Introduction
S100A8 and S100A9 are calcium-binding proteins that
are secreted primarily by granulocytes and monocytes (1). Although S100A8 and S100A9 are able to
form homodimers, they typically exhibit pro- and anti-inflammatory
properties (2) by forming
heterodimers of S100A8/A9, alternatively known as calprotectin
(3,4).
High serum levels of S100A8/A9 correlate with inflammatory response
in a number of chronic diseases, including rheumatic arthritis,
inflammatory bowel disease and atherosclerosis (5). S100A8/A9 is proposed to be a sensitive
biomarker for monitoring inflammatory activities (6). Circulating levels of S100A8/A9 have
additionally been identified to be elevated in several tumors,
including lung, colon, gastric and breast cancer (BC), and may
contribute to cancer cell survival and metastasis (7).
BC may be classified into several molecular subtypes
based on gene expression profiles (8). The estrogen receptor (ER)+
subtypes, known as luminal A and luminal B, are the most
predominant molecular subtypes of BC (8). ER+ BC, which represents ~70%
of all BC cases, presents good prognosis and a better response to
endocrine therapies than ER− BC (9). Human epidermal growth factor receptor 2
(Her2)-amplified and basal-like BC subgroups constitute the
ER− subtypes (8). Other
less characterized molecular subtypes, including normal breast-like
and claudin-low BC, have additionally been categorized as
ER− BC in certain studies (10). Evidence from previous studies strongly
suggests that the molecular subtype influences the systemic therapy
and clinical outcome of BC (11).
Her2-amplified BC accounts for ~20–25% of all invasive BC cases,
and presents a poor overall survival (OS) rate (12). However, following the development of
drugs such as trastuzumab, which selectively targets Her2, an
improved prognosis may be achieved for this subtype of BC (12). Basal-like BC accounts for ~10% of all
BC cases, and presents the worst prognosis, as there is currently
no available endocrine or targeted therapy for this subtype of BC
(13).
In BC, S100A8/A9 has been suggested to be a
potential candidate for the mediation of metastasis of breast
epithelial cells (14). S100A8/A9 is
additionally able to promote the invasion of BC cells by binding to
its receptor, known as receptor for advanced glycation end-product
(RAGE), on the surface of cancer cells (15). However, the molecular mechanisms by
which S100A8/A9 participates in the regulation of BC survival
remain to be elucidated.
In the present study, the expression of S100A8 and
S100A9 was investigated in various subtypes of BC, and its
secretion by BC cells was evaluated. The present study additionally
analyzed the correlation between S100A8/A9 expression and the
expression of other genes, including GATA binding protein 3 (GATA3)
and estrogen receptor 1 (ESR1), using GeneChip® data for BC. The
effects of the recombinant protein S100A8/A9 on the regulation of
GATA3 and ESR1 gene expression were assessed in the MCF-7 human
ER+ BC cell line. The transcriptional levels of S100A8
and S100A9 and their association with the prognosis of BC were
examined using microarray data.
Materials and methods
Microarrays
In the present study, published BC microarray data
from a Netherlands Cancer Institute (NKI) cohort was analyzed
(16). NKI data were generated using
the Affymetrix® platform (Affymetrix, Inc., Santa Clara, CA, USA).
The gene expression profiling and clinical data of the NKI cohort
may be downloaded from http://www.bioconductor.org/packages/release/data/experiment/html/cancerdata.html.
In summary, the NKI dataset contains data measured in 25,000 spot
oligonucleotide arrays obtained from 295 cases of BC. All patients
were <53 years old and exhibited lymph node-negative stage I or
II disease.
Cell culture
The MCF-7 BC cell line was obtained from the
American Type Culture Collection (Manassas, VA, USA), and grown in
Dulbecco's modified Eagle's medium/nutrient mixture F-12 (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
Cells were maintained at 37°C in a humidified incubator with an
atmosphere of 5% CO2.
For investigation of the effects of S100A8/A9 on
gene expression, including ESR1 and GATA3, 1×106 MCF-7
cells were seeded in a 6-well plate (Corning Life Sciences, Lowell,
MA, USA) and treated with 10 ng/ml human recombinant S100A8/A9
(R&D Systems, Inc., Minneapolis, MN, USA) for 24 h.
S100A8/A9 expression was additionally investigated
in breast tumor tissue samples. Breast tumor tissue was obtained
from a 53-year-old patient, who underwent a modified radical
mastectomy at the Second Hospital of Jiaxing (Jiaxing, China)
following a diagnosis of stage I invasive ductal carcinoma of the
left breast. Samples of size 0.5×0.5 cm were disected from breast
tumors and adjacent normal tissue, and were stored in liquid
nitrogen within half an hour of removal.
