Introduction
Gastric cancer is the fifth most common malignancy
worldwide. It is estimated that 952,000 new gastric cancer cases
and 723,000 gastric cancer-associated mortalities occurred in 2012
(1). Over 70% of gastric cancer cases
occur in developing countries, with half of these occurring in
China (1,2). Surgery is the primary method for
treating early-stage disease; however, a high proportion of
patients develop local or distant recurrence even after receiving
surgical treatment. At present, neoadjuvant chemotherapy followed
by surgery is widely used for gastric cancer patients, and this has
been demonstrated to improve survival times (3).
Although the outcomes of patients with gastric
cancer have been improved significantly by platinum-based
chemotherapy, its efficacy and toxicity are highly variable in
different patients (4,5). Recent increasing evidence has indicated
that hereditary factors are crucial in such individual differences
in the response to platinum-based chemotherapy (6–10). Single
nucleotide polymorphisms (SNPs) may lead to changes in the activity
of enzymes and transporters that are involved in platinum-based
drug elimination, and may affect survival and treatment-associated
toxicity (11).
Platinum analogs can bind to DNA, forming adducts
(intrastrand and interstrand crosslinks) and inhibiting DNA
replication (12). DNA repair
mechanisms are, therefore, important factors determining the
response to chemotherapy. Intrastrand crosslinks represent the
majority of chemotherapy-induced DNA damage, and nucleotide
excision repair (NER) is the predominant mechanism involved in
their repair (13). However, as
interstrand crosslinks affect both DNA strands, they are more
cytotoxic compared with intrastrand crosslinks, and their repair
through homologous recombination repair (HRR) and translesion
polymerases is crucial for genomic stability (14–16).
All DNA repair pathways are complex and involve
numerous different enzymes. The NER pathway is primarily important
in helix-distorting DNA lesions and cytotoxic DNA interstrand
crosslinks (17). Two genes encoding
enzymes in the NER pathway are frequently associated with
resistance to platinum-based chemotherapy: Excision repair
cross-complementation group 2 (ERCC2) and ERCC1.
Other complex DNA damage requires alternative
mechanisms, such as HRR, for successful repair. In the HRR pathway,
nibrin (NBN) is one of the complexes involved in the recognition of
DNA damage, while RAD51 recombinase (RAD51) and X-ray complementing
defective repair in Chinese hamster cells 3 (XRCC3) catalyze
homologous search and strand invasion.
The aim of the current study was to investigate the
role of polymorphisms in DNA repair pathways in the clinical
outcome of gastric cancer patients treated with platinum-based
chemotherapy.
Patients and methods
Patients
A total of 380 gastric cancer patients treated with
platinum-based chemotherapy were enrolled at the First Affiliated
Hospital of Zhengzhou University (Zhengzhou, China) between January
2008 and January 2011. All the patients were Han Chinese with newly
diagnosed and histopathologically confirmed gastric cancer. None of
the patients had previously received systemic anticancer
chemotherapy. Bone marrow reserve and normal renal function, liver
function and cardiac function prior to chemotherapy were necessary
for inclusion in the study. Patients who had severe concomitant
systemic disorders and were unable to receive chemotherapy,
presented metastasis with symptoms, lacked comprehensive data or
developed other diseases (such as neural system diseases) were
excluded from the study, as these symptoms or diseases may affect
the safety of patients or the evaluation of the results. All
patients provided written informed consent for blood sample
collection to establish the clinical significance of genetic
polymorphisms in the response to platinum-based chemotherapy. The
study was approved by the review board of the First Affiliated
Hospital of Zhengzhou University.
Assessment of treatment outcome
Demographic, clinical and treatment parameters were
obtained from the medical records. Tumor response to chemotherapy
was evaluated based on the World Health Organization criteria
(18). All the patients were
followed-up until 30th December 2013, with a median follow-up time
of 29.6 months (range, 2–60 months). Patients were followed-up by
telephone every four weeks until mortality or the end of the
study.
The overall survival (OS) was defined as the period
from the beginning of treatment until mortality or the end of the
follow-up period. Complete remission and partial remission were
considered to be responsive, while stable disease and progressive
disease were considered to be non-responsive. Patients without an
event or mortality at the time of the analysis were censored at the
date of the last follow-up.
