Introduction
As the most common neoplasm in the kidney, renal
cell carcinoma (RCC) comprises 3% of all cases of adult cancer. In
the United States, the incidence of RCC is increasing, with an
estimated 63,920 new cases and 13,860 mortalities in 2014 (1). In South Korea, RCC accounts for ∼1% of
all primary malignancies and is the tenth most common cancer in
males (2). RCC may be classified into
a number of subtypes, including papillary, chromophobe and
collecting duct RCC, clear cell RCC (CCRCC) and other rare
subtypes. CCRCC represents 70% of all RCCs and tends to have a
poorer prognosis compared with other RCCs (3).
Epigenetic changes may initiate cancer and promote
progression (4). Epigenetics may be
described as a stable alteration in gene expression potential that
takes place during cell proliferation and development, without any
change in gene sequence (5). In
cancer, DNA hypermethylation is critical in gene expression.
Methylation in cytosine-phosphate-guanine (CpG) islands may inhibit
gene expression, and CpG hypermethylation in promoter regions may
represent one mechanism of carcinogenesis; this may also provide
markers for tumor initiation and progression. Hypomethylation is
the second type of methylation defect that is observed in a wide
range of malignancies. Hypomethylation involves repeated DNA
sequences, including long interspersed nuclear elements (LINEs),
whereas hypermethylation involves CpG islands.
Chemokines regulate cancer cell migration and
contribute to cancer cell proliferation and survival (6). Approximately 45 chemokines and 20
chemokine receptors have been identified, and may be grouped into
four categories: C, CC, CXC and CX3C (7). Chemokine receptors relay their signals
through heterotrimeric guanine nucleotide-binding proteins (G
proteins) (7). CC chemokine receptor
5 (CCR5) belongs to the trimeric G protein-coupled
seven-transmembrane domain receptor superfamily (9), and binds to the RANTES (CCL5),
macrophage inflammatory protein (MIP)-1α (CCL3), and MIP-1β (CCL4)
chemokines (10). CCR5 is involved in
the chemotaxis of leukocytes to sites of inflammation, and is
important in the recruitment of macrophages, T cells and monocytes
(11). In the present study, the
methylation profile of CCR5 was investigated in CCRCC. The
association between tumoral CCR5 immunohistochemical expression and
clinicopathological parameters was also evaluated.
Material and methods
Patients and preparation of DNA
samples
The GoldenGate high-throughput genotyping assay was
adapted to determine the methylation state of 1,505 specific CpG
sites in 807 cancer-related genes (12). Tissue specimens consisted of 62 cancer
tissues and 62 matched adjacent normal tissues from CCRCC patients
at Kyung Hee University Hospital (Seoul, South Korea). The
Institutional Review Board of Kyung Hee University Hospital
approved this study (KHNMC IRB 2013-040). DNA was extracted as
previously described (13).
Methylation profiling and
validation
Bisulfite conversion of all DNA samples was
performed with the EZ-96 DNA Methylation™ kit (Zymo Research,
Orange, CA, USA) according to the manufacturer's instructions.
Following bisulfite treatment, the methylcytosine content was
quantified using Illumina's GoldenGate Methylation Cancer Panel I
microarray (Illumina Inc., San Diego, CA, USA) (12). The raw methylation ratios were
calculated using the Methylation Module in Illumina's BeadStudio
following normalization to a background level derived by averaging
the signals of an internal negative control. The methylation status
of the CpG sites was examined by bisulfite sequencing. The
procedure described previously by Herman et al (14) was adopted, with slight
modification.
Tissue microarray and
immunohistochemistry
For immunohistochemical staining, tissue samples
from 61 CCRCC cases were used. All neoplasms were surgically
resected at Kyung Hee University Hospital between 2006 and 2013.