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from tissues or cells using
TRIzol® reagent purchased from Invitrogen (Thermo Fisher
Scientific, Inc.). The RNA samples were treated with 1 µl DNase (5
U/µl; Toyobo Co., Ltd., Osaka, Japan). A total of l µg RNA from
each sample was reverse transcribed to complementary (c)DNA in a
final volume of 20 µl using a ReverTra Ace® quantitative (q)PCR RT
kit (Toyobo Co., Ltd.). A total of 1 µl of each cDNA sample was
subsequently amplified in a PCR mixture with DNA polymerase (Toyobo
Co., Ltd.) in a final volume of 25 µl. Human β-actin was utilized
as a reference gene. The sequences of the primers used in the
reaction are shown in Table I.
Primers were designed using Primer Express® software version 3.0
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers
were synthesized by Invitrogen™ (Thermo Fisher Scientific, Inc.).
Negative controls (no cDNA and no reverse transcriptase) were run
in parallel.
 | Table I.Primers for reverse
transcription-polymerase chain reaction. |
Table I.
Primers for reverse
transcription-polymerase chain reaction.
| Gene | Primer sequence
(5′-3′) |
|---|
| S100A8 |
|
|
Forward |
TGTCTCTTGTCAGCTGTCTTTCA |
|
Reverse |
CCTGTAGACGGCATGGAAAT |
| S100A9 |
|
|
Forward |
GGAATTCAAAGAGCTGGTGC |
|
Reverse |
TCAGCATGATGAACTCCTCG |
| β-actin |
|
|
Forward |
GTGGCATCCACGAAACTACCTT |
|
Reverse |
GGACTCGTCATACTCCTGCTTG |
The PCR cycling conditions were as follows: PCR was
performed on a thermal cycler (ABI Veriti®; Thermo Fisher
Scientific, Inc.) with an initial denaturation step at 94°C for 2
min, followed by a specific number of cycles (S100A8 and S100A9, 32
cycles; β-actin, 25 cycles) consisting of denaturation at 94°C for
20 sec, annealing at the specified temperature for 25 sec, and
extension at 72°C for 50 sec. A final extension step was conducted
at 72°C for 5 min. PCR products were separated using a 1% agarose
gel (Gene Company Ltd., Hong Kong, China) with a 100 bp DNA ladder
(Toyobo Co., Ltd.) stained with GelRed™ (Biotium, Inc., Hayward,
CA, USA). Image Lab™ version 3.0 software (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) was used for densitometry analysis of the
DNA gels.
qPCR
Relative messenger (m)RNA levels were quantified
using qPCR with Applied Biosystems® StepOnePlus™ (Thermo Fisher
Scientific, Inc.). Primers were designed using Primer Express®
Software. The sequences of the primers used are presented in
Table II. PCR amplification was
performed in duplicate using 96-well plates (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and SYBR® Green RealTime PCR Master
Mix (Toyobo Co., Ltd.). The PCR cycling conditions were as follows:
95°C for 10 min, followed by 40 cycles consisting of 95°C for 15
sec and 60°C for 1 min. The mRNA levels of human β-actin were
measured in an identical manner, and served as the reference gene.
All samples were normalized to β-actin values, and the results are
expressed as fold-changes of Cq value relative to the control using
the 2−ΔΔCq formula (17).
 | Table II.Primers for quantitative polymerase
chain reaction. |
Table II.
Primers for quantitative polymerase
chain reaction.
| Gene | Primer sequence
(5′-3′) |
|---|
| S100A8 |
|
|
Forward |
ATTTCCATGCCGTCTACAGG |
|
Reverse |
TGCCACGCCCATCTTTATCA |
| S100A9 |
|
|
Forward |
CACCCAGACACCCTGAACCA |
|
Reverse |
CCTCGAAGCTCAGCTGCTTG |
| Estrogen receptor
1 |
|
|
Forward |
CCTGATGATTGGTCTCGTCTG |
|
Reverse |
GGCACACAAACTCCTCTCC |
| GATA binding
protein 3 |
|
|
Forward |
ACAAAATGAACGGACAGA |
|
Reverse |
GTGGTGGTCTGACAGTTC |
| β-actin |
|
Forward |
GGATGCAGAAGGAGATCACTG |
|
Reverse |
CGATCCACACGGAGTACTTG |
Serum analyses
Cultured MCF-7 cell medium was collected following
two days of incubation, and frozen at −20°C. The concentration of
S100A8/9 in the medium was measured using a commercially available
enzyme-linked immunosorbent assay (ELISA) kit (Cusabio Biotech Co.,
Ltd., Wuhan, China), according to the manufacturer's protocol.
Statistical analysis
Data are presented as the mean ± standard error.
Differences between means were analyzed using Student's t-test and
one-way analysis of variance. For the purpose of studying
correlations, Pearson's correlation coefficient was determined
using univariate Cox regression analysis. A Kaplan-Meier survival
curve was used to analyze survival, and the P-value was calculated
using the log-rank (Mantel-Cox) method. The graphical
representation of the data and the statistical analyses were
performed using GraphPad Prism version 5.0 software (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression patterns of S100A8 and
S100A9 differ in various subtypes of BC
Initially, the present study investigated the
transcriptional levels of S100A8 and S100A9 in various subtypes of
BC using the NKI BC cohort. A significant difference in the
transcriptional levels of S100A8 was observed among the four
subgroups of BC, with increased expression of S100A8 in the
basal-like and Her2-amplified subgroups, and lower expression in
the luminal A and B subtypes (P<0.001; Fig. 1A). A similar expression pattern was
observed for S100A9 (P<0.001; Fig.
