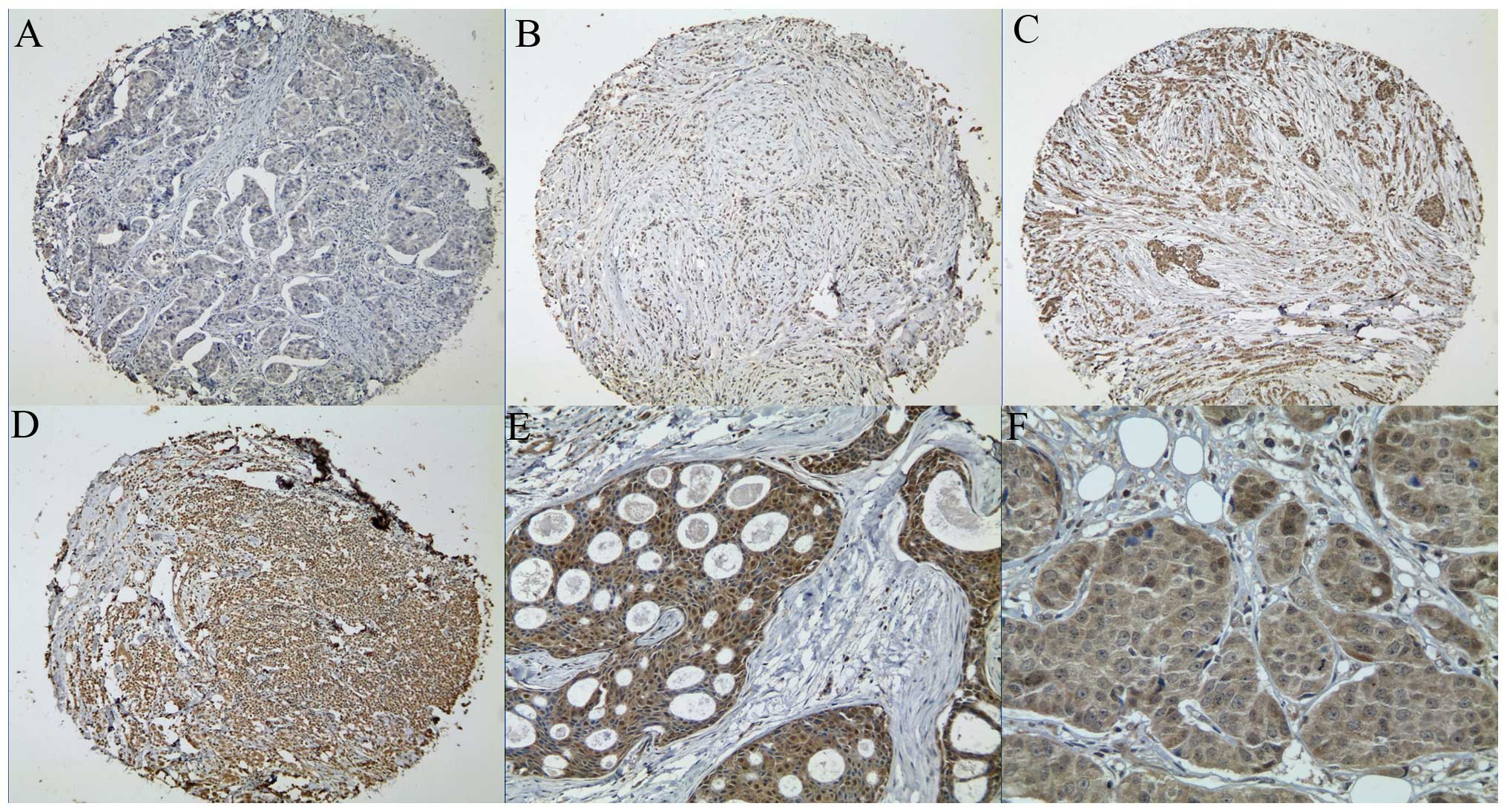
To extend this observation in vivo,
immunohistochemistry was used to evaluate Vav3 expression in the
tissue microarray, which contained 173 human primary breast cancers
and 19 benign breast tissues. Vav3 was mainly located in the
cytoplasm and nucleus of the epithelial cells of the breast
tissues, but not in the stroma (Fig.