The tissue microarrays were assembled using a commercially
available manual tissue microarrayer (Quick-Ray; Unitma Co., Ltd.,
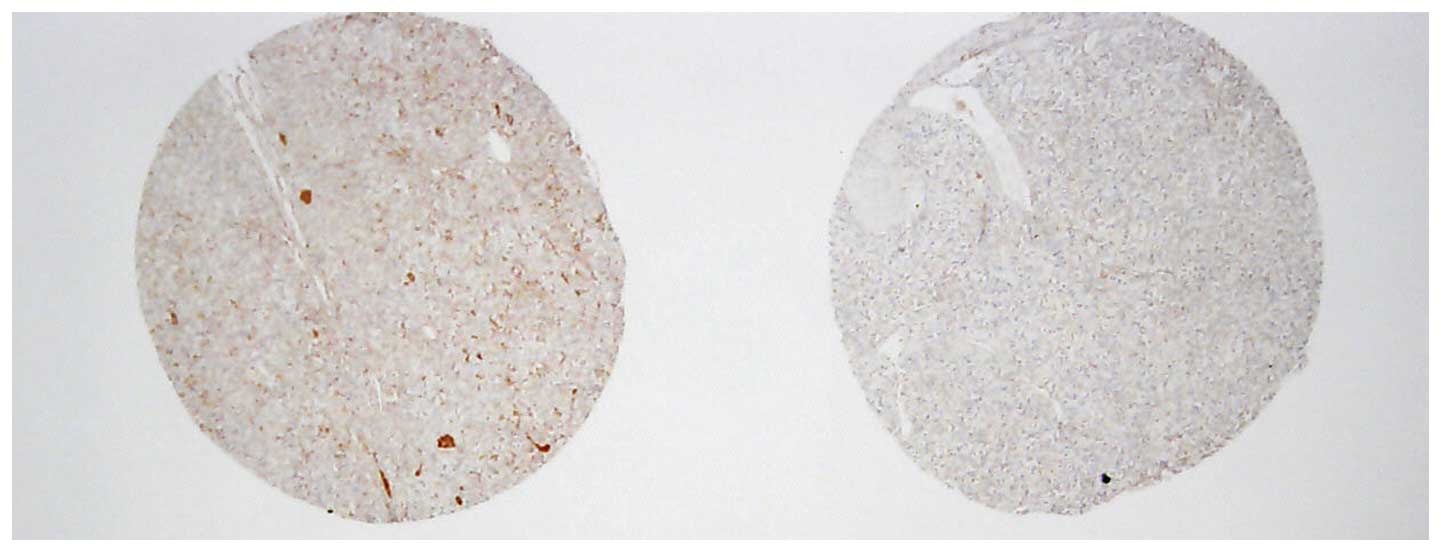
Seoul, South Korea). Three representative tumor cores with
diameters of 2.0 mm were punched from each tumor tissue block. Each
of the tissue microarray blocks contained three normal kidney
tissue cores (Fig. 1).