1B). However, no difference was observed for the expression
levels of RAGE (the S100A8/A9 receptor) in the four subgroups of BC
(Fig. 1C).
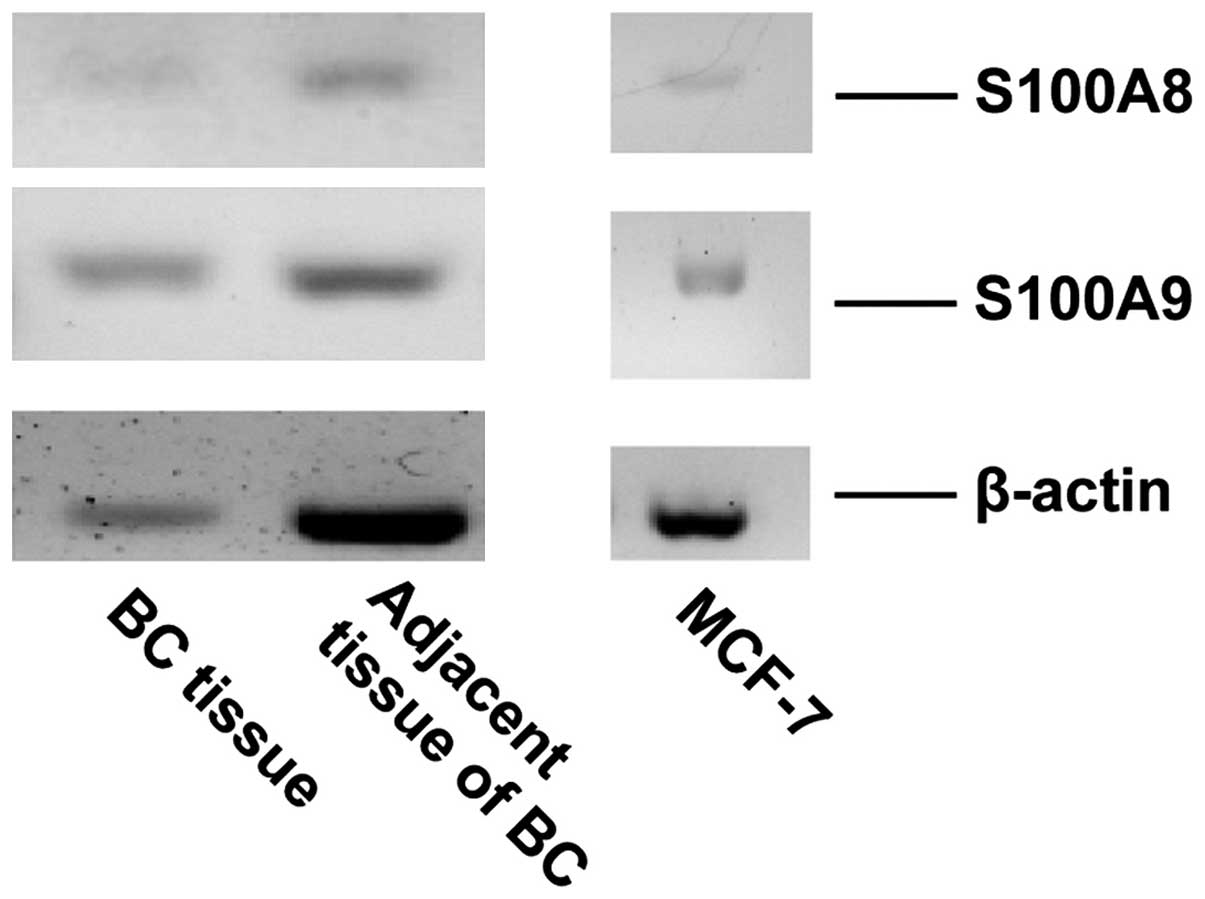
S100A8/A9 is expressed and secreted by
BC cells
RT-PCR was performed to evaluate the gene expression
of S100A8 and S100A9 in MCF-7 cells and breast tumor tissue.
Signals corresponding to S100A8 and S100A9 were detected in MCF-7
cells and in breast tumor tissue (Fig.
2). ELISA was performed to detect the secretion of S100A8/A9 in
the medium of the cultured MCF-7 cells. The concentration of
S100A8/A9 was ~7.18 ng/ml in the medium of cultured MCF-7 cells
following two days of incubation (data not shown).