Individuals who had smoked ≥1 cigarette per week for
more than half a year previously were defined as smokers, whilst
individuals who had consumed alcoholic beverages at least once per
week for more than half a year previously were defined as
drinkers.
DNA extraction and genotyping
Genomic DNA was isolated from peripheral blood
lymphocytes using a QIAamp DNA Blood Mini kit (Qiagen, Hilden,
Germany) according to the manufacturer's instructions. The
genotypes of ERCC1 rs11615 (Asn118Asn) and rs3212986
(*197G>T), ERCC2 rs1799793 (Asn312Asp) and rs13181
(Lys751Gln), NBN rs1805794 (Gln185Gln) and rs1063054
(*1209A>C), RAD51 rs1801321 (−61G>T) and rs12593359
(*502T>G), and XRCC3 rs861539 (Thr241Met) were determined
by polymerase chain reaction (PCR)-restriction fragment length
polymorphism analysis, according to manufacturer's instructions.
The PCR reaction was conducted in a 25 µl reaction solution with 25
mM MgCl2, 1 ng/µl of each primer and 2 mM
deoxynucleotide triphosphates, and 1.25 units of Taq polymerase
(Takara Biotechnology Co., Ltd., Dalian, China) as well as 0.5 µl
5X PCR buffer (Takara Biotechnology Co., Ltd.). The cycling program
of the PCR reaction conditions commenced with a 95°C denaturation
step lasting for 10 min, followed by 40 cycles of 95°C for 30 sec,
62°C for 30 sec and 72°C for 30 sec, and an extension at 72°C for
10 min. A total of ~5% of the samples were repeatedly genotyped,
and the results were 100% concordant.
Statistical analysis
All statistical analyses were conducted using STATA
statistical software version 9.0 (StataCorp LP, College Station,
TX, USA). Frequencies were used to describe the distribution of
categorical variables, while the median and interquartile range
were used for continuous variables. A standard χ2 test
was used to assess deviation from the Hardy-Weinberg equilibrium.
The Cox proportional hazards model was used in the survival
analysis to calculate the hazard ratio (HR) and 95% confidence
interval (CI). Survival distributions were estimated by using the
Kaplan-Meier method. Logistic regression was used to assess the
effect of genetic polymorphisms or clinical variables on binary
treatment outcomes, and odds ratios (ORs) and their 95% CIs were
determined. A dominant genetic model was used in all statistical
analyses. All P-values were two-sided, and a P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient demographic and clinical
characteristics
The demographic and clinical characteristics of the
patients are listed in Table I. The
ages of the patients were between 23 and 82 years at diagnosis
(mean ± standard deviation, 58.7±16.3 years). Among the included
380 gastric cancer patients, 244 (64.21%) patients were men, 136
(35.79%) were women, 166 (43.68%) were smokers, 208 (54.74%)
consumed alcohol, 172 (45.26%) had intestinal type gastric cancer,
164 (43.16%) had signet ring type gastric cancer, 248 (65.26%) had
a poor histological grade, and 234 (61.58%) were of TNM stage
III–IV (19). Using the Cox
proportional hazards regression analysis, patients with stage
III–IV tumors were found to have a significantly higher risk of
mortality associated with gastric cancer compared with those with
stage I–II disease, with a HR of 2.54 (95% CI, 1.46–3.28;
P=0.007).
 | Table I.Association between demographic and
clinical characteristics and overall survival of gastric
cancer. |
Table I.
Association between demographic and
clinical characteristics and overall survival of gastric
cancer.