2). Vav3 was detected in 3 of 19 (15.8%) normal breast tissues,
including one case of moderate intensity and two cases of weak
intensity. In the breast carcinoma tissues, 149 of 173 (86.1%)
tumor specimens stained positive for Vav3 (P<0.05). These in
vitro and in vivo data demonstrated that Vav3 was
overexpressed in the breast cancer cells.
Correlation between Vav3 expression
and clinicopathological features
Next, the clinicopathological features of
Vav3-positive and Vav3-negative breast cancers were analyzed
(Table II). The expression of Vav3
was significantly correlated with the clinical
tumor-node-metastasis (TNM) phase (P=0.0309), pathological type
(P=0.007), ER status (P=0.038) and axillary lymph node involvement
(P=0.045). There was no correlation between Vav3 expression and
tumor size, HER2 overexpression, PR status, age at diagnosis or p53
status (P>0.05).
 | Table II.Association of Vav3 expression status
with clinicopathological and molecular characteristics. |
Table II.
Association of Vav3 expression status
with clinicopathological and molecular characteristics.
| Characteristic | Number of
patients | Vav3-positive, n | % | P-value |
|---|
| Age at diagnosis,
years |
|
|
|
|
| ≤35 | 6 | 5 | 83.3 | 0.3860 |
|
>35 | 167 | 144 | 86.2 |
|
| Tumor size, cm |
|
|
|
|
| ≤2 | 97 | 85 | 87.6 | 0.2680 |
| 2–5 | 66 | 55 | 83.3 |
|
|
>5 | 10 | 9 | 90.0 |
|
| Lymph node |
|
|
|
|
|
Negative | 94 | 76 | 80.9 | 0.0450 |
|
Positive | 79 | 73 | 86.7 |
|
| Histological
subtype |
|
|
|
|
|
Invasive ductal carcinoma | 151 | 133 | 88.1 | 0.0070 |
|
Invasive lobular
carcinoma | 12 | 11 | 91.7 |
|
| Ductal
carcinoma in situ | 4 | 1 | 25.0 |
|
|
Othera | 6 | 4 | 66.7 |
|
| Estrogen
receptor |
|
|
|
|
|
Positive | 109 | 89 | 69.0 | 0.0380 |
|
Negative | 64 | 60 | 93.8 |
|
| Progesterone
receptor |
|
|
|
|
|
Positive | 119 | 99 | 83.2 | 0.1530 |
|
Negative | 54 | 50 | 92.6 |
|
| HER2 |
|
|
|
|
|
Positive | 46 | 43 | 93.5 | 0.1340 |
|
Negative | 127 | 106 | 83.5 |
|
| TNM staging |
|
|
|
|
|
I–II | 137 | 114 | 83.2 | 0.0309 |
|
III–IV | 36 | 35 | 97.2 |
|
Prognostic value of Vav3 in breast
cancer patients
The median follow-up period was 59 months (range,
46–85 months). The OS rate at the end of the follow-up period was
87.3%. At the end of the follow-up, 140 (80.9%) patients were free
of disease. Among the 33 (19.1%) patients with events, six
presented with local recurrence, 14 with distant metastasis
(including 12 mortalities) and three with contralateral metastasis,
and 10 succumbed to unknown causes.
