Introduction
Prostate cancer (PCa) is the second leading cause of
cancer-related mortality in men (1).
During the progression of PCa, a number of factors play important
roles, including genetic and epigenetic alternations and the tumor
microenvironment. Previous studies have indicated that a single
oncogenic event is not sufficient to initiate prostate neoplasia,
and that cooperating oncogenic events are required (2). Accordingly, a detailed understanding of
how oncogenic aberrations in PCa may cooperate with one another to
augment malignant progression and produce an aggressive,
metastasis-prone tumor are critical.
MicroRNAs (miRNAs) are small non-coding RNAs of
18–25 nucleotides, which negatively regulate gene expression by
binding to regulatory sites predominantly located in the
3′-untranslated region (3-UTR) of a transcript (3). Numerous miRNAs have been demonstrated to
be involved in a wide range of biological functions, including
cellular proliferation, migration, differentiation and apoptosis
(4). The dysregulation of these
miRNAs has been implicated in cancer development and metastasis,
and may be useful in predicting the outcome of various
malignancies, including PCa (5).
Previous studies, including our own, have
demonstrated that miRNAs and components of the intracellular miRNA
machinery exhibit widespread dysregulation in PCa biology (6,7). We have
also shown that microRNA-30c (miR-30c) acts as a tumor suppressor
gene in PCa cells by inhibiting proliferation, migration, invasion
and metastasis (8). Accordingly, our
results further demonstrated the involvement of miR-30c in PCa
progression and suggested its potential role as an independent
predictor of biochemical recurrence (BCR)-free survival (8). miR-30c has been demonstrated to be
involved in inhibiting the self-renewal capacity of breast
tumor-initiating cells via reducing ubiquitin carrier protein 9, in
inducing cellular senescence via regulating B-Myb, and in promoting
an invasive phenotype via reducing epithelial-to-mesenchymal
transition (EMT) (9–11). However, whilst accumulating evidence
indicates an involvement of aberrant miR-30c expression in
carcinogenesis, there are still limited data available regarding
the functional role and mechanism of miR-30c in PCa.
The canonical Wnt/β-catenin pathway has been
implicated in the pathogenesis of a broad range of human cancers
(12), and its components have
emerged as targets for cancer therapy (13). Numerous coactivators for β-catenin
transcription have been identified, including B-cell lymphoma 9
(BCL9), its homolog BCL9-like, Pygopus and others (14,15). The
human BCL9 gene was first identified by cloning the
t(1;14)(q21;q32) translocation from a patient with precursor B-cell
acute lymphoblastic leukemia (16).
BCL9 is overexpressed in a variety of malignancies and, as a
component of the aberrantly activated Wnt signaling pathway,
promotes cell proliferation, migration, invasion and metastasis of
tumor cells (17,18). Previous studies have identified BCL9
as a direct target of miR-30c in a number of cancer types (19,20).
However, a functional link between miR-30c and the Wnt pathway
coactivator BCL9, and their association with clinicopathological
variables in PCa have not been established.
The present study aimed to investigate the effects
of miR-30c overexpression on BCL9 in PCa cells, and whether BCL9 is
a direct target of miR-30c. In addition, the effects of ectopic
expression of miR-30c on the expression of Wnt pathway downstream
targets, including c-Myc, CD44 and SOX9 were explored. Furthermore,
the association between BCL9 expression and various
clinicopathological factors, including Gleason score and BCR, were
assessed. The findings indicate that there is a functional link
between miR-30c and BCL9 in PCa, and that the small molecule
miR-30c may serve as a potential therapy by inhibiting the
BCL9/β-catenin interaction and selectively suppressing oncogenic
Wnt transcription.
Materials and methods
Patients and tissues samples
Approval for the present study was obtained from the
Research Ethics Committee of Guangzhou First People's Hospital
Affiliated to Guangzhou Medical University (Guangzhou, China). For
miRNA extraction and immunohistochemistry, 98 tumor tissue samples
were obtained from PCa patients who underwent radical prostatectomy
at Guangzhou First People's Hospital between January 2002 and
August 2012. In addition, 20 benign prostate hyperplasia (BPH)
specimens obtained by transurethral resection of the prostate were
collected for subsequent experiments. Patients with PCa who
received radiotherapy or hormonal treatment prior to surgery were
excluded. PCa cases were classified by the World Health
Organization criteria (21) and
staged according to the tumor node metastasis classification
(22) and the Gleason grading system
(23). The detailed characteristics
of these patients are presented in Table
I. BCR was defined as a postoperative serum prostate-specific
antigen (PSA) level of ≥0.2 ng/ml.
 | Table I.Characteristics of prostate cancer
patients (n=98). |
Table I.
Characteristics of prostate cancer
patients (n=98).
| Clinicopathological
feature | Value |
|---|
| Age |
|
| Median,
years | 63 |
| Range,
years | 54–83 |
| Preoperative
prostate-specific antigen (ng/ml), n |
|
| ﹤4 | 17 |
| 4–10 | 63 |
| ≥10 | 18 |
| Gleason score, n |
|
| 6 | 33 |
| 7 | 54 |
| 8 | 6 |
| 9 | 5 |
| Pathological tumor
stage, n |
|
|
pT2 | 68 |
|
pT3 | 25 |
|
pT4 | 5 |
| Follow-up,
months |
|
|
Median | 45 |
|
Range | 1–128 |
| Biochemical
recurrence (months), n | 18 |
|
1–12 | 7 |
|
13–24 | 6 |
|
>24 | 5 |
Cell culture and reagents
The DU145 prostate cancer cell line (American Type
Culture Collection, Rockville, MD, USA) was cultured in RPMI 1640
medium (GE Healthcare Life Sciences, Logan, UT, USA), supplemented
with fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and 100 U/ml penicillin and 100 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) and
maintained at 37°C in an atmosphere of 5% CO2.