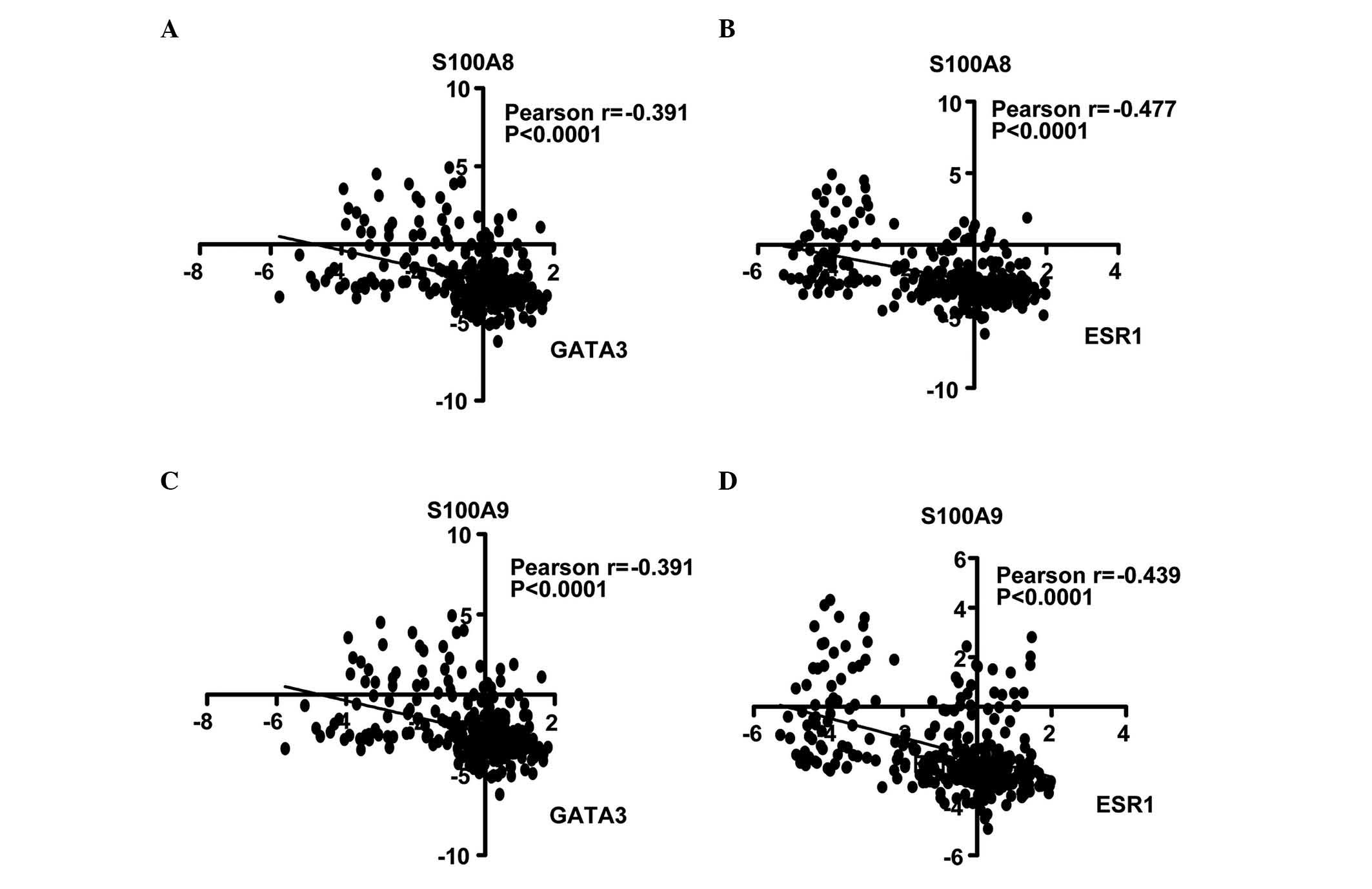
S100A8/A9 expression negatively
correlates with ESR1 and GATA3 expression in BC
Calculation of the Pearson's correlation coefficient
is a method of measuring the correlation (linear dependence)
between two variables (X and Y), whereby a value between +1 and −1
is assigned (18). In the present
study, the correlation of S100A8 and S100A9 with ESR1 and GATA3
genes was analyzed. In the NKI cohort, it was identified that
S100A8 expression was inversely correlated with GATA3 (r=−0.391;
Fig. 3A) and ESR1 expression
(r=−0.477; Fig. 3B). Similarly,
S100A9 was negatively correlated with GATA3 (r=−0.391; Fig. 3C) and ESR1 (r=−0.439; Fig. 3D). Univariate Cox regression analysis
revealed that the correlations were significant (P<0.0001).
