Introduction
The balance between cell growth and death is
critical for the maintenance of normal tissue architecture
(1). However, the disorder of these
processes has been implicated in numerous pathological conditions,
including the pathogenesis of cancer (2,3).
Therefore, inhibition of cell proliferation and induction of
apoptosis may result in the effective treatment of various types of
cancer. Survivin is a member of the inhibitor of apoptosis family
of proteins, which inhibits apoptosis and significantly promotes
cell proliferation (4). Caspase-3 is
a key mediator of apoptosis that regulates programmed cell death
via two main pathways: The mitochondrial pathway (intrinsic
pathway) and the death receptor pathway (extrinsic pathway)
(5).
Evodia rutaecarpa has previously been used in
Traditional Chinese medicine and was reported to have various
biological functions, including the inhibition of influenza
virus-induced inflammation (6) as
well as type I and II topoisomerases (7); in addition, Evodia rutaecarpa is
a source of natural larvicides (8).
Evodiamine was identified as one of the major active substances of
Evodia rutaecarpa (5,9). Previous studies have reported the
antitumor activity of evodiamine; one study demonstrated that
evodiamine was able to inhibit proliferation of human thyroid
cancer cells through cell cycle arrest at M phase and the induction
of apoptosis (10). In addition,
evodiamine was found to inhibit the growth of prostate cancer cells
via the induction of apoptosis (11).
However, the antitumor effect of evodiamine in gastric cancer cells
remains to be elucidated.
In the present study, SGC7901 human gastric cancer
cells were treated with various concentrations of evodiamine for 24
h in order to assess the effect of evodiamine on the regulation of
cell proliferation and apoptosis as well as to identify the
molecular mechanism involved in its antitumor effects.
Materials and methods
Cell lines
The SGC7901 human gastric cancer cell line was
purchased from the Cell Bank of the Chinese Academy of Medical
Science (Beijing, China) and cells were cultured in RPMI 1640
medium (Gibco-BRL, Carlsbad, CA, USA) supplemented with 10% fetal
bovine serum (FBS; Gibco-BRL). Cells were maintained at 37°C in a
humidified 5% CO2 atmosphere and passaged every 2–4
days.
MTT assay for cell proliferation
SGC7901 cells were seeded at a density of
5×103 cells per well in a 96-well plate containing 0.2
ml RPMI 1640 medium with 10% FBS and cultured for 24 h at 37°C in a
humidified atmosphere of 5% CO2. Cells were then treated
with various concentration of evodiamine (0, 3, 6, 12, 24 and 48
µmol/l) in 200 µl RPMI 1640 medium and incubated for a further 24
h. Following incubation, 20 µl freshly prepared and filtered MTT
(Sigma-Aldrich, St Louis, MO, USA) was added to each well at a
final concentration of 5 mg/ml and incubated for 3 h. The medium
was then removed and cells in each well were dissolved in 100 µl
dimethyl sulfoxide (Sangon Biological Engineering Technology and
Services Co., Ltd, Shanghai, China). Absorbance values were
measured at 570 nm using a microplate reader (680; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The following formula was
used to calculate the inhibitory rate: Cells inhibition rate = [1 -
average optical density (OD) value of treatment group/average OD
value of control group] × 100%.
Morphological observation of
apoptosis
SGC7901 cells were seeded at a density of
5×104 cells per well on coverslips in six-well plates.
Once cells had reached the logarithmic growth phase, evodiamine was
administered at concentrations of 0, 3, 6, 12, 24 and 48 µmol/l and
cells were cultured for 24 h at 37°C in a humidified atmosphere of
5% CO2. Morphological changes were observed under an
inverted phase contrast microscope (DMI4000; Leica, Wetzlar,
Germany). Supernatant was collected and centrifuged at 200 × g for
5 min. Cell pellets was resuspended in 4% paraformaldehyde
(Sigma-Aldrich) and fixed for 5 min. Following washing three times
in PBS (Gibco-BRL), cells were incubated with 10 µl Hochest 33258
(Sigma-Aldrich) for 30 min. Cells were then added to slides and
observed under a fluorescence microscope (LSM710; Carl Zeiss
Microscopy GmbH, Jena, Germany). For each sample, 500 cells were
counted in order to calculate the percentage of apoptotic cells.