Transfection of mature miRNA
For transient transfection, 0.5×105 DU145
cells were plated in 24 well plates, 12 h prior to transfection.
The pGCMV/EGFP/Neo vector (catalog no., C05001) with overexpression
of miR-30c was purchased from Shanghai GenePharma Co., Ltd.
(Shanghai, China). The blank vector was used as negative control.
DU145 human prostate carcinoma cells were transiently transfected
with mature miR-30c or control (miR-NC) using Invitrogen™
Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Following the
transfection of cells, BCL9 and several Wnt pathway downstream
targets were detected by western blot analysis.
Western blot analysis
Following the standard protocol, total proteins were
extracted from cultured cells using 200 µl M-PER Mammalian Protein
Extraction Reagent (Thermo Fisher Scientific, Inc.). Protein
concentration was measured using a bicinchoninic acid protein assay
kit (Thermo Fisher Scientific, Inc.). Total protein concentration
was calculated by measuring the absorbance at a wavelength of 260
nm (NanoDrop 2000c Spectrophotometer; Thermo Fisher Scientific
Inc., Wilmington, DE, USA). Next, the lysates were separated by
sodium dodecyl sulfate-polyacrylamide gel electrophoresis (NuPAGE™
Novex 4–12% Bis-Tris Gel; catalog no., NP0322BOX; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and blotted onto polyvinylidene
fluoride (PVDF) membranes (catalog no., IPFL00010; EMD Millipore,
Billerica, MA, USA). The PVDF membranes were blocked with 5% skim
milk in phosphate-buffered saline (PBS)-Tween 20 and probed with
rabbit polyclonal IgG antibodies against human BCL9 (catalog no.,
ab37305; dilution, 1:200; Abcam, Cambridge, MA, USA), SOX9 (catalog
no., sc-20095; dilution, 1:200; Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA), c-Myc (catalog no., sc-764; dilution, 1:200;
Santa Cruz Biotechnology) or CD44 (catalog no., ab41478; dilution,
1:1,000; Abcam), or a mouse monoclonal IgG1 antibody
against human β-actin (catalog no., sc-47778; dilution, 1:100;
Santa Cruz Biotechnology) overnight at 4°C. The membranes were then
washed with Tris-Buffered Saline with Tween 20 and incubated with
alkaline phosphatase-conjugated goat anti-rabbit (catalog no.,
SAB3700852; dilution, 1:2,000; Sigma-Aldrich, St. Louis, MO, USA)
or anti-mouse IgG secondary antibody (catalog no., A2179; dilution,
1:2,000; Sigma-Aldrich) for 1 h at room temperature. Signals were
visualized using SuperSignal West Pico Chemiluminescent Substrate
(Thermo Fisher Scientific, Inc.) and the STORM 860 Molecular Imager
(Amersham Biosciences, Uppsala, Sweden).
Luciferase reporter assay
The putative miR-30c complementary site in the
3′-UTR of BCL9 mRNA, or mutant sequence, were cloned into the pGL3
luciferase reporter vector (Promega Corporation, Madison, WI, USA)
(Fig. 1A). Cultured DU145 cells were
cotransfected with 10–24 ng Firefly reporter wild-type BCL9
constructs or the mutant vector (3′-UTR-BCL9wt FLuci or
3′-UTR-BCL9mut FLuci vectors; Promega Corporation), 3 ng
Renilla reporter plasmid pGL3 and 30 nM miR-30c mimic or NC
mimic. Following 48 h of transfection, PCa cells were harvested,
and reporter assays were performed using a Dual-Glo® Luciferase
Assay System (Promega Corporation) based on the manufacturer's
protocol. The results of the relative reporter activity were
normalized to the activity of the Renilla luciferase second
reporter (internal control), according to the manufacturer's
protocol. The experiment was conducted in triplicate.
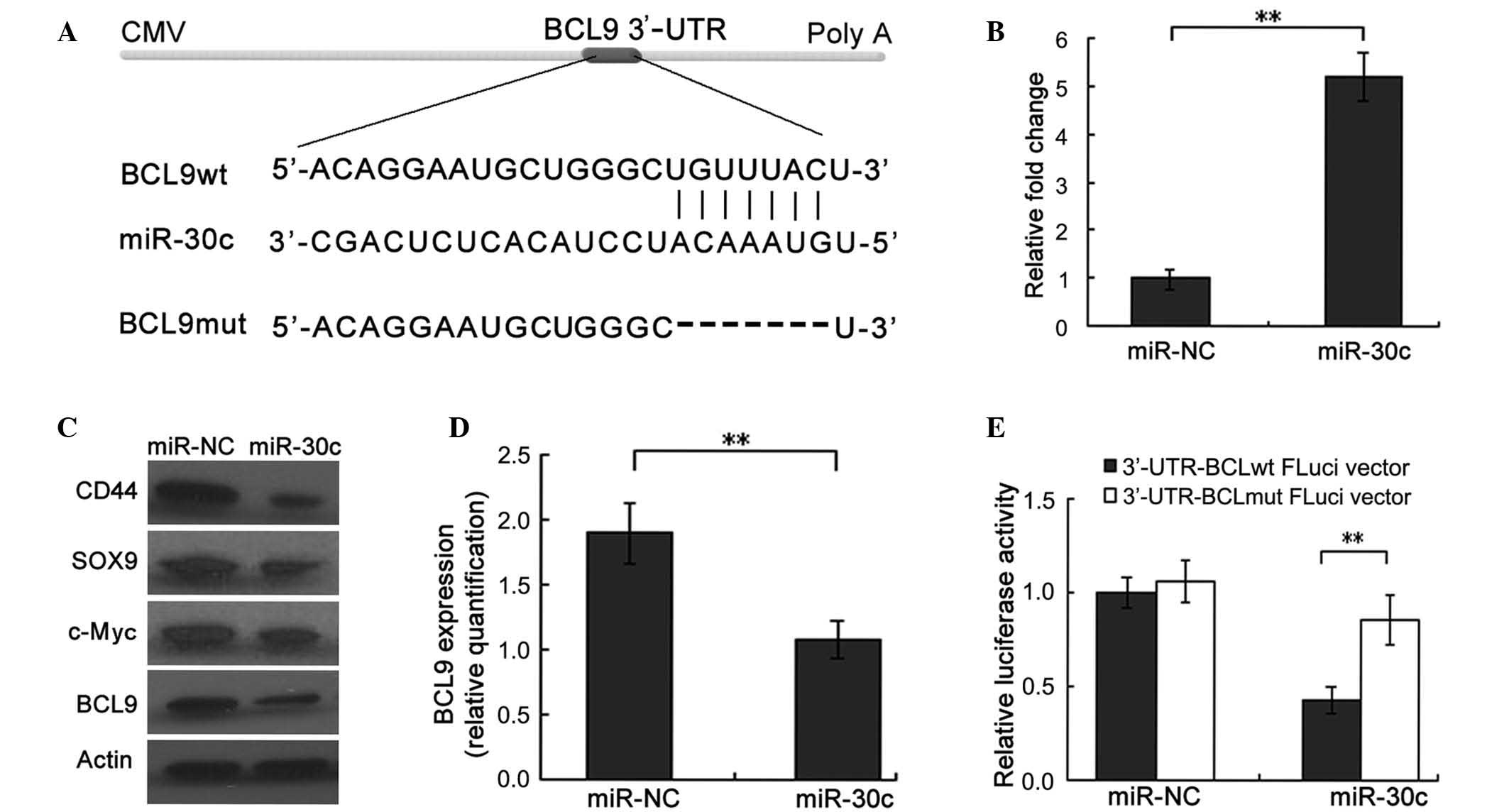 | Figure 1.BCL9 is a direct target of miR-30c in