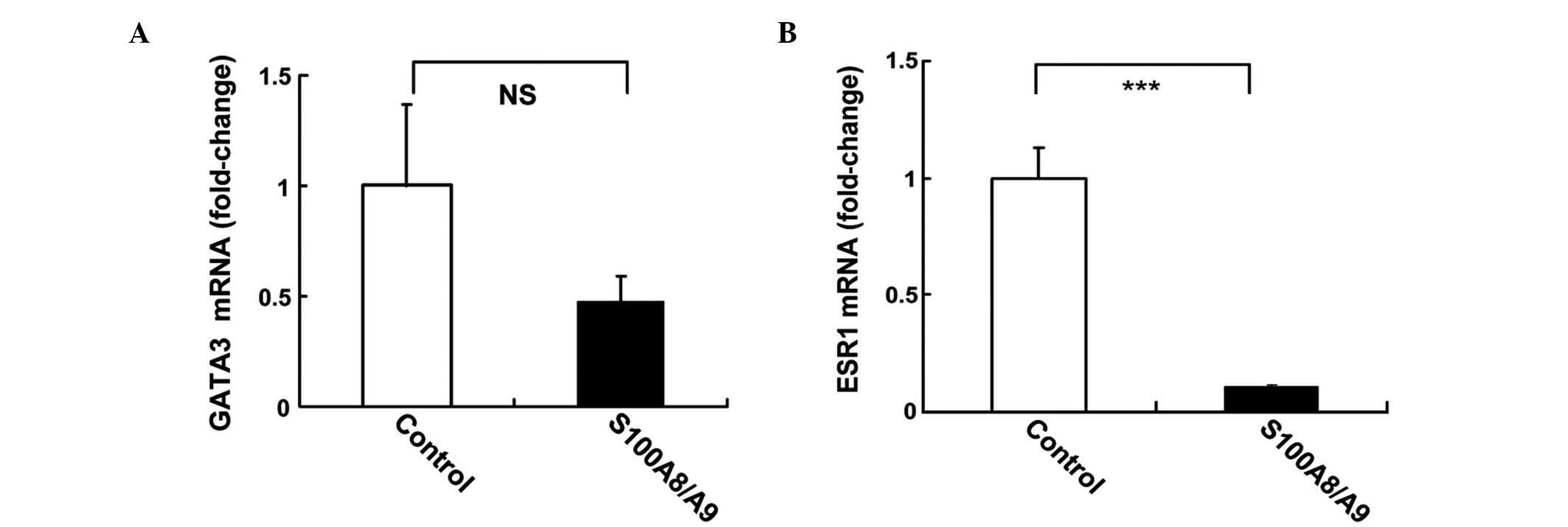
Treatment with S100A8/A9 regulates
ESR1 in MCF-7 cells
The regulation of ESR1 and GATA3 gene expression by
recombinant S100A8/A9 protein in MCF-7 cells was investigated using
qPCR. MCF-7 cells were treated with 10 ng/ml S100A8/A9 for 24 h.
S100A8/A9 induced 50% inhibition of GATA3, although this was not
statistically significant (P=0.078; Fig.
4A). S100A8/A9 induced a 10-fold reduction in the mRNA levels
of ESR1 (P<0.001; Fig. 4B).
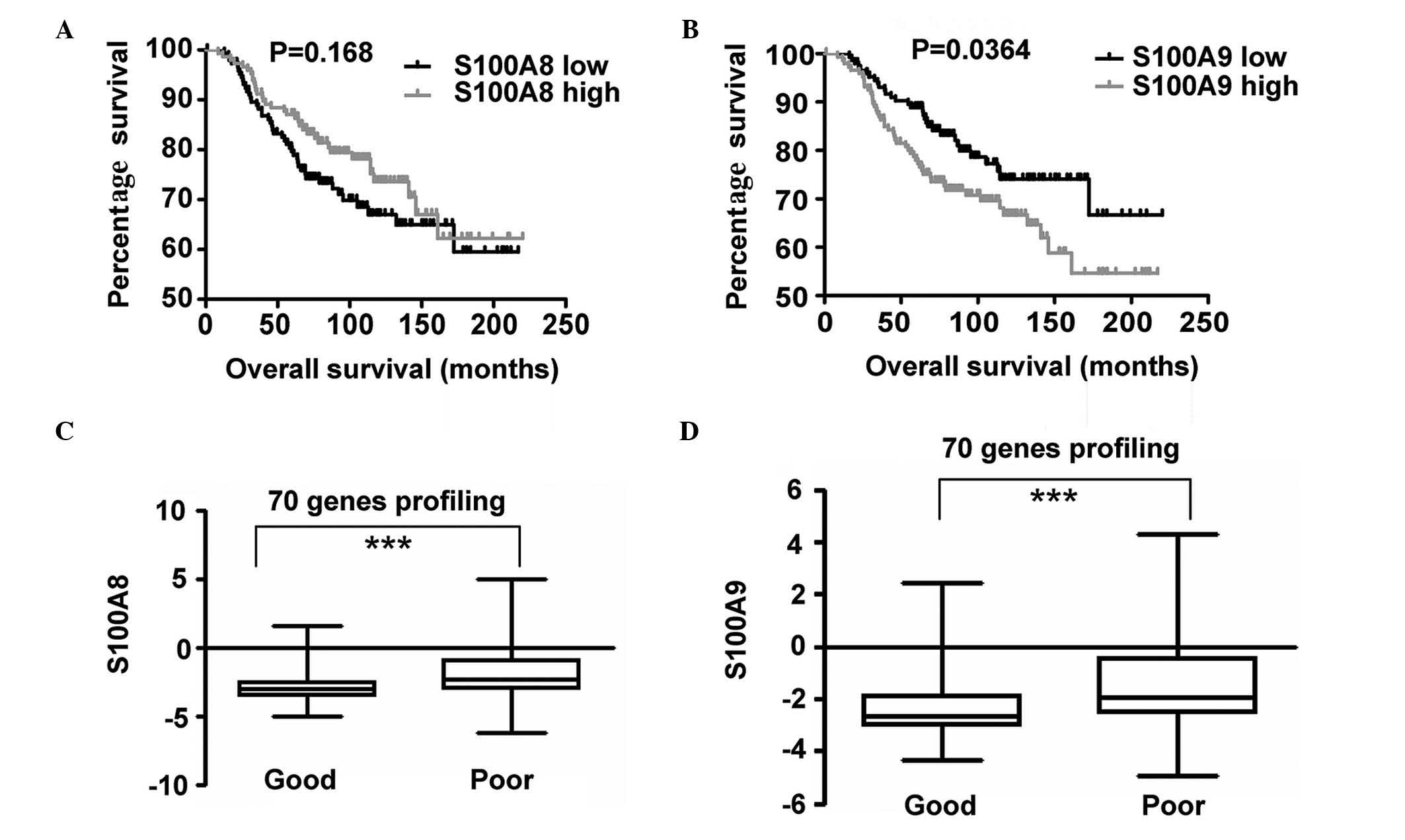
Increased expression of S100A9
correlates with BC prognosis
In order to investigate the expression of S100A8 and
S100A9 and its association with OS, S100A8 and S100A9 expression
profiling and clinical data provided by the NKI cohort was
analyzed. Patients were divided into two groups based on the
expression levels of S100A8 in the top 50th percentile and in the
bottom 50th percentile. Survival curves of OS in the two groups
were calculated using the Kaplan-Meier method, and compared via the
log-rank test. An identical measurement was performed on the basis
of S100A9 gene expression. A statistically significant difference
was observed between the two groups, and increased S100A9
expression was identified to be associated with poor OS. However,
no statistically significant difference was observed for S100A8
expression (Fig. 5). P-values were
calculated using the log-rank test.
Discussion
In the present study, microarray data obtained from
a published database was analyzed. Increased levels of S100A8 and
S100A9 transcripts were observed to be present in
Her2+/basal-like BC subgroups, whereas they were lower
in luminal A/B subtypes. This observation indicated that the
expression levels of S100A8/A9 may be associated with the
expression levels of hormone receptors. Subsequently, it was
identified that S100A8/A9 exhibited a significant inverse
correlation with ESR1 and GATA3 expression.
ERs consist of two isoforms, ERα and ERβ, which have
been identified to be expressed in multiple tissues, including the
mammary gland (19). ERα is the