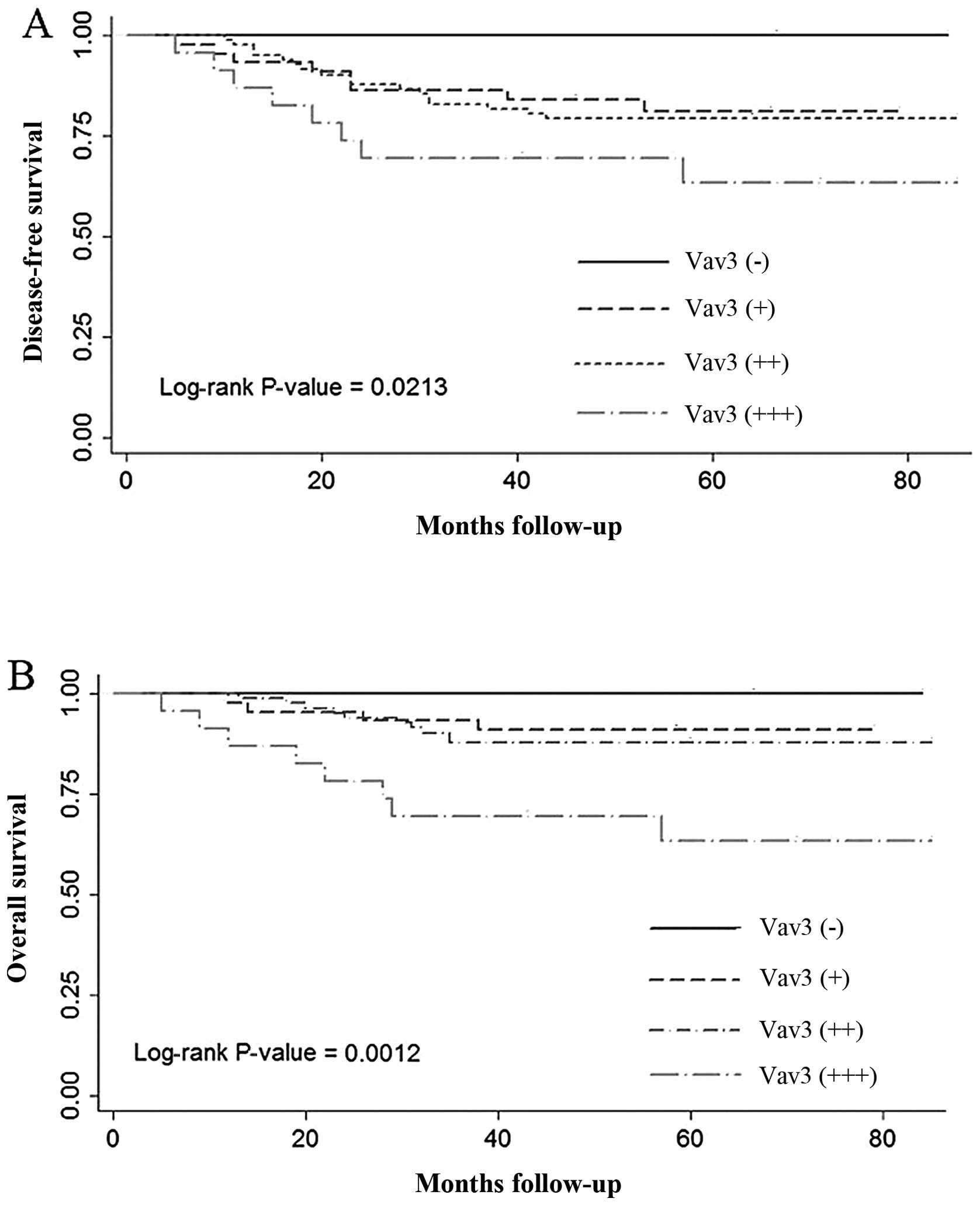
When all breast cancer patients were divided into
groups based on Vav3 expression (negative, weak, moderate or
strong), using the log-rank test, it was observed that patients
with strong positive expression of Vav3 experienced the shortest
DFS (P=0.0213) and OS (P=0.0012) times (Fig. 3). In the multivariate Cox regression
analysis, overexpression of Vav3 was associated with poor DFS
(P=0.019) and OS (P=0.004) when the age at diagnosis, tumor size,
TNM stage and lymph node status were adjusted.