PCa cells. (A) The sequence alignment of miR-30c, with the ‘seed’
binding sequences on the 3′-UTR region of BCL9 mRNA. (B) Reverse
transcription-quantitative polymerase chain reaction verification
of induced ectopic expression of miR-30c in DU145 cells following
transduction of miR-30c or miR-NC (negative control). (C and D)
Western blot analysis confirmed that proteins of multiple target
genes of the Wnt/β-catenin pathway, including c-Myc, CD44, SOX9 and
BCL9, were substantially downregulated in miR-30c-expressing DU145
cells. β-actin was used as an internal loading control. (D)
Quantification of western blot revealed that ectopic expression of
miR-30c significantly inhibited BCL9 protein levels in DU145 cells.
(E) Luciferase activity was detected following transfection of
FLuci vector (3-UTR-BCL9wt FLuci or 3-UTR-BCL9mut FLuci vectors)
into miR-30c- or miR-NC-transfected DU145 cells. **P<0.01. CMV,
cytomegalovirus; BCL9, B-cell lymphoma 9; miR, microRNA; PCa,
prostate cancer; 3-UTR, 3-untranslated region; wt, wild type; mut,
mutated. |
Immunohistochemical analysis
BCL9 expression was detected by immunohistochemistry
assays performed on formalin-fixed, paraffin-embedded slides of PCa
and BPH tissues. The tissues were cut into 5 µm-thick sections.
Using a Dako EnVision system (Dako Diagnostics AG, Zug,
Switzerland), the slides were deparaffinized with xylene and
rehydrated for further hematoxylin and eosin and
immunohistochemical staining. Following proteolytic digestion
(Trypsin Enzymatic Antigen Retrieval Solution; catalog no., ab970;
Abcam) and peroxidase blocking with hydrogen peroxide blocking
reagent (catalog no., ab94666; Abcam) of tissue slides, the slides
were incubated overnight at 4°C with the primary antibody against
BCL9 protein (ab37305; Abcam) at a dilution of 1:150. After
washing, the staining was visualized with a peroxidase-labeled
polymer (EnVision; Dako, Glostrup, Denamark) and DAB
substrate-chromogen system (Dako) using an Olympus AX70 microscope
(Olympus Corporation, Tokyo, Japan).
The stained slides were scored independently by two
experienced pathologists in a blinded manner. If any discrepant
scores were generated, the pathologists simultaneously re-examined
the slide to achieve a consensus score. The percentages of
positively staining cells exhibiting immunoreactivity in the cell
nucleus and cytoplasm in 10 representative microscopic fields were
calculated and a score of 0–4 was assigned, as follows: 0, 0%; 1,
1–25%; 2, 26–50%; 3, 51–75%; or 4, 76–100%. Meanwhile, the staining
intensity of the cells was calculated and scored as follows: 0, no
staining; 1, weakly positive; 2, moderately positive; or 3,
strongly positive. The sum of the two scores was calculated to
determine a final staining score. Tumor specimens with an overall
score of ≥3 were considered to be positive.
miRNA reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
This assay was conducted as previously described
(8). miRNA was extracted from 98 PCa
frozen tissues and transfected DU145 cells using an miRNA Fast
Extraction Kit (BioTeke Corporation, Beijing, China). The primers
specific for miR-30c and the internal control, RNU6B, were
purchased from Thermo Fisher Scientific, Inc. The primer sequences
were as follows: Forward, 5′-TGTGTTTTTATTGTTTTTGTTGTCCCA-3′ and
reverse, 5′-GGGACAGAACAGGTTAATGGGAA-3′ for miR-30c; and forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′ for
RNU6B. cDNA synthesis and amplification were performed according to
the instructions of the All-in-One™ miRNA qRT-PCR detection kit
(GeneCopoeia, Inc., Guangzhou, China) with the specific primers.