| Variable | Patients, n (%) | Median survival time,
months | HR (95% CI) | P-value |
|---|
| Age, years |
|
|
|
|
|
<60 | 161 (42.37) | 18.5 | 1.00 (ref.) | - |
| ≥60 | 219 (57.63) | 21.2 | 0.87 (0.62–1.15) | 0.18 |
| Gender |
|
|
|
|
| Male | 244 (64.21) | 19.8 | 1.00 (ref.) | - |
|
Female | 136 (35.79) | 20.6 | 0.95 (0.76–1.22) | 0.16 |
| Smoking status |
|
|
|
|
|
Never | 214 (56.32) | 23.2 | 1.00 (ref.) | - |
| Yes | 166 (43.68) | 15.6 | 1.42 (0.95–1.73) | 0.11 |
| Drinking status |
|
|
|
|
|
Never | 172 (45.26) | 22.1 | 1.00 (ref.) | - |
| Yes | 208 (54.74) | 16.8 | 1.36 (0.86–1.52) | 0.08 |
| Histological
type |
|
|
|
|
|
Intestinal | 172 (45.26) | 22.5 | 1.00 (ref.) | - |
| Signet
ring | 164 (43.16) | 20.2 | 1.15 (0.83–1.25) | 0.25 |
|
Other | 44
(11.58) | 17.9 | 1.27 (0.72–1.39) | 0.12 |
| Histological
grade |
|
|
|
|
|
Moderate-poor | 132 (34.74) | 23.4 | 1.00 (ref.) | - |
| Poor | 248 (65.26) | 18.6 | 1.22 (0.81–1.32) | 0.17 |
| TNM stage |
|
|
|
|
|
I–II | 146 (38.42) | 24.3 | 1.00 (ref.) | - |
|
III–IV | 234 (61.58) | 15.6 | 2.54
(1.46–3.28) | 0.007 |
Association between SNPs and response
to chemotherapy
The association between the investigated SNPs and
response to chemotherapy in gastric cancer patients are listed in
Table II. Among the 380 gastric
cancer patients, 223 (58.68%) exhibited a good response to
chemotherapy. Following adjustment for clinical variables, the
TC+CC genotypes of ERCC1 rs11615 were correlated with a
significantly improved response to chemotherapy compared with that
of the reference genotype (TT), with an adjusted OR of 1.66 (95%
CI, 1.07–2.56; P=0.02). Furthermore, the GA+AA genotypes of
ERCC2 rs1799793 were associated with a significantly better
response to chemotherapy (OR, 1.61; 95% CI, 1.05–2.49) compared
with that of the GG genotype (P=0.02).
 | Table II.Association between the included nine
SNPs and response to chemotherapy. |
Table II.
Association between the included nine
SNPs and response to chemotherapy.
|
| All patients | Good response
(n=223) | Poor response
(n=157) |
|
|
|---|
|
|
|
|
|
|
|
|---|