Immunohistochemistry was performed on 4-µm tissue sections from
each tissue microarray block using the Bond Polymer Intense
Detection System (Vision BioSystems, Victoria, Australia). Sections
were incubated for 15 min at ambient temperature with primary
rabbit polyclonal antibodies to CCR5 (dilution, 1:100; Novus
Biologicals, Cambridge, UK), using a biotin-free polymeric
horseradish peroxidase-linked antibody conjugate system in a
Bond-max automatic slide stainer (Vision BioSystems). Nuclei were
counterstained with hematoxylin. The negative control was treated
in an identical manner using mouse immunoglobulin G instead of
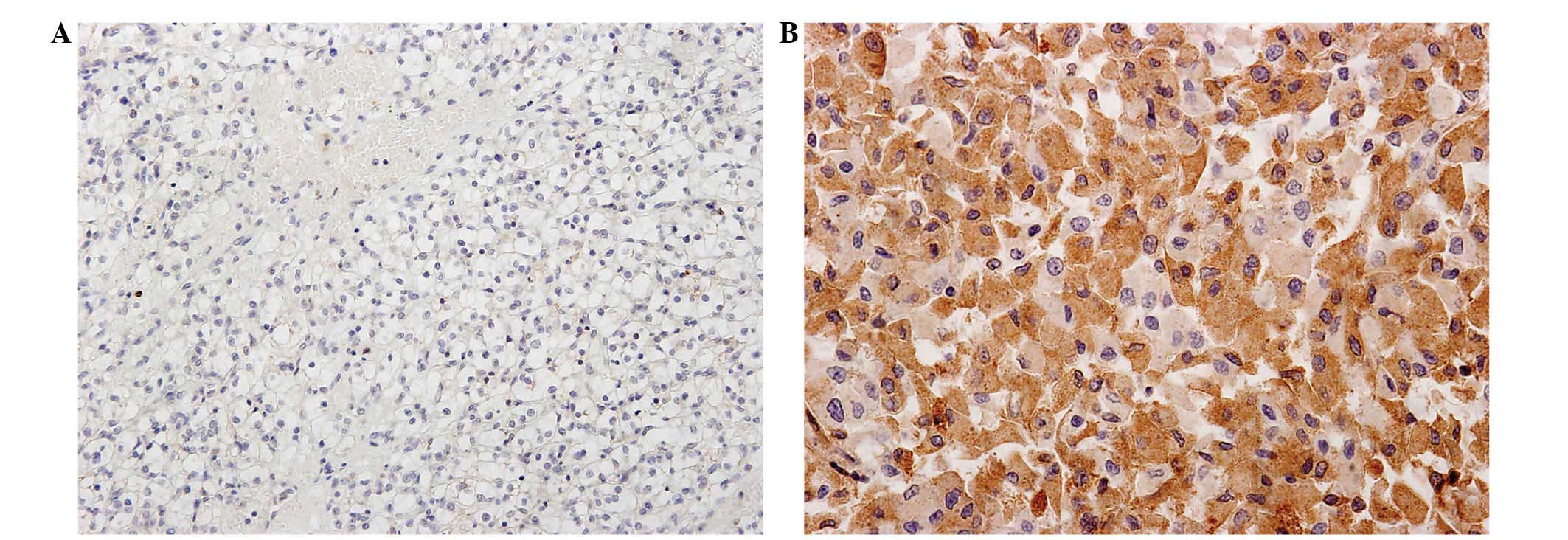
primary antibody. The degree of expression based on
immunohistochemistry was classified by three pathologists.
Semiquantitative analysis of immunoreactivity was performed
according to intensity and proportion: The intensity score was as
follows: 0, no staining; 1, weak but detectable staining; 2,
distinct staining; or 3, strong staining. The proportion score was
as follows: 0, 0% stained cells; 1, 1–33% stained cells; 2, 34–66%
stained cells; or 3, 67–100% stained cells. These two scores were
multiplied together for a total score, categorized as follows: 0–4,
low expression; and 5–9, high expression (Fig. 2).
Statistical analysis
Statistical analyses were performed using SPSS
software (version 15.0; SPSS, Inc., Chicago, IL, USA). A χ2 test
and linear-by-linear association were used to evaluate the
association of the degree of expression by immunohistochemistry
with clinicopathological variables. The overall survival was
defined as the time interval between the primary radical or partial
nephrectomy and the last follow-up or mortality. The
recurrence-free survival period was defined as the time interval
between the primary radical or partial nephrectomy and the last
follow-up or evidence of recurrence. Survival was estimated using
the Kaplan-Meier method. All statistical tests were two sided, and
P<0.05 was considered to indicate a statistically significant
difference.
Results
Methylation status of the CCR5 gene in
CCRCC
The methylation status of CCR5 in the 62 cancer
tissues and 62 matched adjacent normal tissues was analyzed using a
GoldenGate high-throughput genotyping assay. The methylation status
is represented by the β-value (15).