Experiments were performed in triplicate.
Flow cytometric analysis
SGC7901 cells were treated with evodiamine at
concentrations of 0, 3, 6, 12, 24 and 48 µmol/l for 24 h. Cells
were then collected and adjusted to a density of 1×106
cells/ml. Following centrifugation at 200 × g for 5 min, the
supernatant was discarded and cells were resuspended in 100 µl
precooled PBS. Annexin V-fluorescein isothiocyanate (FITC; 5 µl;
Nanjing Keygen Biotech., Co., Ltd., Nanjing, China) and propidium
iodide (PI; 10 µl; Nanjing Keygen Biotech., Co., Ltd.) were then
added to the cells and incubated in the dark for 15 min at room
temperature. Following which, 400 µl PBS was added and the
apoptotic cells were analyzed by flow cytometry (BD FACSCanto II;
BD Biosciences, Franklin Lakes, NJ, USA).
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was extracted from the cells treated with
various concentration of evodiamine using TRIzol® (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer's
instructions and stored at −80°C until further use. Following
denaturation in diethylpyrocarbonate-treated water (Sangon
Biological Engineering Technology and Services Co., Ltd.) for 10
min at 70°C, 2 µg total RNA was reverse-transcribed into
complementary (c)DNA in a reaction volume of 20 µl, which contained
1X RT buffer, 20 Units RNase inhibitor, 50 mM deoxynucleotides
(dNTPs), 200 Units Moloney murine leukemia virus reverse
transcriptase and 0.5 µg oligo (deoxythymine)18 primer
(all obtained from Promega Corporation, Madison, WI, USA). The
reactions was incubated at 42°C for 60 min and then inactivated at
95°C for 10 min.
Polymerase chain reaction
amplification
Each experiment included samples containing no
reverse transcriptase, as negative controls, to exclude
amplification from contaminated genomic DNA. Primers (Sangon
Biological Engineering Technology and Services Co., Ltd) used for
PCR amplification were as follows: Survivin forward,
5′-TTTCTCAAGGACCACCGCA-3′ and reverse, 5′-CAACCGGACGAATGCTTTTT-3′;
Caspase-3 forward, 5′-TGCTTCTGAGCCATGGTGAA-3′ and reverse,
5′-TGGCACAAAGCGACTGGAT-3′; and GADPH forward,
5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′.
PCR amplification was performed using a PTC 225 thermal cycler (MJ
Research, St. Bruno, QC, Canada) in a volume of 50 µl, which
consisted of 1 µl reaction cDNA mixture, 10 pmol each primer, 200
mM each dNTP, 2 mM MgCl2, 10 mM Tris-HCl (pH 8.3), 50 mM
KCl and 2 Units Taq DNA polymerase (Sangon Biological Engineering
Technology and Services Co., Ltd.). The amplification was composed
of 35 cycles of 94°C for 40 sec, 57°C for 40 sec and 72°C for 1
min, followed by a full extension cycle of 72°C for 5 min. In each
experiment, a negative control was included (0 µmol/l evodiamine).
PCR products were then electrophoresed on 1.2% agarose gels (Sangon
Biological Engineering Technology and Services Co., Ltd.), stained
with ethidium bromide (Sangon Biological Engineering Technology and
Services Co., Ltd.) and images were captured using an ultraviolet
transilluminator (CUV20; Cell Biosciences, Inc., Santa Clara, CA,
USA). Results were expressed for each sample as band intensity
relative to that of GAPDH.
Statistical analysis
Data are presented as the mean ± standard deviation.
All data were analyzed using a two-tailed unpaired Student's t-test
and all statistical analyses were performed using SPSS 16.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
Evodiamine inhibits gastric cancer
cell proliferation
The growth of SGC7901 cells was demonstrated to be
significantly inhibited following evodiamine treatment compared
with that of the control group (P<0.05), with the inhibition
rate gradually increasing in a dose-dependent manner (Table I).
 | Table I.Evodiamine inhibits SGC7901 cell
growth. |
Table I.