| SNP | n | % | n | % | n | % | Adjusted OR (95%
CI) | P-value |
|---|
|
ERCC1rs11615 |
|
|
|
|
|
|
| 0.02 |
| TT | 173 | 45.53 | 90 | 40.4 | 83 | 52.87 | 1.00 (Ref.) |
|
|
TC+CC | 207 | 54.47 | 133 | 59.64 | 74 | 47.13 | 1.66
(1.07–2.56) |
|
|
ERCC1rs3212986 |
|
|
|
|
|
|
| 0.05 |
| GG | 202 | 53.16 | 105 | 47.09 | 90 | 57.32 | 1.00 (Ref.) |
|
|
GT+TT | 178 | 46.84 | 118 | 52.91 | 67 | 42.68 | 1.48
(0.96–2.29) |
|
|
ERCC2rs1799793 |
|
|
|
|
|
|
| 0.02 |
| GG | 165 | 43.42 | 86 | 38.57 | 79 | 50.32 | 1.00 (Ref.) |
|
|
GA+AA | 215 | 56.58 | 137 | 61.43 | 78 | 49.68 | 1.61
(1.05–2.49) |
|
|
ERCC2rs13181 |
|
|
|
|
|
|
| 0.82 |
| AA | 160 | 42.11 | 92 | 41.26 | 67 | 42.68 | 1.00 (Ref.) |
|
|
AC+CC | 220 | 57.89 | 131 | 58.74 | 90 | 57.32 | 1.05
(0.68–1.62) |
|
|
NBNrs1805794 |
|
|
|
|
|
|
| 0.39 |
| GG | 119 | 31.32 | 75 | 33.63 | 47 | 29.94 | 1.00 (Ref.) |
|
|
GC+CC | 261 | 68.68 | 148 | 66.37 | 110 | 70.06 | 0.82
(0.52–1.31) |
|
|
NBNrs1063054 |
|
|
|
|
|
|
| 0.48 |
| AA | 171 | 45.00 | 105 | 47.09 | 68 | 43.31 | 1.00 (Ref.) |
|
|
AC+CC | 209 | 55.00 | 118 | 52.91 | 89 | 56.69 | 0.86
(0.56–1.33) |
|
|
RAD51rs1801321 |
|
|
|
|
|
|
| 0.79 |
| GG | 135 | 35.53 | 81 | 36.32 | 55 | 35.03 | 1.00 (Ref.) |
|
|
GT+TT | 245 | 64.47 | 142 | 63.68 | 102 | 64.97 | 0.94
(0.60–1.48) |
|
|
RAD51rs12593359 |
|
|
|
|
|
|
| 0.63 |
| TT | 147 | 38.68 | 90 | 40.36 | 59 | 37.58 | 1.00 (Ref.) |
|
|
TG+GG | 233 | 61.32 | 133 | 59.64 | 98 | 62.42 | 0.90
(0.58–1.40) |
|
|
XRCC3rs861539 |
|
|
|
|
|
|
| 0.51 |
| CC | 242 | 63.68 | 147 | 65.92 | 98 | 62.42 | 1.00 (Ref.) |
|
|
CT+TT | 138 | 36.32 | 76 | 34.08 | 59 | 37.58 | 0.87
(0.55–1.36) |
|
Association between SNPs and
survival
The Kaplan-Meier method and Cox regression analysis
were conducted to assess the role of the 9 investigated SNPs in the
OS of gastric cancer patients (Table
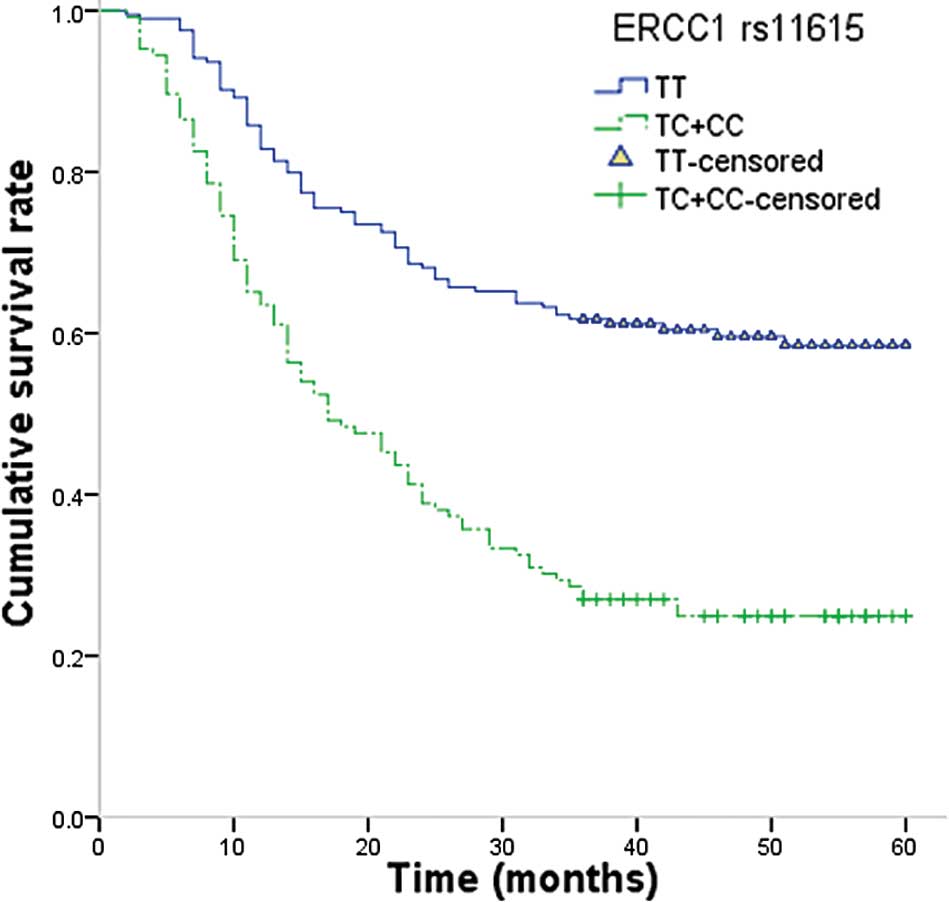
III). This analysis revealed that ERCC1 rs11615 was
associated with OS of gastric cancer: Patients with the TC+CC
genotypes had a longer survival time compared with those with the
common TT genotype (33.20 vs. 23.30 months; log-rank P=0.017).