primary type of ER in breast tissues, and is encoded by the ESR1
gene (20). High expression levels of
ERα correlate with increased mRNA levels of ESR1 (19). A well-established role of ERα is the
maintenance of a differentiated epithelial phenotype in the mammary
gland (21).
GATA3 is the most extensively studied GATA
transcription factor (22). Normal
differentiation of duct epithelia may be terminated with a
tissue-specific knockout of the GATA3 gene in mice (23), which indicates that GATA3 has a
significant role in ductal epithelial cell differentiation during
the development of the mammary gland. GATA3 expression has been
reported to possess a positive correlation with ERα expression
(24), and cooperates with ERα in the
driving of luminal ductal epithelial cell differentiation during
mammary gland maturation (25). It is
considered that GATA3 is able to mediate ERα expression by binding
to the promoter region of the ERα gene (26). Furthermore, increased expression of
GATA3 appears to inhibit BC metastasis (27). It remains to be elucidated whether
S100A8 and S100A9 have a role in development of the normal mammary
gland. A previous study reported that a S100A9−/-
knockout mouse model demonstrated no obvious cancer phenotype and
was fertile; however, this study primarily focused on myeloid cell
function (28). Whether S100A8/A9 has
a role in ductal epithelial cell differentiation may be elucidated
if breast tissue-specific knockout mice are generated.
The inverse correlation of ESR1 and GATA3 with
S100A8 and S100A9 expression observed in the present study
indicated that a negative regulation loop may exist between these
genes. To address this issue, recombinant S100A8/A9 protein was
used to treat MCF-7 cells, and strong inhibition of ESR1 gene
expression was observed. By contrast, the mRNA levels of GATA3
demonstrated no significant change. It remains to be elucidated
whether a positive feedback loop exists between ERα and GATA3, as a
previous study revealed that GATA3 gene expression exhibited no
response following administration of estradiol, which activates ER
(29).