Evodiamine inhibits SGC7901 cell
growth.
| Evodiamine
(µmol/l) | Inhibition (%) |
|---|
| 0 |
0.00±4.57 |
| 3 |
17.12±4.13a |
| 6 |
39.37±5.61a |
| 12 |
61.21±5.66a |
| 24 |
77.40±3.69a |
| 48 |
80.01±5.39a |
Evodiamine induces gastric cancer cell
apoptosis
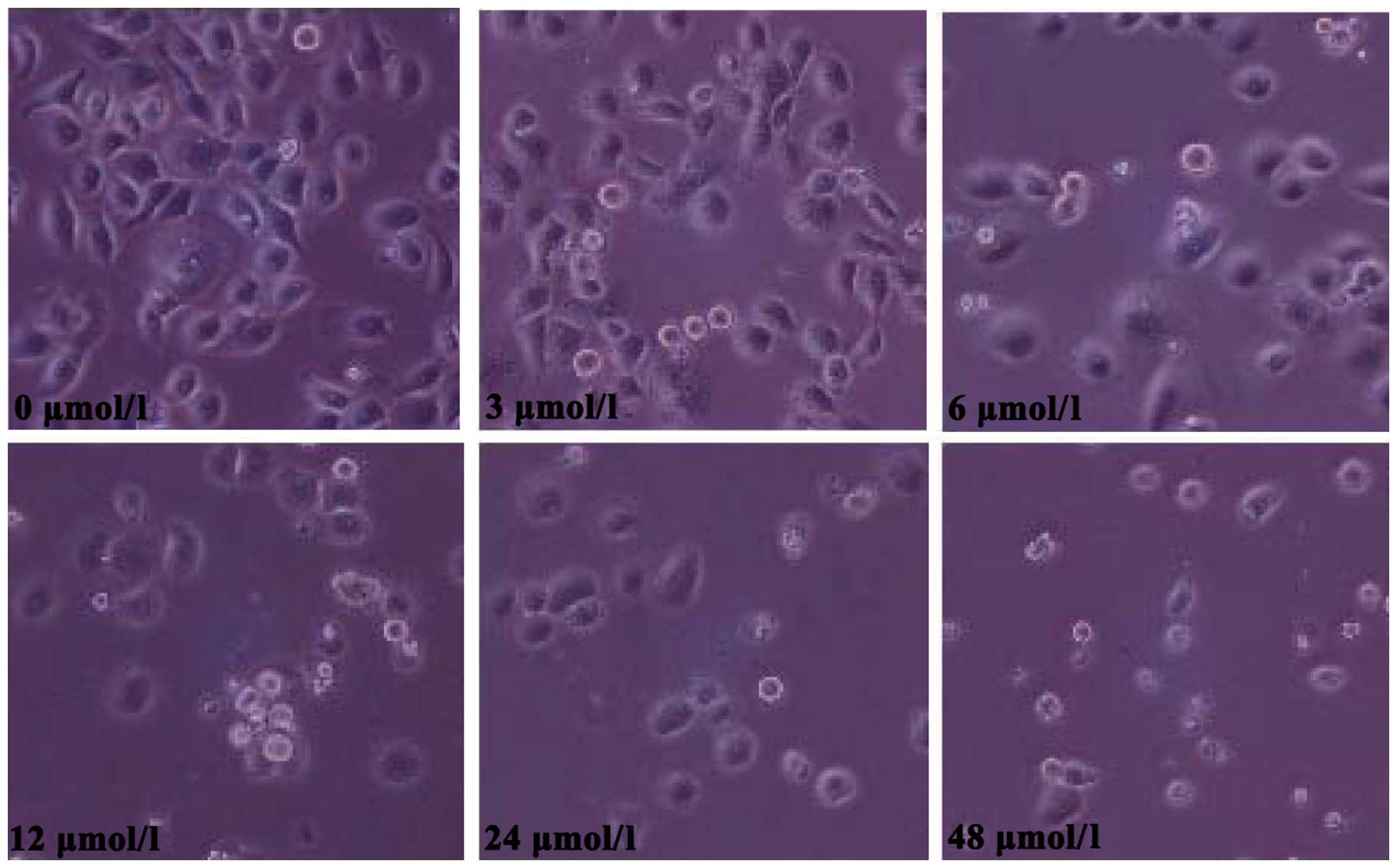
Morphological changes observed in SGC7901 cells
following evodiamine treatment are shown in Fig. 1. Treated cells exhibited a rounded
shape; in addition, shrinkage, nuclear condensation and
fragmentation were observed in evodiamine-treated cells, while the
negative control cells had an irregular shape (fusiform or
polygonal) and certain cells were integrated to form colonies. The
apoptosis of SGC7901 cells was also observed under a fluorescent