PCR was performed using the MyiQ2 Two-Color Real-Time PCR Detection
System (Bio-Rad Laboratories, Inc.) under the following conditions:
Initial denaturation step at 95°C for 5 min, followed by 40 cycles
at 95°C for 10 seconds and 60°C for 20 sec, with a final extension
step at 72°C for 20 sec. All assays were conducted in triplicate.
Relative quantification of miR-30c was performed using IQ5 Standard
Edition Optical System version 2.0 software (Bio-Rad Laboratories,
Inc.) and the comparative quantification cycle (Cq) method
(24), and normalized to the
expression of RNU6B. For the analysis of the possible correlations
between miR-30c expression levels and BCR, the 98 PCa patients were
divided into a negative and a positive expression group according
to median relative expression of miR-30c.
Statistical analysis
All statistical analyses were performed using SPSS
version 17.0 for Windows (SPSS, Inc., Chicago, IL, USA). Continuous
variables were compared using a Students t-test. The associations
between miR-30c and BCL9 expression were assessed by Spearman's
rank correlation coefficient. Associations between BCL9 and
clinicopathological variables were evaluated by Fisher's exact or
Pearsons χ2 tests. Kaplan-Meier survival curves were
generated to evaluate the effect of miR-30c and BCL9 expression
levels on survival rate. A Cox proportional hazards regression
model was used to establish independent factors associated with
BCR. P<0.05 was considered to indicate a statistically
significant difference.
Results
BCL9 is a direct target of miR-30c in
PCa cells
The present study first investigated whether BCL9 is
modulated by miR-30c in PCa cells. DU145 cells were transiently
transfected with miR-30c vector or blank vector (miR-NC).
Subsequent RT-qPCR analysis confirmed that the miR-30c expression
level was significantly increased in cells transfected with miR-30c
compared with those transfected with miR-NC (P<0.001) (Fig. 1B). The cells were then used for
further assays. Western blot analysis revealed that ectopic
expression of miR-30c was associated with a significant reduction
in the expression of BCL9 protein (P=0.007; Fig. 1C and D). Consistent with the role of
BCL9 as a transcriptional coactivator of the Wnt signaling pathway,
proteins of multiple target genes of the Wnt/β-catenin pathway,
including c-Myc, CD44 and SOX9, were observed to be substantially
downregulated in miR-30c-transfected DU145 cells compared with
their expression in miR-NC-transfected cells (all P<0.001;
Fig. 1C) (17,25).
To further determine whether miR-30c may interact
directly with BCL9 protein, a luciferase reporter assay was
conducted. Using the databases TargetScan (http://targetscan.org/), PicTar (http://pictar.mdc-berlin.de/) and miRDB (http://www.mirdb.org/miRDB/), the 3′-UTR of BCL9
containing a sequence motif matching with the ‘seed’ sequence of
miR-30c was identified (Fig. 1A). The
wild-type and mutant BCL9 3′-UTR reporter vectors (Fig. 1A) were cotransfected into DU145 cells,
together with miR-30c or miR-NC. The luciferase activity of
wild-type, but not mutant, BCL9 3′-UTR reporters was significantly