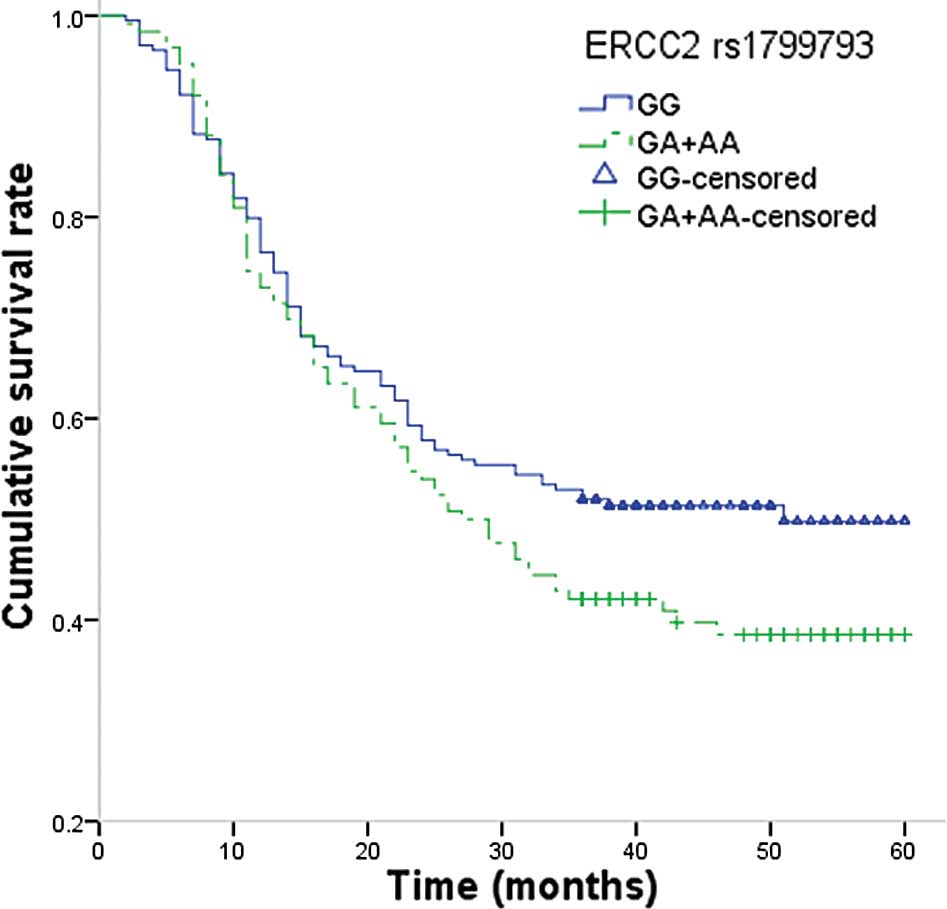
Furthermore, the GA+AA genotypes were associated with a longer
survival time compared with that of the GG genotype (34.80 vs.
23.10 months; log-rank P=0.005).
 | Table III.Association between included nine
SNPs and overall survival in gastric cancer patients. |
Table III.
Association between included nine
SNPs and overall survival in gastric cancer patients.