The results revealed that the mean β-values for CCR5 were 0.44 in
the normal tissues and 0.23 in the CCRCC tissues; the mean β-value
difference was −0.21. The methylation status of the CpG sites was
examined by bisulfite sequencing, revealing that CCR5
hypomethylation occurred to a greater extent in the cancer tissues
than the normal tissues. Methylation profiling revealed ∼10
significant genes, including CCR5. However, the other genes
revealed no significant results using immunohistochemical
staining.
CCR5 expression in CCRCC
Normal glomeruli and tubules exhibited no
immunoreactivity for CCR5. However, 17 cases out of a total of 61
CCRCCs exhibited high CCR5 expression. CCR5 was strongly expressed
in the cytoplasm of CCRCC cells.
Association between CCR5 expression
and clinicopathological parameters
Patients with low CCR5 expression (n=44) were
compared with those with high expression (n=17). The low expression
group was composed of 28 males and 16 females, while 12 males and 5
females formed the high expression group (P=0.766). The mean age (±
standard deviation) was 61.75±8.35 years in the low expression
group and 56.74±18.91 years in the high expression group (P=0.156).
The tumor (T) stage was significantly lower in the low CCR5
expression group compared with the high expression group (P=0.047).
In addition, the low expression group was associated with a
significantly lower Fuhrman grade compared with that of the high
expression group (P=0.044). Although the node (N) stage and
metastasis (M) stage of the low CCR5 expression group were lower
than those of the high expression group, this difference was not
statistically significant (N stage, P=0.632; M stage; P=0.896;
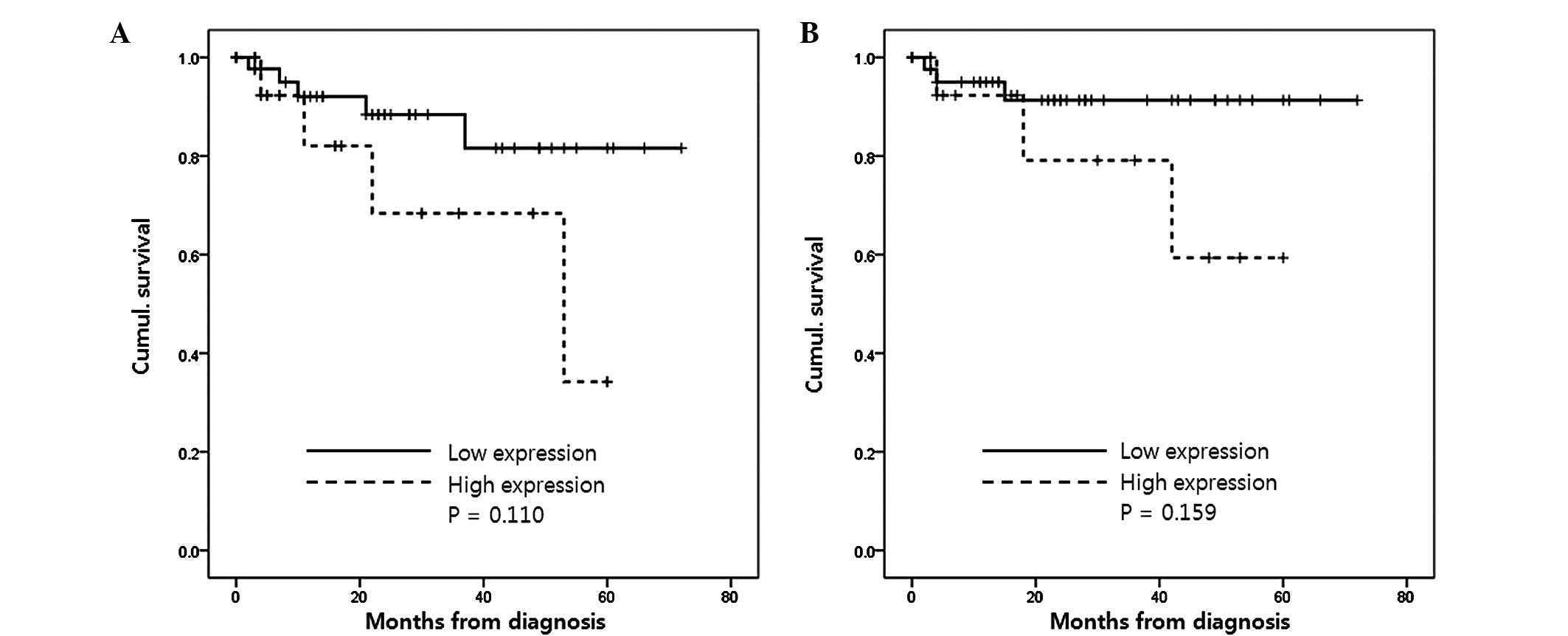
Table I). The low expression group
also had lower risks of post-operative tumor recurrence (P=0.110)
and mortality from CCRCC (P=0.159), however, these results were not
statistically significant (Fig.