downregulated when cotransfected with miR-30c compared with that of
miR-NC-transfected cells, confirming that BCL9 is a direct target
of miR-30c (P=0.004; Fig. 1E).
BCL9 is upregulated in PCa tissues,
and its expression is inversely correlated with miR-30c
expression
Immunohistochemical analyses were conducted on
tissue samples from a cohort comprising 98 PCa and 20 BPH cases,
using an antibody specific for BCL9. Staining was predominantly
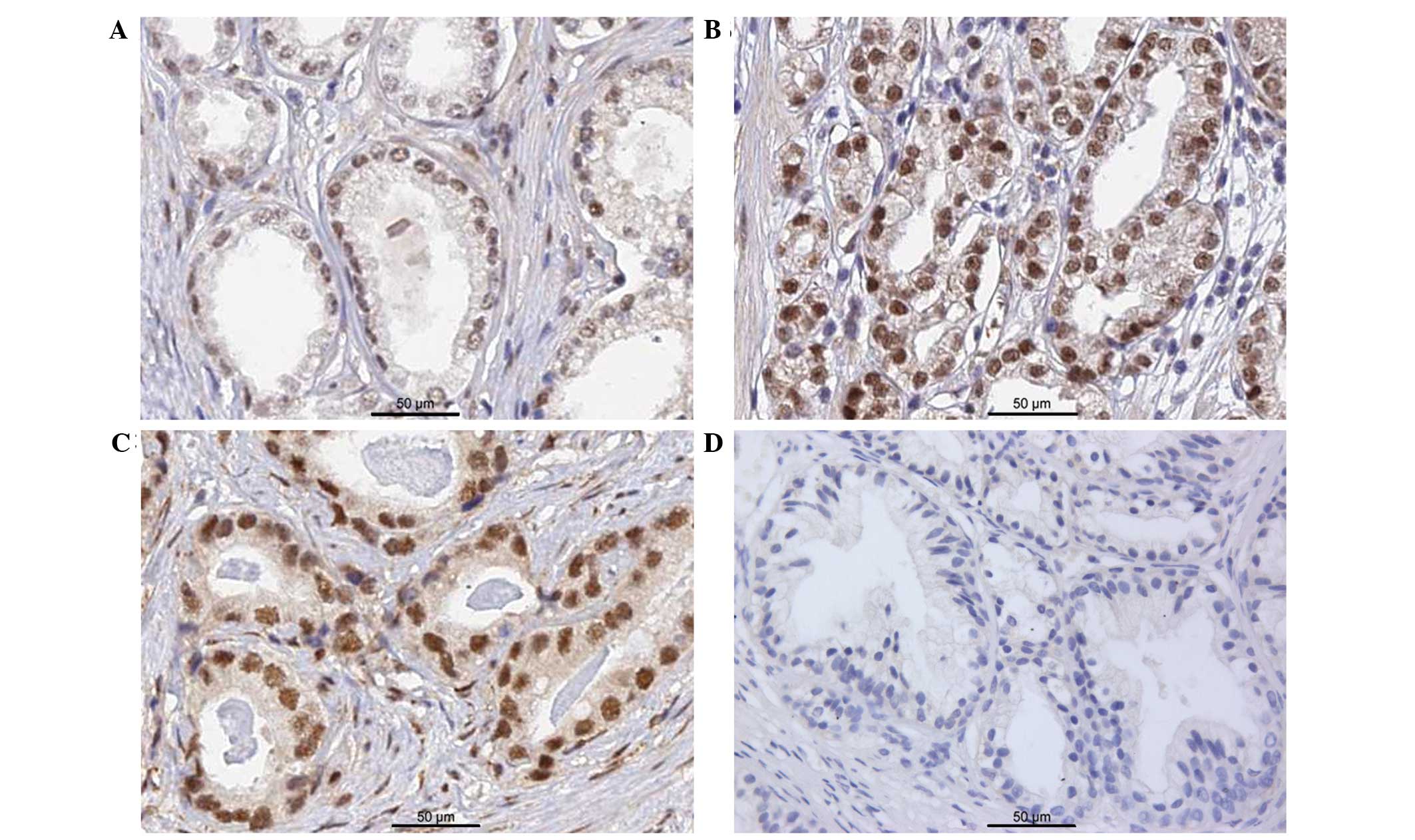
distributed in the nuclei of PCa cells (Fig. 2A–C). In some sections of BPH tissues,
very faint BCL9 staining was observed when compared to PCa tissues,
and was considered in the negative range (Fig. 2D). The positive expression rate of
BCL9 in the tissues from PCa patients (52/98; 53.1%) was
significantly higher than that in normal prostate tissues (5/20;
25.0%) (P=0.022).
To further investigate the association between BCL9
expression and miR-30c levels, RT-qPCR was conducted to quantify
the expression of miR-30c in the samples from the 98 PCa patients.
A non-parametric Spearman's rank correlation coefficient
(rS) analysis was then conducted using the cases with
both miR-30c and BCL9 protein quantification data. The results
indicated that BCL9 and miR-30c expression across these cases was
significantly inversely correlated (rS=0.38;
P<0.001).
BCL9 is associated with
clinicopathological features
The possible correlation between BCL9 and
clinicopathological features was further investigated in 98 PCa
patients. As shown in Table II,
increased expression levels of BCL9 were frequently observed in PCa
patients with a high Gleason score (P=0.016) and BCR (P=0.020).
However, a statistically significant correlation was not identified
between BCL9 expression and other features, including preoperative
PSA levels (P=0.326), pathological stage (P=0.073) and surgical
margin (P=0.111).
 | Table II.BCL9 expression and
clinicopathological features. |
Table II.
BCL9 expression and
clinicopathological features.
|
|
| BCL9 expression, n
(%) |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | No. of
patients | Negative | Positive | P-value |
|---|
| Preoperative PSA
(ng/ml) |
|
|
| 0.326 |
|
<10 | 79 | 39 (49.4) | 40 (50.6) |
|
|
≥10 | 19 | 7
(36.8) | 12 (63.2) |
|
| Gleason score |
|
|
| 0.016 |
| ≤6 | 33 | 22 (66.7) | 11 (33.3) |
|
| 7 | 54 | 21 (38.9) | 33 (61.1) |
|
| ≥8 | 11 | 3
(27.3) | 8
(72.7) |
|
| Pathological tumor
stage |
|
|
| 0.073 |
|
pT2 | 68 | 36 (52.9) | 32 (47.1) |
|
|
pT3-pT4 | 30 | 10 (33.3) | 20 (66.7) |
|
| Surgical margin
status |
|
|
| 0.111 |
|
Negative | 81 | 41 (50.6) | 40 (49.4) |
|
|
Positive | 17 | 5
(29.4) | 12 (70.6) |
|
| Biochemical
recurrence |
|
|
| 0.020 |
|
Negative | 80 | 42 (52.5) | 38 (47.5) |
|
|
Positive | 18 | 4
(22.2) | 14 (77.8) |
|
Coexpression status of miR-30c and
BCL9 predicts BCR
Our previous data indicated the involvement of
miR-30c in PCa progression and suggested its potential role as an
independent predictor of BCR in PCa (8). In the present study, the combined
utility of these two biomarkers was investigated with regard to
predicting BCR in patients who had undergone radical prostatectomy.