|
|
|
| Patients, n
(%) |
|
|
|---|
|
|
|
|
|
|
|
|---|
| SNP | Median survival
time, months | P-value
(log-rank) | Survived
(n=204) | Succumbed
(n=176) | HR (95% CI) | Adjusted
P-value |
|---|
|
ERCC1rs11615 |
|
|
|
|
| 0.02 |
| TT | 23.30 |
| 82
(40.20) | 91
(51.70) | 1.00 (Ref.) |
|
|
TC+CC | 33.20 | 0.017 | 122 (59.80) | 85
(48.30) | 1.71
(1.06–2.72) |
|
|
ERCC1rs3212986 |
|
|
|
|
| 0.04 |
| GG | 26.40 |
| 100 (49.02) | 102 (50.00) | 1.00 (Ref.) |
|
|
GT+TT | 31.70 | 0.07 | 104 (50.98) | 74
(36.27) | 1.48
(0.96–2.29) |
|
|
ERCC2rs1799793 |
|
|
|
|
| 0.001 |
| GG | 23.10 |
| 73
(35.78) | 92
(52.27) | 1.00 (Ref.) |
|
|
GA+AA | 34.80 | 0.005 | 131 (64.22) | 84
(47.73) | 1.97
(1.28–3.03) |
|
|
ERCC2rs13181 |
|
|
|
|
| 0.82 |
| AA | 28.90 |
| 89
(43.63) | 71
(34.80) | 1.00 (Ref.) |
|
|
AC+CC | 29.50 | 0.52 | 115 (56.37) | 105 (51.47) | 1.05
(0.68–1.62) |
|
|
NBNrs1805794 |
|
|
|
|
| 0.39 |
| GG | 30.40 |
| 76
(37.25) | 43
(21.08) | 1.00 (Ref.) |
|
|
GC+CC | 28.80 | 0.19 | 128 (62.75) | 133 (65.20) | 0.82
(0.52–1.31) |
|
|
NBNrs1063054 |
|
|
|
|
| 0.48 |
| AA | 31.10 |
| 104 (50.98) | 67
(32.84) | 1.00 (Ref.) |
|
|
AC+CC | 28.90 | 0.13 | 100 (49.02) | 109 (53.43) | 0.86
(0.56–1.33) |
|
|
RAD51rs1801321 |
|
|
|
|
| 0.79 |
| GG | 32.30 |
| 85
(41.67) | 50
(24.51) | 1.00 (Ref.) |
|
|
GT+TT | 27.60 | 0.21 | 119 (58.33) | 126 (61.76) | 0.94
(0.60–1.48) |
|
|
RAD51rs12593359 |
|
|
|
|
| 0.63 |
| TT | 30.90 |
| 91
(44.61) | 56
(27.45) | 1.00 (Ref.) |
|
|
TG+GG | 27.10 | 0.25 | 113 (55.39) | 120 (58.82) | 0.90
(0.58–1.40) |
|
|
XRCC3rs861539 |
|
|
|
|
| 0.51 |
| CC | 30.60 |
| 138 (67.65) | 104 (50.98) | 1.00 (Ref.) |
|
|
CT+TT | 28.30 | 0.43 | 66
(32.35) | 72
(35.29) | 0.87
(0.55–1.36) |
|
The Cox regression analysis revealed that patients
with TC+CC genotypes of ERCC1 rs11615 exhibited a
significantly reduced risk of mortality compared with patients with
the TT genotype (HR, 1.71; 95% CI, 1.06–2.7; P=0.02; Fig. 1). For ERCC2 rs1799793, the
GA+AA genotypes were found to be significantly associated with a
lower risk of mortality from gastric cancer compared with that of
the GG genotype (HR, 1.97; 95% CI, 1.28–3.03; P=0.001; Fig. 2).
Discussion
In the present study, the effect of SNPs in genes
involved in DNA repair mechanisms on the response to treatment and
survival in gastric cancer patients treated with platinum-based
chemotherapy was investigated. The genotypes of ERCC1
rs11615 and ERCC2 rs1799793 were revealed to affect the
response to chemotherapy, and were associated with the OS in
gastric cancer patients.