microscope. Following 24 h of evodiamine incubation, SGC7901 cells
demonstrated signs of shrinkage, cytoplasmic concentration, nuclei
pyknosis, chromatin margination and the formation of apoptotic
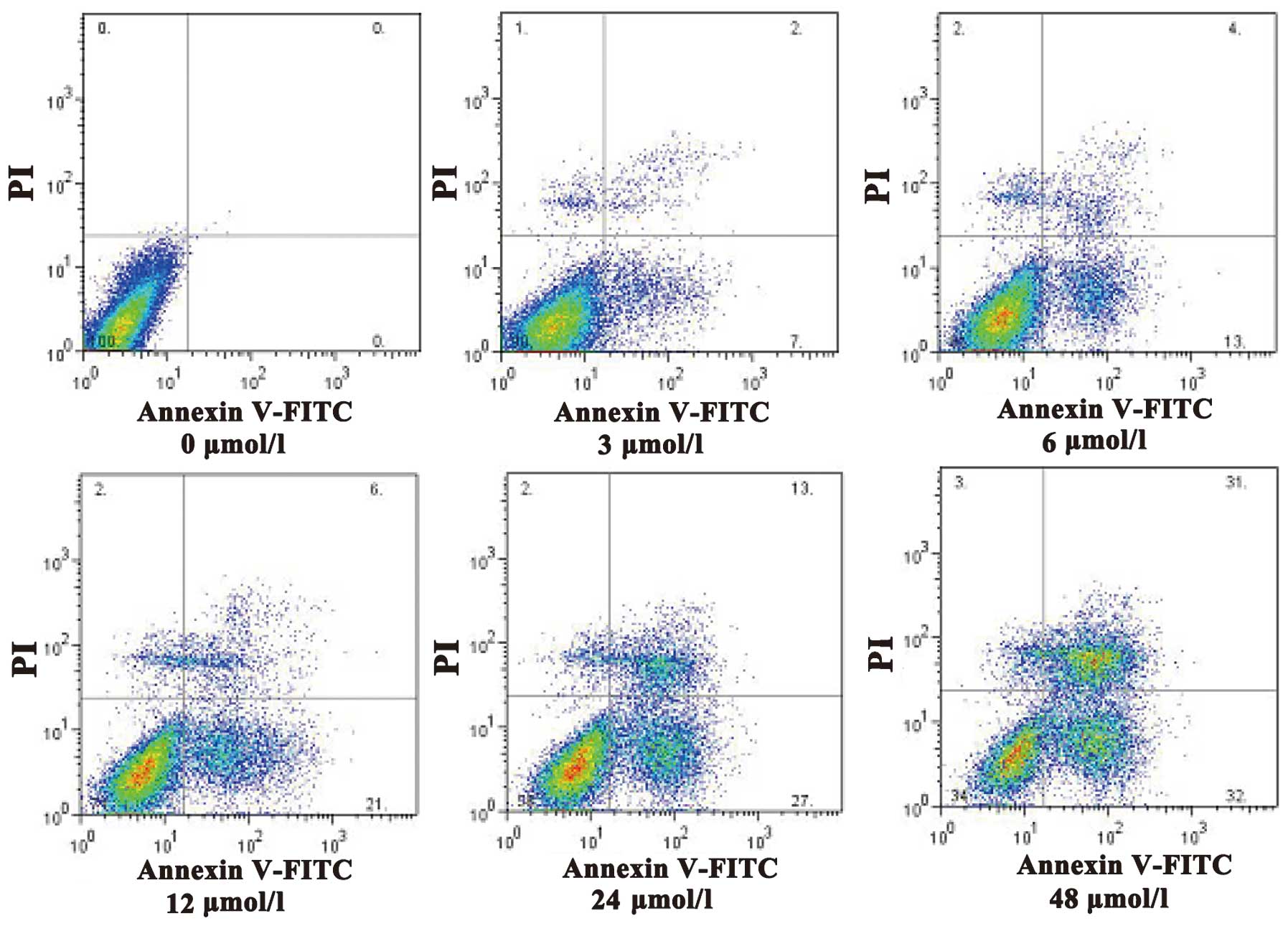
bodies (Fig. 2). Apoptotic cells were
quantified and the results demonstrated that evodiamine
significantly increased the rate of early and late apoptosis or
necrosis in SGC7901 cells in a dose-dependent manner (P<0.05)
(Table II). In addition, as shown in
Fig. 3, increasing concentrations of
evodiamine resulted in an increase in the early apoptotic rate of
SGC7901 cells, as determined by Annexin V-FITC/PI double staining
(Fig. 3).
 | Table II.Evodiamine induces SGC7901 cell
apoptosis. |