S100A8 and S100A9 are primarily released by myeloid
cells. In the present study, S100A8 and S100A9 gene expression was
not only present in BC cells, but S100A8 and S100A9 were
additionally secreted by BC cells (30). Cells in the tumor microenvironment,
including endothelial, stromal and infiltrating immune cells,
exhibit crosstalk and affect the proliferation and survival of
cancer cells (31). Myeloid-derived
suppressor cells (MDSCs) are a population of myeloid cells that are
trafficked to the primary or metastatic sites of a tumor (32). MDSCs display immunosuppressive
activities by reducing the proliferation and function of effector T
cells (32). Blocking S100A8/A9 and
its receptor RAGE on the surface of MDSCs restores T cell
proliferation (7). A previous study
demonstrated that overexpression of chemokine (C-X-C motif) ligand
1/2 in BC cells attracted MDSCs to accumulate in the tumor
microenvironment (33). MDSCs release
S100A8/A9, which enhances cancer cell survival and induces
additional MDSC accumulation in tumor sites (33). The tumor-stroma paracrine axis
provides a survival benefit for cancer cells in the tumor
microenvironment (33). In the
present study, it was difficult to clarify the primary source of
elevated S100A8/A9 in ER− BC. S100A8/A9 may exert
autocrine and/or paracrine roles, mediating downregulation of the
expression of ESR1. The increased concentration of S100A8/A9 in the
tumor microenvironment may contribute to resistance to endocrine
therapy in ER+ BC, as a consequence of suppressing ERα
expression.
Using a Kaplan-Meier survival curve, increased mRNA
levels of S100A9 were observed to be associated with decreased OS
and poor prognosis by analyzing the NKI data. It has been proposed
that a 70-gene prognostic signature, the intrinsic subtypes and the
recurrence score may be strongly concordant in the evaluation of BC
outcome (34). In the present study,
increased S100A8 and S100A9 expression was observed to be
associated with poor prognosis and the 70-gene prognostic
signature. The results of the present study suggested that the
detection of the transcript levels of S100A9 may serve as an
indicator for prediction of prognosis of BC.
In conclusion, the present study revealed increased
mRNA expression levels of S100A8 and S100A9 in
Her2+/basal-like BC. S100A8 and S100A9 genes are
expressed in BC cells, and their expression is inversely correlated
with ESR1 and GATA3 expression. Furthermore, treatment of S100A8/A9
in vitro repressed ESR1 expression in MCF-7 BC cells.
Increased mRNA levels of S100A9 were associated with decreased OS
in BC. Therefore, the results of the present study suggested that
enhanced S100A8/A9 expression in the BC microenvironment may reduce
the sensitivity of BC cells to endocrine therapy, possibly due to a
loss of ER, and S100A8/A9 may serve as a biomarker for prediction
of prognosis of BC.
Acknowledgements
The present study was supported by grants from the
Chinese National Science Fund for Young Scholars (Beijing, China;
grant no. 81101707), the Zhejiang Traditional Chinese Medicine
Foundation Project (Hangzhou, China; grant no. 2014ZB119), the
Natural Science Foundation of Zhejiang Province (Hangzhou, China;
grant no. LY16H070007) and the Science and Technology Bureau of
Jiaxing (Jiaxing, China; grant no. 2012AY1071–2).
References
|
1
|
Hessian PA, Edgeworth J and Hogg N: MRP-8
and MRP-14, two abundant Ca(2+)-binding proteins of
neutrophils and monocytes. J Leukoc Biol. 53:197–204.
1993.PubMed/NCBI
|
|
2
|
Sinha P, Okoro C, Foell D, Freeze HH,
Ostrand-Rosenberg S and Srikrishna G: Proinflammatory S100 proteins
regulate the accumulation of myeloid-derived suppressor cells. J
Immunol. 181:4666–4675. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Heizmann CW, Fritz G and Schäfer BW: S100
proteins: Structure, functions and pathology. Front Biosci.
7:d1356–d1368. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Srikrishna G: S100A8 and S100A9: New
insights into their roles in malignancy. J Innate Immun. 4:31–40.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chan JK, Roth J, Oppenheim JJ, Tracey KJ,
Vogl T, Feldmann M, Horwood N and Nanchahal J: Alarmins: Awaiting a
clinical response. J Clin Invest. 122:2711–2719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vogl T, Eisenblätter M, Völler T, Zenker
S, Hermann S, van Lent P, Faust A, Geyer C, Petersen B, Roebrock K,
et al: Alarmin S100A8/S100A9 as a biomarker for molecular imaging
of local inflammatory activity. Nat Commun. 5:45932014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang L, Chang EW, Wong SC, Ong SM, Chong
DQ and Ling KL: Increased myeloid-derived suppressor cells in
gastric cancer correlate with cancer stage and plasma S100A8/A9
proinflammatory proteins. J Immunol. 190:794–804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reis-Filho JS and Pusztai L: Gene
expression profiling in breast cancer: Classification,
prognostication, and prediction. Lancet. 378:1812–1823. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Holst F, Stahl PR, Ruiz C, Hellwinkel O,
Jehan Z, Wendland M, Lebeau A, Terracciano L, Al-Kuraya K, Jänicke
F, et al: Estrogen receptor alpha (ESR1) gene amplification is
frequent in breast cancer. Nat Genet. 39:655–660. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prat A, Parker JS, Karginova O, Fan C,
Livasy C, Herschkowitz JI, He X and Perou CM: Phenotypic and
molecular characterization of the claudin-low intrinsic subtype of
breast cancer. Breast Cancer Res. 12:R682010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sonnenblick A, Fumagalli D, Sotiriou C and
Piccart M: Is the differentiation into molecular subtypes of breast
cancer important for staging, local and systemic therapy, and
follow up? Cancer Treat Rev. 40:1089–1095. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gajria D and Chandarlapaty S:
HER2-amplified breast cancer: Mechanisms of trastuzumab resistance
and novel targeted therapies. Expert Rev Anticancer Ther.