3).
 | Table I.Association between CCR5 expression
and clinicopathological characteristics in clear cell renal cell
carcinoma. |
Table I.
Association between CCR5 expression
and clinicopathological characteristics in clear cell renal cell
carcinoma.
|
| CCR5, n |
|
|---|
|
|
|
|
|---|
| Clinicopathological
variable | Low expression | High expression | P-value |
|---|
| Patients | 44 | 17 |
|
| Gender |
|
| 0.766 |
| Male | 28 | 12 |
|
|
Female | 16 | 5 |
|
| Fuhrman nuclear
grade |
|
| 0.044 |
| I | 1 | 0 |
|
| II | 23 | 4 |
|
| III | 17 | 11 |
|
| IV | 3 | 2 |
|
| T stage |
|
| 0.047 |
| T1 | 36 | 9 |
|
| T2 | 2 | 3 |
|
| T3 | 6 | 5 |
|
| N stage |
|
| 0.632 |
| N0 | 42 | 16 |
|
| N1 | 2 | 1 |
|
| M stage |
|
| 0.896 |
| M0 | 41 | 16 |
|
| M1 | 3 | 1 |
|
Discussion
DNA hypomethylation was the first epigenetic
abnormality recognized in human malignancy (16). However, the hypermethylation of
promoters of tumor-suppressor genes is focused in carcinogenesis
(17). Recent high-resolution
genome-wide studies confirm that DNA hypomethylation frequently
co-occurs with hypermethylation of the genome in cancer. DNA
hypomethylation may be detected early in carcinogenesis, however,
it is also often associated with cancer progression (18). Hypomethylation may promote
carcinogenesis by causing an increase in DNA recombination, and via
direct and indirect effects on protein expression (19). Repeated DNA sequences that are
frequently hypomethylated in cancer tissue may act as tumor markers
for cancer diagnosis and prognosis (17). Hypomethylated DNA sequences are more
sensitive markers than unique sequences that are subject to
cancer-linked DNA hypermethylation.
DNA hypomethylation is a common methylation defect
that is observed in a wide variety of malignancies (20); it is common in solid tumors, including
prostate, cervical and metastatic hepatocellular cancer (21–23).
Global hypomethylation, such as in breast, cervical and brain
cancer, has been shown to progressively increase with grade of
malignancy (17). Patients with
immunodeficiency, centromeric instability or facial abnormalities,
as well as numerous other malignancies, exhibit severely
hypomethylated pericentric heterochromatin regions on chromosomes 1
and 16 (24). DNA hypomethylation has
been hypothesized to contribute to oncogenesis by the activation of
oncogenes, by the activation of latent retrotransposons or by
chromosome instability (25).