Table II.
Evodiamine induces SGC7901 cell
apoptosis.
| Evodiamine
(µmol/l) | Early apoptosis
(%) | Late apoptosis and
necrosis (%) |
|---|
| 0 |
0.67±0.16 |
1.49±0.23 |
| 3 |
8.54±1.28a |
4.36±0.64a |
| 6 |
13.69±2.71a |
10.43±2.01a |
| 12 |
20.42±3.45a |
16.79±3.06a |
| 24 |
31.66±5.13a |
23.83±4.11a |
| 48 |
44.35±7.34a |
32.54±5.48a |
Evodiamine attenuates gastric cancer
cell growth through deregulation of survivin and caspase-3
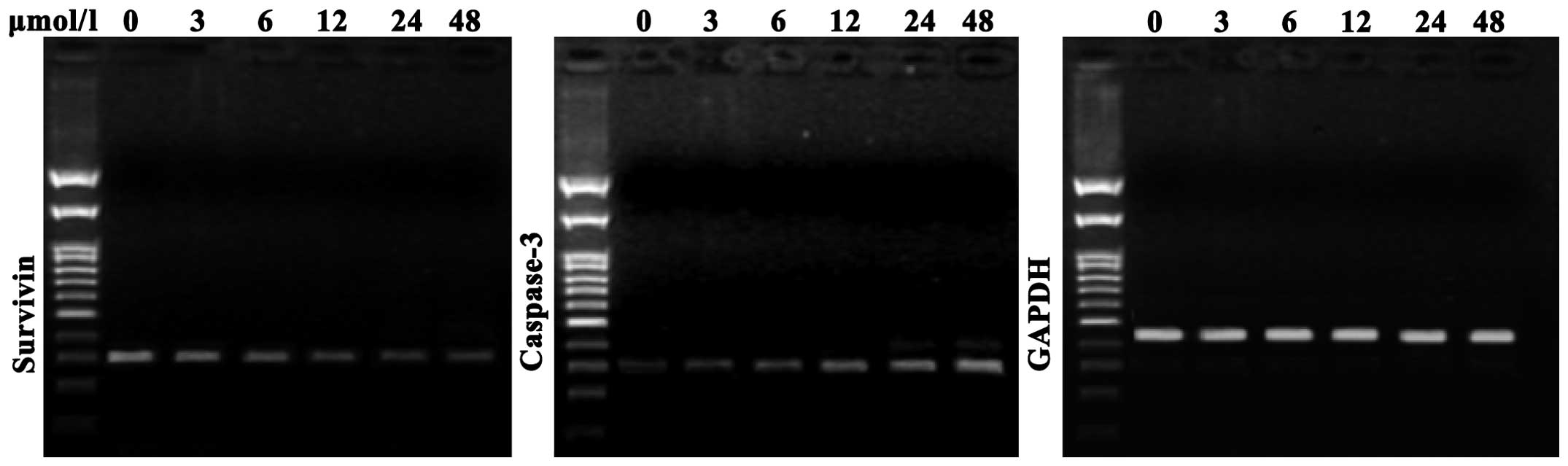
As shown in Fig. 4,
compared with the negative control cells, SGC7901 cells treated
with evodiamine exhibited a decreased expression of survivin
messenger (m)RNA; in addition, survivin expression was further
decreased with increasing concentrations of evodiamine.