Platinum-based chemotherapy is a commonly used
chemotherapeutic strategy that exerts a cytotoxic effect primarily
through the formation of various types of DNA lesions. DNA repair
mechanisms may, therefore, be important in determining the response
of tumors to platinum-based chemotherapy (20). NER enzymes are among the most
important factors that are able to modify susceptibility to
platinum-based chemotherapy; however, other mechanisms involved in
the repair of complex forms of DNA damage, including HRR, may also
contribute to differences in response between individual patients
(21). The current study revealed
that ERCC1 rs11615 is associated with the outcome in cases
of gastric cancer treated with chemotherapy. A previous study has
also reported that ERCC1 rs11615 was associated with the
response to chemotherapy and clinical outcome in cases of gastric
cancer (22). Li et al
(23) reported that individuals
carrying ERCC1 rs11615 polymorphisms had an increased risk
of mortality from gastric cancer compared with the risk of
individuals carrying the common genotype, and this genetic
polymorphism may contribute substantially to the future design of
individualized treatments for gastric cancer patients. Another
study has previously investigated the association between three
functional SNPs in DNA repair pathways and the clinical outcome of
940 cases of gastric cancer in a Chinese population, and reported
that ERCC1 rs11615 did not affect the OS of gastric cancer
patients (23). However, a study by
Lu et al (24) reported the
opposite conclusions. The authors conducted a study including 447
patients in China, and reported that the T allele of ERCC1
rs11615 decreased the risk of mortality from gastric cancer
(19). Such discrepancies among the
reports may be explained by differences in the ethnicities of the
studied patients, sample size and study design, and also by chance.
Further studies are required in order to confirm the association
between ERCC1 rs11615 and clinical outcome in cases of
gastric cancer.
The present study also revealed that ERCC2
rs1799793 is an important factor influencing the response to
chemotherapy and OS in cases of gastric cancer, following
adjustment for multiple potential risk factors. A previous study
reported that the non-synonymous ERCC2 rs1799793 SNP is
associated with a reduced DNA repair capacity compared with that of
the common genotype (25). The
current results are in concordance with the proposed biological
effect of ERCC2 rs1799793, as a lower repair capacity may
lead to increased DNA damage and therefore to greater efficacy of
chemotherapy, but also increased toxicity (25). Considering these data, ERCC2
SNPs may serve as predictors of chemotherapy response in gastric
cancer patients (26). Furthermore,
two previous studies reported an association between ERCC2
rs1799793 and survival in gastric cancer patients (17,18).
However, only Li et al (23)
reported that the rs1799793 AA genotype had a significant impact on
OS in gastric cancer, which is consistent with the results of the
present study.
Several limitations must be considered in the
current study. Firstly, the study was conducted in a single
hospital in China, and this may not sufficiently represent the
entire Chinese population; selection bias may therefore have
affected the results. In addition, several DNA repair pathways that
may modify response to chemotherapy were analyzed; however, certain
different genetic factors may influence the treatment outcome, and
interaction may exist between these genes. Therefore, the results
of this study require validation with larger samples. Finally, the
limited sample size may also limit the statistical power to
identify differences between groups. Further large sample size and
multicentre studies are required to investigate the role of DNA
repair genes in the clinical outcome of gastric cancer.
In conclusion, the results of the present study
suggest that the ERCC1 rs11615 and ERCC2 rs1799793
SNPs in DNA repair pathways may be utilized as predictive factors
of the clinical outcome in gastric cancer patients. In the future,
these SNPs may contribute to the identification of patients who are
less likely to achieve a favorable response to platinum-based
chemotherapy. Translation of these pharmacogenetic predictors into
clinical practice may lead to improved gastric cancer treatment
planning and outcome.
References
|
1
|
World Health Organization; International
Agency for Research on Cancer, . GLOBOCAN 2012. Stomach Cancer.
Estimated Incidence, Mortality and Prevalence Worldwide in 2012.
http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspxAccessed:December
1–2014.
|
|
2
|
No authors listed: Schistosomes, liver
flukes, and Helicobacter pylori, . IARC Working Group on the
Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994.
IARC Monogr Eval Carcinog Risks Hum. 61:1–241. 1994.PubMed/NCBI
|
|
3
|
Macdonald JS: Clinical overview: Adjuvant
therapy of gastrointestinal cancer. Cancer Chemother Pharmacol. 54
(Suppl 1):S4–S11. 2004.PubMed/NCBI
|
|
4
|
Alberts DS, Garcia D and Mason-Liddil N:
Cisplatin in advanced cancer of the cervix: An update. Semin Oncol.