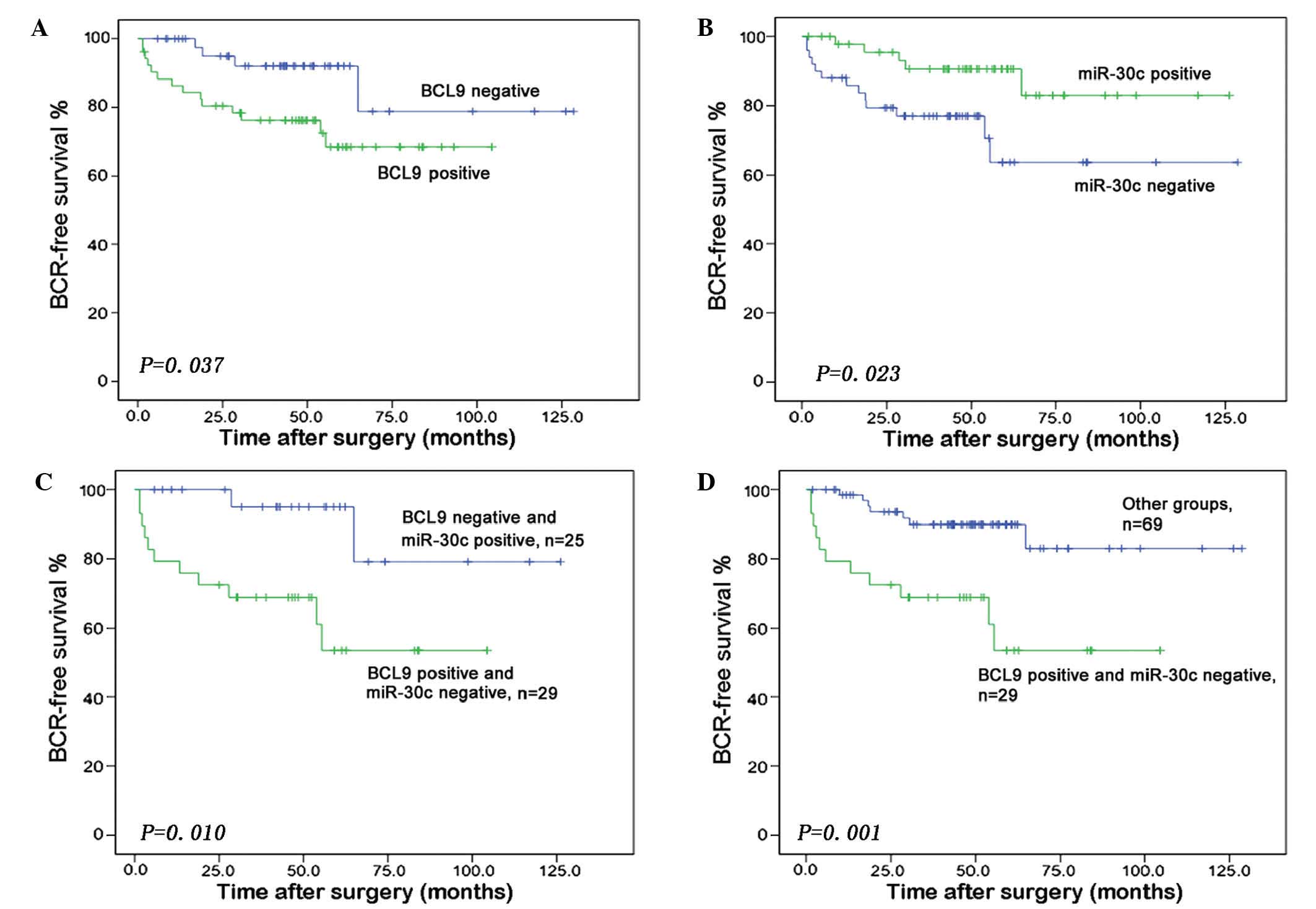
A Kaplan-Meier analysis revealed that increased BCL9 expression
(P=0.037) and reduced miR-30c expression (P=0.023) were significant
predictors of shorter BCR-free survival time (Fig. 3A and B). Furthermore, the group of PCa
patients with miR-30c-negative and BCL9-positive expression had a
significantly lower BCR-free survival relative to patients with a
miR-30c-positive and BCL9-negative combined expression status
(P=0.010) (Fig. 3C) or to those with
any other combined expression status (P=0.001) (Fig. 3D).
When a univariate Cox proportional hazards
regression model was applied, the pathological stage (P=0.001),
Gleason score (P<0.001), PSA level (P=0.001) and surgical margin
(P=0.003) were identified to be significant predictors of BCR.
Furthermore, a high hazard ratio (HR) for BCR was observed in
patients with miR-30c-negative and BCL9-positive expression (HR,
5.79; 95% confidence interval, 1.28–26.19; P=0.023) (Table III). On multivariate analysis,
miR-30c-negative and BCL9-positive expression (HR, 5.08; P=0.048)
and a higher Gleason score (HR, 6.89; P=0.006) were revealed to be
independent prognostic factors for poor BCR-free survival (Table III).
 | Table III.Univariate and multivariate analyses
with Cox proportional hazards regression model for biochemical
recurrence-free survival. |
Table III.
Univariate and multivariate analyses
with Cox proportional hazards regression model for biochemical
recurrence-free survival.
| Variables | HR (95% CI) | P-value |
|---|
| Univariate |
|
|
|
miR-30c/BCL9 status | 5.79
(1.28–26.19) |
0.023 |
| Gleason
score | 3.39
(2.10–5.47) | <0.001 |
|
Preoperative PSA | 1.01
(1.01–1.04) |
0.001 |
|
Pathological tumor stage | 4.78
(1.84–12.37) |
0.001 |
|
Surgical margin status | 4.14
(1.63–10.53) |
0.003 |
| Multivariate |
|
|
|
miR-30c/BCL9 status | 5.08
(1.02–25.39) |
0.048 |
| Gleason
score | 6.89
(1.75–27.13) |
0.006 |
|
Preoperative PSA | 0.97
(0.89–1.05) |
0.449 |
|
Pathological tumor stage | 1.29
(0.20–5.79) |
0.744 |
|
Surgical margin status | 2.10
(0.60–7.31) |
0.246 |
Discussion
Our previous study demonstrated that miR-30c serves
as a tumor suppressor, and its aberrant expression is associated
with PCa progression (8). However,
the underlying mechanisms of the tumor-suppressive role of miR-30c
have not been well described, particularly its role in the
Wnt/β-catenin pathway. The canonical Wnt/β-catenin pathway
constitutes a receptor-mediated signal transduction network during
normal embryonic development and adult tissue homeostasis, and
plays a critical role in multiple types of human cancer, including
PCa (26). Although the Wnt/β-catenin
pathway is constitutively activated in PCa, mutations that have
frequently been shown to activate the pathway in other malignancies
(e.g., β-catenin mutation or adenomatous polyposis coli truncation
in colon cancers) have rarely been observed in PCa (27,28).
Therefore, further investigation of the Wnt/β-catenin signaling
components is merited.