11:263–275. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dey N, Smith BR and Leyland-Jones B:
Targeting basal-like breast cancers. Curr Drug Targets.
13:1510–1524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moon A, Yong HY, Song JI, Cukovic D,
Salagrama S, Kaplan D, Putt D, Kim H, Dombkowski A and Kim HR:
Global gene expression profiling unveils S100A8/A9 as candidate
markers in H-ras-mediated human breast epithelial cell invasion.
Mol Cancer Res. 6:1544–1553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yin C, Li H, Zhang B, Liu Y, Lu G, Lu S,
Sun L, Qi Y, Li X and Chen W: RAGE-binding S100A8/A9 promotes the
migration and invasion of human breast cancer cells through actin
polymerization and epithelial-mesenchymal transition. Breast Cancer
Res Treat. 142:297–309. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van't Veer LJ, Dai H, van de Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ,
Witteveen AT, et al: Gene expression profiling predicts clinical
outcome of breast cancer. Nature. 415:530–536. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Campbell MJ and Machin D: Medical
Statistics: A Commonsense Approach (3rd). London, UK: Wiley.
1999.
|
|
19
|
Kerdivel G, Flouriot G and Pakdel F:
Modulation of estrogen receptor alpha activity and expression
during breast cancer progression. Vitam Horm. 93:135–160. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miyoshi Y, Murase K, Saito M, Imamura M
and Oh K: Mechanisms of estrogen receptor-α upregulation in breast
cancers. Med Mol Morphol. 43:193–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mueller SO, Clark JA, Myers PH and Korach
KS: Mammary gland development in adult mice requires epithelial and
stromal estrogen receptor α. Endocrinology. 143:2357–2365. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fang SH, Chen Y and Weigel RJ: GATA-3 as a
marker of hormone response in breast cancer. J Surg Res.
157:290–295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kouros-Mehr H, Slorach EM, Sternlicht MD
and Werb Z: GATA3 maintains the differentiation of the luminal cell
fate in the mammary gland. Cell. 127:1041–1055. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen JQ, Litton J, Xiao L, Zhang HZ,
Warneke CL, Wu Y, Shen X, Wu S, Sahin A, Katz R, et al:
Quantitative immunohistochemical analysis and prognostic
significance of TRPS-1, a new GATA transcription factor family
member, in breast cancer. Horm Cancer. 1:21–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kouros-Mehr H, Kim JW, Bechis SK and Werb
Z: GATA3 and the regulation of the mammary luminal cell fate. Curr
Opin Cell Biol. 20:164–170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eeckhoute J, Keeton EK, Lupien M, Krum SA,
Carroll JS and Brown M: Positive cross-regulatory loop ties GATA3
to estrogen receptor alpha expression in breast cancer. Cancer Res.
67:6477–6483. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan W, Cao QJ, Arenas RB, Bentley B and
Shao R: GATA3 inhibits breast cancer metastasis through the
reversal of epithelial-mesenchymal transition. J Biol Chem.
285:14042–14051. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hobbs JA, May R, Tanousis K, McNeill E,
Mathies M, Gebhardt C, Henderson R, Robinson MJ and Hogg N: Myeloid
cell function in MRP-14 (S100A9) null mice. Mol Cell Biol.
23:2564–2576. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hoch RV, Thompson DA, Baker RJ and Weigel
RJ: GATA3 is expressed in association with estrogen receptor in
breast cancer. Int J Cancer. 84:122–128. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gebhardt C, Németh J, Angel P and Hess J:
S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol.
72:1622–1631. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liotta LA and Kohn EC: The
microenvironment of the tumour-host interface. Nature. 411:375–379.
2001. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lindau D, Gielen P, Kroesen M, Wesseling P
and Adema GJ: The immunosuppressive tumour network: Myeloid-derived
suppressor cells, regulatory T cells and natural killer T cells.
Immunology. 138:105–115. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Acharyya S, Oskarsson T, Vanharanta S,
Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N,
Seshan VE, et al: A CXCL1 paracrine network links cancer
chemoresistance and metastasis. Cell. 150:165–178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fan C, Oh DS, Wessels L, Weigelt B, Nuyten
DS, Nobel AB, van't Veer LJ and Perou CM: Concordance among
gene-expression- based predictors for breast cancer. N Engl J Med.
355:560–569. 2006. View Article : Google Scholar : PubMed/NCBI
|