Furthermore, compared with the negative control group, evodiamine
gradually upregulated the expression of caspase-3 mRNA in SGC7901
cells in a dose-dependent manner following 24 h of incubation
(Fig. 4).
Discussion
The present study aimed to investigate the role of
evodiamine, a natural active ingredient of the Traditional Chinese
medicine Evodia rutaecarpa, in gastric cancer cells. The
results revealed that evodiamine inhibited the proliferation of
SGC7901 cells. In addition, using inverted phase contrast
microscopy and fluorescence microscopy, cell shrinkage,
concentrated cytoplasm, pyknotic nuclei, chromatin margination,
nuclear fragmentation and the formation of apoptotic bodies were
observed in evodiamine-treated SGC7901 cells.
Previous studies have reported a close association
between apoptosis and tumor development (12,13).
Therefore, the exploration of the mechanisms underlying
drug-induced tumor cell apoptosis is essential for improving drug
efficacy and the development of novel anticancer drugs. In the
present study, it was demonstrated that gastric cancer cells
treated with evodiamine exhibited a concentration-dependent
increase in the number of apoptotic and necrotic cells, as
determined using Annexin V-FITC/PI double staining.
Survivin is a member of the inhibitors of apoptosis
proteins gene family, which has been reported to have important
roles in the inhibition of apoptosis and is predominantly expressed
in the mitotic cycle of cells in S and G2/M phase
(4). Survivin was demonstrated to be
widely expressed in a variety of tumor tissues; in addition, the
expression of survivin was found to be deregulated in cancer cells
(14,15). Survivin expression in gastric cancer
was reported to be negatively correlated with apoptosis index
(16), which is concurrent with the
results of the present study that showed a gradual decrease in
survivin mRNA expression with increased concentrations of
evodiamine. This therefore suggested that evodiamine may decrease
survivin mRNA expression. Caspases are cysteine proteases, which
were named due to their strict specificity for cleaving peptide
sequences at the C-terminal of aspartic acid residues (17). Once activated, effector caspases
induce a series of hydrolysis reactions, which ultimately lead to
the initiation of cell death (18).
It is widely recognized that caspase-3 is a key protease and an
essential effector agent involved in hydrolysis, which acts alone
or in combination with the apoptosis-associated proteins (19,20).
Activation of caspase-3 inevitably leads to the initiation of
cascades, which result in the induction of apoptosis (21). The results of the present study
demonstrated that mRNA levels of caspase-3 in SGC7901 cells were
gradually increased with increasing concentrations of
evodiamine.
The results of the present study indicated that
evodiamine-induced SGC7901 cell apoptosis may be associated with
decreased expression levels of survivin and increased expression
levels of caspase-3. Thus, evodiamine may present a potential
treatment for gastric cancer. In addition, further studies are
required to investigate the combined therapeutic effect of
chemotherapy and evodiamine for the treatment of gastric
cancer.
Acknowledgements
The present study was supported by grants from
Zhejiang Provincial Natural Science Foundation of China (nos.
LY14H160027 and LQ12H16009) and the Science and Technology Bureau
of Zhejiang Province (no. 2013C33137).