18 (1 Suppl 3):11–24. 1991.PubMed/NCBI
|
|
5
|
Zhou F, Yu Z, Jiang T, et al: Genetic
polymorphisms of GSTP1 and XRCC1: prediction of clinical outcome of
platinum-based chemotherapy in advanced non-small cell lung cancer
(NSCLC) patients. Swiss Med Wkly. 141:w132752011.PubMed/NCBI
|
|
6
|
Xu J, Ma J, Zong HT, et al:
Pharmacogenetic role of XRCC1 polymorphisms on the clinical outcome
of gastric cancer patients with platinum-based chemotherapy: A
systematic review and meta-analysis. Genet Mol Res. 13:1438–1446.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Jiang LP, Yin Y and Wang YD:
XRCC1 and XPD genetic polymorphisms and clinical outcomes of
gastric cancer patients treated with oxaliplatin-based
chemotherapy: A meta-analysis. Tumour Biol. 35:5637–5645. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu ZM, Luo TH, Nie MM, et al: Influence of
ERCC1 and ERCC4 polymorphisms on response to prognosis in gastric
cancer treated with FOLFOX-based chemotherapy. Tumour Biol.
35:2941–2948. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang X, Zhu H, Wu X, et al: A genetic
polymorphism in TOX3 is associated with survival of gastric cancer
in a Chinese population. PLoS One. 8:e721862013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Liu Z, Liu H, et al: ERCC1 and ERCC2
variants predict survival in gastric cancer patients. PLoS One.
8:e719942013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hasmats J, Kupershmidt I, Rodríguez-Antona
C, et al: Identification of candidate SNPs for drug induced
toxicity from differentially expressed genes in associated tissues.
Gene. 506:62–68. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Marsh S, McLeod H, Dolan E, et al:
Platinum pathway. Pharmacogenet Genomics. 19:563–564. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Deans AJ and West SC: DNA interstrand
crosslink repair and cancer. Nat Rev Cancer. 11:467–480. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Enoiu M, Jiricny J and Schärer OD: Repair
of cisplatin-induced DNA interstrand crosslinks by a
replication-independent pathway involving transcription-coupled
repair and translesion synthesis. Nucleic Acids Res. 40:8953–8964.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sharma S and Canman CE: REV1 and DNA
polymerase zeta in DNA interstrand crosslink repair. Environ Mol
Mutagen. 53:725–740. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li X and Heyer WD: Homologous
recombination in DNA repair and DNA damage tolerance. Cell Res.
18:99–113. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Costa RM, Chiganças V, Galhardo Rda S, et
al: The eukaryotic nucleotide excision repair pathway. Biochimie.
85:1083–1099. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Miller AB, Hoogstraten B, Staquet M and
Winkler A: Reporting results of cancer treatment. Cancer.
47:207–214. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Edge SB, Byrd DR, Compton CC, et al:
StomachAJCC Cancer Staging Manual. 7th. Springer; New York, NY: pp.
1202010
|
|
20
|
Zhou J, Liu ZY, Li CB, et al: Genetic
polymorphisms of DNA repair pathways influence the response to
chemotherapy and overall survival of gastric cancer. Tumour Biol.
36:3017–3023. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Booton R, Ward T, Heighway J, et al:
Xeroderma pigmentosum group D haplotype predicts for response,
survival, and toxicity after platinum-based chemotherapy in
advanced nonsmall cell lung cancer. Cancer. 106:2421–2427. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chu H, Gu D, Xu M, et al: A genetic
variant in ERCC2 is associated with gastric cancer prognosis in a
Chinese population. Mutagenesis. 28:441–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li J, Zuo X, Lv X, et al: Association of
DNA repair gene polymorphisms with response to chemotherapy and
prognosis of gastric cancer in a Chinese population. Tumour Biol.
35:7569–7574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu ZM, Luo TH, Nie MM, et al: Influence of
ERCC1 and ERCC4 polymorphisms on response to prognosis in gastric
cancer treated with FOLFOX-based chemotherapy. Tumour Biol.
35:2941–2948. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Caronia D, Patiño-García A, Milne RL, et
al: Common variations in ERCC2 are associated with response to
cisplatin chemotherapy and clinical outcome in osteosarcoma
patients. Pharmacogenomics J. 9:347–353. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Spitz MR, Wu X, Wang Y, et al: Modulation
of nucleotide excision repair capacity by XPD polymorphisms in lung
cancer patients. Cancer Res. 61:1354–1357. 2001.PubMed/NCBI
|