Jürgens et al (26) reported an association between
hypomethylation and urothelial carcinoma. In the study, DNA
methylation alterations were analyzed in 6 urothelial carcinoma
cell lines and 13 tumor tissues. L1 LINE sequence methylation was
reduced in the majority of tumors compared with that of normal
bladder mucosa. DNA hypermethylation of the calcitonin gene CpG
islands was restricted to the cell lines and was not detected in
the primary tumor tissues. L1 LINE hypomethylation appears to be
frequent in urothelial carcinoma and may be useful for diagnostic
or prognostic purposes.
A number of studies have previously investigated
CCR5 expression and solid tumor carcinogenesis. A study by
Zimmermann et al (27)
revealed that a low expression level of CCR5 in human colorectal
cancer is associated with lymphatic dissemination and reduced CD8+
T-cell infiltration. CCR5 expression that was weak or absent was
also significantly associated with lymph node metastasis and
advanced stage. The study hypothesized that T-cell retention at the
tumor site may be mediated by CCR5-dependent mechanisms of immune
and tumor cells, and concluded that CCR5 may play a role during the
progression of colorectal carcinoma, possibly preventing cancer
progression. However, van Deventer et al (28) revealed that CCR5 expression in
stromal, and not hematopoietic cells, contributes to tumor
metastasis. The study reported that mice expressing CCR5 exhibited
enhanced local tumor growth and an impaired response to vaccine
therapy compared with CCR5-knockout mice. Lin et al
(29) reported that CCR5 and CCL5
were highly expressed in breast cancer lymph node metastasis, and
that CCR5-CCL5 interaction promotes cancer cell migration under
hypoxic conditions.
In the present study, the methylation profile of
CCR5 in CCRCC, and the association between tumoral CCR5
immunohistochemical expression and clinicopathological parameters
were investigated. Patients with low CCR5 expression were compared
with those with high CCR5 expression, and the results revealed that
T stage was significantly lower in the low expression group
compared with the high expression group. The Fuhrman grades of the
low expression group were reduced compared with those of the high
expression group. The low CCR5 expression group tended to be of
lower N and M stages compared with the high expression group, and
the low expression group also tended to have lower risks of
post-operative tumor recurrence and mortality from CCRCC, however,
these differences were not statistically significant.
In summary, the present study indicates that the
CCR5 gene is hypomethylated to a greater extent in cancer tissue
compared with in normal tissue. Although the biological function of
CCR5 in CCRCC remains unclear, the CCRCC patients with low CCR5
expression exhibit a low T stage and low Fuhrman grade, however,
this is not statistically significant. Determining the expression
of mRNA in tumor cells is required as this may aid in determining
the diagnosis and prognosis of CCRCC cases. This study reveals CCR5
as a potential new tumor marker for kidney cancer.
Acknowledgements
This study was supported by a grant from the Kyung
Hee University in 2013 (no. KHU-20130367).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim H, Cho NH, Kim DS, et al:
Genitourinary Pathology Study Group of the Korean Society of
Pathologists: Renal cell carcinoma in south Korea: a multicenter
study. Hum Pathol. 35:1556–1563. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Prasad SR, Humphrey PA, Catena JR, et al:
Common and uncommon histologic subtypes of renal cell carcinoma:
imaging spectrum with pathologic correlation. Radiographics.
26:1795–1806. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taby RI and Issa JP: Cancer epigenetics.
CA Cancer J Clin. 60:376–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Balkwill F: Cancer and the chemokine
network. Nat Rev Cancer. 4:540–550. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Koizumi K, Hojo S, Akashi T, Yasumoto K
and Saiki I: Chemokine receptors in cancer metastasis and cancer
cell-derived chemokines in host immune response. Cancer Sci.
98:1652–1658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
O'Hayre M, Salanga CL, Handel TM and Allen
SJ: Chemokines and cancer: migration, intracellular signalling and
intercellular communication in the microenvironment. Biochem J.