Discussion
High levels of Vav3 have been observed in various
types of cancer, including glioblastoma (16) and prostate cancer (8). In the present study, Vav3 was observed
to be significantly upregulated in breast cancers compared with
benign breast diseases. Furthermore, Vav3 was identified to be a
biomarker of a poor prognosis in breast cancer.
The Vav3 oncogene induces cell transformation
(17) and mediates receptor protein
tyrosine kinase signaling, including that of the epidermal growth
factor, insulin and insulin-like growth factor I receptors. Vav3
suppresses apoptosis by activating the Ras/Raf/MEK/ERK/Elk-1
signaling pathway (18). Furthermore,
while neovascularization is inherent to the growth and metastasis
of tumors, Vav3 has been found to enhance tumor angiogenesis by
stimulating the activation of the EphA2 receptor-mediated signaling
pathway, which has a crucial role in the growth of vascular
endothelial cells (19,20). Thus, the progression and maintenance
of tumors may depend on the deregulation of Vav3.
In the present study, Vav3 protein levels were
elevated in the human breast cancer cells in comparison with the
human breast epithelial cells. Immunohistochemical analysis
revealed that Vav3 was expressed in 86.1% of breast cancers, but in
only 15.8% of benign breast diseases. Most significantly, a close
association was noted between Vav3 expression and several
indicators of a poor prognosis in breast cancer, including ER
negativity, axillary lymph node involvement and advanced TNM
stage.
Previous studies reported that ER was a sensitive
prognostic factor and that patients that were ER-negative had poor
DFS (21,22). The present study observed that Vav3
expression levels were significantly higher in ER-negative
patients. In addition, the Vav3-positive group contained more
patients with axillary lymph node involvement, which is also a
significant prognostic factor (23),
suggesting that patients with increased Vav3 may have a poorer
outcome.
In the present study, the expression rate of Vav3 in
the patients with stage III–IV breast cancer (97.2%) was
significantly higher than that in the patients with stage I–II
breast cancer (83.2%). However, no significant association was
noted among the four TNM staging subgroups, possibly due to the
small sample size. It was previously reported that patients younger
than 35 years frequently presented with high-grade breast cancers,
which predicted a poorer outcome in these young patients (24). However, in the present study, Vav3
expression was not associated with age, which may be explained by
the small population of patients aged less than 35 years.
Results of the Kaplan-Meier survival analysis
demonstrated that patients with the highest expression of Vav3 had
the poorest DFS and OS times, and this was further supported by the
multivariate analysis. Together, these data suggest that Vav3 is an
independent factor in the prognosis of breast cancer.
The current study is limited by the relatively small
sample size, which may lead to selection bias. Furthermore, as
there is no international standard to define the Vav3 expression
level, large population-based studies are required to determine a
reference for evaluating Vav3 expression. Finally, the study was
based on a retrospective analysis. Prospective studies are required
to further investigate Vav3 expression in breast cancer.
In conclusion, the present study demonstrated that
Vav3 was upregulated in breast cancer and associated with poor
survival, suggesting that Vav3 is a biomarker for the prognosis of
breast cancer.
Acknowledgements
This study was supported by the Priority Academic
Program Development of Jiangsu Higher Education Institutions.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bustelo XR: The VAV family of signal
transduction molecules. Crit Rev Oncog. 7:65–88. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Aelst L and D'Souza-Schorey C: Rho
GTPases and signaling networks. Genes Dev. 11:2295–2322. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katzav S, Martin-Zanca D and Barbacid M:
vav, a novel human oncogene derived from a locus ubiquitously
expressed in hematopoietic cells. EMBO J. 8:2283–2290.