References
|
1
|
O'Brien LE and Bilder D: Beyond the niche:
tissue-level coordination of stem cell dynamics. Annu Rev Cell Dev
Biol. 29:107–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bissell MJ and Radisky D: Putting tumours
in context. Nat Rev Cancer. 1:46–54. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Potter JD: Morphogens, morphostats,
microarchitecture and malignancy. Nat Rev Cancer. 7:464–474. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Altieri DC: Survivin, cancer networks and
pathway-directed drug discovery. Nat Rev Cancer. 8:61–70. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiao BY, Mao SJ and Li XD: Variations in
the composition of Fructus Evodiae after processing with Radix
Glycyrrhizae extract. Chin J Integr Med. 18:782–787. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chiou WF, Ko HC and Wei BL: Evodia
rutaecarpa and three major alkaloids abrogate influenza a virus
(H1N1)-induced chemokines production and cell migration. Evid Based
Complement Alternat Med. 2011:7505132011.PubMed/NCBI
|
|
7
|
Wiart C: A note on Evodia rutaecarpa.
Phytomedicine. 19:12442012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu ZL, Liu QZ, Du SS and Deng ZW:
Mosquito larvicidal activity of alkaloids and limonoids derived
from Evodia rutaecarpa unripe fruits against Aedes albopictus
(Diptera: Culicidae). Parasitol Res. 111:991–996. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cai GX, Huang D, Li SX, Xu F, Wang L, et
al: Comparative analysis of essential oil components of Evodia
rutaecarpa (Juss.) Benth. var. officinalis (Dode) Huang and Evodia
rutaecarpa (Juss.) Benth. Nat Prod Res. 26:1796–1798. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen MC, Yu CH, Wang SW, Pu HF, Kan SF, et
al: Anti-proliferative effects of evodiamine on human thyroid
cancer cell line ARO. J Cell Biochem. 110:1495–1503. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Wu LJ, Tashiro S, Onodera S and
Ikejima T: Evodiamine induces tumor cell death through different
pathways: Apoptosis and necrosis. Acta Pharmacol Sin. 25:83–89.
2004.PubMed/NCBI
|
|
12
|
Cotter TG: Apoptosis and cancer: the
genesis of a research field. Nat Rev Cancer. 9:501–507. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ashkenazi A: Targeting the extrinsic
apoptotic pathway in cancer: lessons learned and future directions.
J Clin Invest. 125:487–489. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carter BZ, Milella M, Altieri DC and
Andreeff M: Cytokine-regulated expression of survivin in myeloid
leukemia. Blood. 97:2784–2790. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Adida C, Recher C, Raffoux E, Daniel MT,
Taksin AL, et al: Expression and prognostic significance of
survivin in de novo acute myeloid leukaemia. Br J Haematol.
111:196–203. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu CD, Altieri DC and Tanigawa N:
Expression of a novel antiapoptosis gene, survivin, correlated with
tumor cell apoptosis and p53 accumulation in gastric carcinomas.
Cancer Res. 58:1808–1812. 1998.PubMed/NCBI
|
|
17
|
Shalini S, Dorstyn L, Dawar S and Kumar S:
Old, new and emerging functions of caspases. Cell Death Differ.
22:526–539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vandenabeele P, Galluzzi L, Vanden Berghe
T and Kroemer G: Molecular mechanisms of necroptosis: an ordered
cellular explosion. Nat Rev Mol Cell Biol. 11:700–714. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
D'Amelio M, Cavallucci V and Cecconi F:
Neuronal caspase-3 signaling: not only cell death. Cell Death
Differ. 17:1104–1114. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wakeyama H, Akiyama T, Kadono Y, Nakamura
M, Oshima Y, Nakamura K and Tanaka S: Posttranslational regulation
of Bim by caspase-3. Ann NY Acad Sci. 1116:271–280. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lakhani SA, Masud A, Kuida K, et al:
Caspases 3 and 7: key mediators of mitochondrial events of
apoptosis. Science. 311:847–851. 2006. View Article : Google Scholar : PubMed/NCBI
|