Earlier studies revealed that dysregulation of BCL9
expression is an oncogenic mechanism of Wnt pathway activation
(29). BCL9 serves fundamental roles
in tumor progression by increasing tumor load, metastasis,
angiogenesis and invasion through regulation of Wnt target genes
(30,31). The current investigation identified
significant overexpression of BCL9 in PCa tissues compared with
that of BPH tissues. This finding is consistent with a number of
previous studies that have reported elevated levels of BCL9 in
other cancer types (17,32). Further investigation in the present
study indicated that increased expression of BCL9 was associated
with pathological predictors of PCa aggressiveness. Furthermore,
Kaplan-Meier survival curves indicated that BCL9 expression was
closely associated with BCR in PCa patients; those with high levels
of BCL9 and low levels of miR-30c experienced earlier BCR following
radical prostatectomy. Meanwhile, the Cox proportional hazards
regression model revealed that BCL9 and miR-30c coexpression could
serve as an independent predictor of BCR-free survival in PCa
patients. These findings support the notion that BCL9 may have
potential as a biomarker for PCa, and that therapeutic approaches
targeting aberrant levels of BCL9 should be explored as a potential
approach to improve clinical outcomes.
The current study identified a new signaling pathway
in PCa connecting miR-30c to BCL9/Wnt/β-catenin transcriptional
activity. The connection is achieved by the silencing of the
miR-30c locus, which targets BCL9 at its 3′-UTR. These results are
in agreement with those of previous studies documenting a link
between miR-30c (also denoted as miR-30c-5p) and BCL9 in other
cancer types, including ovarian carcinoma and multiple myeloma
(19,20). More importantly, the current analysis
of expression patterns in PCa patient samples indicated that
expression levels of miR-30c and BCL9 are inversely correlated,
corroborating the physiological significance of a miR-30c-mediated
BCL9 inhibitory mechanism in PCa biology. The other well-known
functions of miR-30c include its ability to regulate the expression
of several targets. For example, a previous study found that
miR-30c directly targeted and downregulated Rab18 expression and
inhibited the proliferation of non-small cell lung cancer cells
(33). miR-30c also regulates
invasion of breast cancer by targeting the cytoskeletal network
genes encoding twinfilin 1 and vimentin, important for the EMT
(34). Additional studies are likely
to further highlight the association between miR-30c and these
identified targets in PCa.
In conclusion, the present study demonstrated that
the tumor suppressor miR-30c is involved in PCa pathogenesis,
possibly by targeting BCL9, which is a known transcriptional
coactivator of the Wnt/β-catenin signaling pathway. Furthermore,
the coexpression status of BCL9 and miR-30c is associated with PCa
progression. Further studies are required to validate the clinical
utility of BCL9 and miR-30c as prognostic biomarkers in large and
multicenter PCa patient cohorts.
Acknowledgements
The present study was supported by grants from the
Key Projects of Huizhou Municipal Central People's Hospital [no.
(2014)#115], the National Natural Science Foundation of China (nos.
81170699, 81272813, 81200550), the Science and Technology Project
of Guangdong (no. 2014A020212035) and the Medical Research Fund of
Guangdong (no. A2012489).
References
|
1
|
Kakehi Y: Watchful waiting as a treatment
option for localized prostate cancer in the PSA era. Jpn J Clin
Oncol. 33:1–5. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heng HH, Bremer SW, Stevens JB, Ye KJ, Liu
G and Ye CJ: Genetic and epigenetic heterogeneity in cancer: A
genome-centric perspective. J Cell Physiol. 220:538–547. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu Q, Song R, Ortogero N, Zheng H, Evanoff
R, Small CL, Griswold MD, Namekawa SH, Royo H, Turner JM, et al:
The RNase III enzyme DROSHA is essential for microRNA production
and spermatogenesis. J Biol Chem. 287:25173–25190. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Plaisier CL, Pan M and Baliga NS: A
miRNA-regulatory network explains how dysregulated miRNAs perturb
oncogenic processes across diverse cancers. Genome Res.
22:2302–2314. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kent OA and Mendell JT: A small piece in
the cancer puzzle: microRNAs as tumor suppressors and oncogenes.
Oncogene. 25:6188–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin
ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al: Global analysis of the
differentially expressed miRNAs of prostate cancer in Chinese
patients. BMC Genomics. 14:7572013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rane JK, Scaravilli M, Ylipää A, Pellacani
D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T and
Maitland NJ: MicroRNA Expression Profile of Primary Prostate Cancer
Stem Cells as a Source of Biomarkers and Therapeutic Targets. Eur
Urol. 67:7–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ling XH, Han ZD, Xia D, He HC, Jiang FN,
Lin ZY, Fu X, Deng YH, Dai QS, Cai C, et al: MicroRNA-30c serves as
an independent biochemical recurrence predictor and potential tumor
suppressor for prostate cancer. Mol Biol Rep. 41:2779–2788. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu F, Deng H, Yao H, Liu Q, Su F and Song
E: Mir-30 reduction maintains self-renewal and inhibits apoptosis
in breast tumor-initiating cells. Oncogene. 29:4194–4204. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martinez I, Cazalla D, Almstead LL, Steitz
JA and DiMaio D: miR-29 and miR-30 regulate B-Myb expression during
cellular senescence. Proc Natl Acad Sci USA. 108:522–527. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhong Z, Xia Y, Wang P, Liu B and Chen Y:
Low expression of microRNA-30c promotes invasion by inducing
epithelial mesenchymal transition in non-small cell lung cancer.