1989.PubMed/NCBI
|
|
5
|
Zugaza JL, Lopez-Lago MA, Caloca MJ, Dosil
M, Movilla N and Bustelo XR: Structural determinants for the
biological activity of Vav proteins. J Biol Chem. 277:45377–45392.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bustelo XR: Regulatory and signaling
properties of the Vav family. Mol Cell Biol. 20:1461–1477. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lyons LS and Burnstein KL: Vav3, a Rho
GTPase guanine nucleotide exchange factor, increases during
progression to androgen independence in prostate cancer cells and
potentiates androgen receptor transcriptional activity. Mol
Endocrinol. 20:1061–1072. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong Z, Liu Y, Lu S, et al: Vav3 oncogene
is overexpressed and regulates cell growth and androgen receptor
activity in human prostate cancer. Mol Endocrinol. 20:2315–2325.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rosenblatt AE, Garcia MI, Lyons L, et al:
Inhibition of the Rho GTPase, Rac1, decreases estrogen receptor
levels and is a novel therapeutic strategy in breast cancer. Endoc
Relat Cancer. 18:207–219. 2011.
|
|
10
|
Loss LA, Sadanandam A, Durinck S, et al:
Prediction of epigenetically regulated genes in breast cancer cell
lines. BMC Bioinformatics. 11:3052010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sauter G, Simon R and Hillan K: Tissue
microarrays in drug discovery. Nat Rev Drug Discov. 2:962–972.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Torhorst J, Bucher C, Kononen J, et al:
Tissue microarrays for rapid linking of molecular changes to
clinical endpoints. Am J Pathol. 159:2249–2256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nocito A, Bubendorf L, Tinner EM, et al:
Microarrays of bladder cancer tissue are highly representative of
proliferation index and histological grade. J Pathol. 194:349–357.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moch H, Schraml P, Bubendorf L, et al:
High-throughput tissue microarray analysis to evaluate genes
uncovered by cDNA microarray screening in renal cell carcinoma. Am
J Pathol. 154:981–986. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kononen J, Bubendorf L, Kallioniemi A, et
al: Tissue microarrays for high-throughput molecular profiling of
tumor specimens. Nat Med. 4:844–847. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Salhia B, Tran NL, Chan A, et al: The
guanine nucleotide exchange factors trio, Ect2 and Vav3 mediate the
invasive behavior of glioblastoma. Am J Pathol. 173:1828–1838.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng L, Sachdev P, Yan L, et al: Vav3
mediates receptor protein tyrosine kinase signaling, regulates
GTPase activity, modulates cell morphology and induces cell
transformation. Mol Cell Biol. 20:9212–9224. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Palmby TR, Abe K, Karnoub AE and Der CJ:
Vav transformation requires activation of multiple GTPases and
regulation of gene expression. Mol Cancer Res. 2:702–711.
2004.PubMed/NCBI
|
|
19
|
Hunter SG, Zhuang G, Brantley-Sieders D,
Swat W, Cowan CW and Chen J: Essential role of Vav family guanine
nucleotide exchange factors in EphA receptor-mediated angiogenesis.
Mol Cell Biol. 26:4830–4842. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fang WB, Brantley-Sieders DM, Hwang Y, Ham
AJ and Chen J: Identification and functional analysis of
phosphorylated tyrosine residues within EphA2 receptor tyrosine
kinase. J Biol Chem. 283:16017–16026. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vollenweider-Zerargui L, Barrelet L, Wong
Y, Lemarchand-Beraud T and Gomez F: The predictive value of
estrogen and progesterone receptors' concentrations on the clinical
behavior of breast cancer in women. Clinical correlation on 547
patients. Cancer. 57:1171–1180. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Viale G, Regan MM, Maiorano E, et al:
Prognostic and predictive value of centrally reviewed expression of
estrogen and progesterone receptors in a randomized trial comparing
letrozole and tamoxifen adjuvant therapy for postmenopausal early
breast cancer: BIG 1–98. J Clin Oncol. 25:3846–3852. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Altan E and Altundag K: Clinical and
pathological characteristics of occult breast cancer and review of
the literature. J BUON. 16:434–436. 2011.PubMed/NCBI
|
|
24
|
Colleoni M, Rotmensz N, Robertson C, et
al: Very young women (<35 years) with operable breast cancer:
features of disease at presentation. Ann Oncol. 13:273–279. 2002.
View Article : Google Scholar : PubMed/NCBI
|