409:635–649. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Raport CJ, Gosling J, Schweickart VL, Gray
PW and Charo IF: Molecular cloning and functional characterization
of a novel human CC chemokine receptor (CCR5) for RANTES, MIP-1beta
and MIP-1alpha. J Biol Chem. 271:17161–17166. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Samson M, Labbe O, Mollereau C, Vassart G
and Parmentier M: Molecular cloning and functional expression of a
new human CC-chemokine receptor gene. Biochemistry. 35:3362–3367.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spagnolo P, Renzoni EA, Wells AU, et al:
C-C chemokine receptor 5 gene variants in relation to lung disease
in sarcoidosis. Am J Respir Crit Care Med. 172:721–728. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bibikova M, Lin Z, Zhou L, et al:
High-throughput DNA methylation profiling using universal bead
arrays. Genome Res. 16:383–393. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoo KH, Park YK, Kim HS, Jung WW and Chang
SG: Epigenetic inactivation of HOXA5 and MSH2 gene in clear cell
renal cell carcinoma. Pathol Int. 60:661–666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bibikova M, Lin Z, Zhou L, et al:
High-throughput DNA methylation profiling using universal bead
arrays. Genome Res. 16:383–393. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ehrlich M: DNA hypomethylation in cancer
cells. Epigenomics. 1:239–259. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ehrlich M: DNA methylation in cancer: too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
de Capoa A, Musolino A, Della Rosa S, et
al: DNA demethylation is directly related to tumour progression:
evidence in normal, pre-malignant and malignant cells from uterine
cervix samples. Oncol Rep. 10:545–549. 2003.PubMed/NCBI
|
|
19
|
Brothman AR, Swanson G, Maxwell TM, et al:
Global hypomethylation is common in prostate cancer cells: a
quantitative predictor for clinical outcome? Cancer Genet
Cytogenet. 156:31–36. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Das PM and Singal R: DNA methylation and
cancer. J Clin Oncol. 22:4632–4642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim YI, Giuliano A, Hatch KD, et al:
Global DNA hypomethylation increases progressively in cervical
dysplasia and carcinoma. Cancer. 74:893–899. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin CH, Hsieh SY, Sheen IS, et al:
Genome-wide hypomethylation in hepatocellular carcinogenesis.
Cancer Res. 61:4238–4243. 2001.PubMed/NCBI
|
|
23
|
Bedford MT and van Helden PD:
Hypomethylation of DNA in pathological conditions of the human
prostate. Cancer Res. 47:5274–5276. 1987.PubMed/NCBI
|
|
24
|
Okano M, Bell DW, Haber DA and Li E: DNA
methyltransferases Dnmt3a and Dnmt3b are essential for de novo
methylation and mammalian development. Cell. 99:247–257. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Feinberg AP and Vogelstein B:
Hypomethylation of ras oncogenes in primary human cancers. Biochem
Biophys Res Commun. 111:47–54. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jürgens B, Schmitz-Dräger BJ and Schulz
WA: Hypomethylation of L1 LINE sequences prevailing in human
urothelial carcinoma. Cancer Res. 56:5698–5703. 1996.PubMed/NCBI
|
|
27
|
Zimmermann T, Moehler M, Gockel I, et al:
Low expression of chemokine receptor CCR5 in human colorectal
cancer correlates with lymphatic dissemination and reduced CD8+
T-cell infiltration. Int J Colorectal Dis. 25:417–424. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
van Deventer HW, O'Connor W Jr, Brickey
WJ, Aris RM, Ting JP and Serody JS: C-C chemokine receptor 5 on
stromal cells promotes pulmonary metastasis. Cancer Res.
65:3374–3379. 2005.PubMed/NCBI
|
|
29
|
Lin S, Wan S, Sun L, et al: Chemokine C-C
motif receptor 5 and C-C motif ligand 5 promote cancer cell
migration under hypoxia. Cancer Sci. 103:904–912. 2012. View Article : Google Scholar : PubMed/NCBI
|