Mol Med Rep. 10:2575–2579. 2014.PubMed/NCBI
|
|
12
|
Yun SI, Kim HH, Yoon JH, Park WS, Hahn MJ,
Kim HC, Chung CH and Kim KK: Ubiquitin specific protease 4
positively regulates the WNT/β-catenin signaling in colorectal
cancer. Mol Oncol. 9:1834–1851. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carotenuto M, De Antonellis P, Liguori L,
Benvenuto G, Magliulo D, Alonzi A, Turino C, Attanasio C, Damiani
V, Bello AM, et al: H-Prune through GSK-3β interaction sustains
canonical WNT/β-catenin signaling enhancing cancer progression in
NSCLC. Oncotarget. 5:5736–5749. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kramps T, Peter O, Brunner E, Nellen D,
Froesch B, Chatterjee S, Murone M, Züllig S and Basler K:
Wnt/wingless signaling requires BCL9/legless-mediated recruitment
of pygopus to the nuclear beta-catenin-TCF complex. Cell.
109:47–60. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Townsley FM, Cliffe A and Bienz M: Pygopus
and Legless target Armadillo/beta-catenin to the nucleus to enable
its transcriptional co-activator function. Nat Cell Biol.
6:626–633. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Willis TG, Zalcberg IR, Coignet LJ,
Wlodarska I, Stul M, Jadayel DM, Bastard C, Treleaven JG, Catovsky
D, Silva ML, et al: Molecular cloning of translocation
t(1;14)(q21;q32) defines a novel gene (BCL9) at chromosome 1q21.
Blood. 91:1873–1881. 1998.PubMed/NCBI
|
|
17
|
Mani M, Carrasco DE, Zhang Y, Takada K,
Gatt ME, Dutta-Simmons J, Ikeda H, Diaz-Griffero F, Pena-Cruz V,
Bertagnolli M, et al: BCL9 promotes tumor progression by conferring
enhanced proliferative, metastatic, and angiogenic properties to
cancer cells. Cancer Res. 69:7577–7586. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deka J, Wiedemann N, Anderle P,
Murphy-Seiler F, Bultinck J, Eyckerman S, Stehle JC, André S,
Vilain N, Zilian O, et al: Bcl9/Bcl9l are critical for Wnt-mediated
regulation of stem cell traits in colon epithelium and
adenocarcinomas. Cancer Res. 70:6619–6628. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao JJ, Lin J, Zhu D, Wang X, Brooks D,
Chen M, Chu ZB, Takada K, Ciccarelli B, Admin S, et al: miR-30-5p
functions as a tumor suppressor and novel therapeutic tool by
targeting the oncogenic Wnt/β-catenin/BCL9 pathway. Cancer Res.
74:1801–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jia W, Eneh JO, Ratnaparkhe S, Altman MK
and Murph MM: MicroRNA-30c-2* expressed in ovarian cancer cells
suppresses growth factor-induced cellular proliferation and
downregulates the oncogene BCL9. Mol Cancer Res. 9:1732–1745. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
World Health Organization. HO (2010)
International statistical classification of diseases and related
health problems. 10th revision. 2:2010.http://www.who.int/classifications/icd/ICD10Volume2_en_2010.pdfAccessed.
August 10–2012
|
|
22
|
International Union Against Cancer (UICC):
Urological Tumors, Prostate. TNM Classification of Malignant
Tumours (6th). Sobin LH and Wittekind Ch: (New York, NY).
Wiley-Liss. 184–187. 2002.
|
|
23
|
Montironi R, Mazzuccheli R, Scarpelli M,
Lopez-Beltran A, Fellegara G and Algaba F: Gleason grading of
prostate cancer in needle biopsies or radical prostatectomy
specimens: Contemporary approach, current clinical significance and
sources of pathology discrepancies. BJU Int. 95:1146–1152. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ,
Lee KH, Yeh SD, Hong TM and Chen YL: MicroRNA-320 suppresses the
stem cell-like characteristics of prostate cancer cells by
downregulating the Wnt/beta-catenin signaling pathway.
Carcinogenesis. 34:530–538. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sun Y, Campisi J, Higano C, Beer TM,
Porter P, Coleman I, True L and Nelson PS: Treatment-induced damage
to the tumor microenvironment promotes prostate cancer therapy
resistance through WNT16B. Nat Med. 18:1359–1368. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Femia AP, Dolara P, Giannini A, Salvadori
M, Biggeri A and Caderni G: Frequent mutation of Apc gene in rat
colon tumors and mucin-depleted foci, preneoplastic lesions in
experimental colon carcinogenesis. Cancer Res. 67:445–449. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yardy GW, Bicknell DC, Wilding JL,
Bartlett S, Liu Y, Winney B, Turner GD, Brewster SF and Bodmer WF:
Mutations in the AXIN1 gene in advanced prostate cancer. Eur Urol.
56:486–494. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
de la Roche M, Worm J and Bienz M: The
function of BCL9 in Wnt/beta-catenin signaling and colorectal
cancer cells. BMC Cancer. 8:1992008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Adachi S, Jigami T, Yasui T, Nakano T,
Ohwada S, Omori Y, Sugano S, Ohkawara B, Shibuya H, Nakamura T, et
al: Role of a BCL9-related beta-catenin-binding protein, B9L, in
tumorigenesis induced by aberrant activation of Wnt signaling.
Cancer Res. 64:8496–8501. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mieszczanek J, de la Roche M and Bienz M:
A role of Pygopus as an anti-repressor in facilitating
Wnt-dependent transcription. Proc Natl Acad Sci USA.
105:19324–19329. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hyeon J, Ahn S, Lee JJ, Song DH and Park
CK: Prognostic Significance of BCL9 Expression in Hepatocellular
Carcinoma. Korean J Pathol. 47:130–136. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhong K, Chen K, Han L and Li B:
MicroRNA-30b/c inhibits non-small cell lung cancer cell
proliferation by targeting Rab18. BMC Cancer. 14:7032014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bockhorn J, Yee K, Chang YF, Prat A, Huo
D, Nwachukwu C, Dalton R, Huang S, Swanson KE, Perou CM, et al:
MicroRNA-30c targets cytoskeleton genes involved in breast cancer
cell invasion. Breast Cancer Res Treat. 137:373–382. 2013.
View Article : Google Scholar : PubMed/NCBI
|