Introduction
Cancer of the head and neck (HN), primarily squamous
cell carcinoma (SCC) of the oral cavity, and cancer of the pharynx
and larynx account for 6% of all malignancies (1). In the case of pharyngolaryngeal cancer,
radiotherapy and chemotherapy are currently accepted as an
alternative approach to surgery for patients with advanced HNSCC,
since it enables organ preservation without compromising patient
survival (2). However, the response
to chemotherapy and radiotherapy is heterogeneous, and a large
proportion of patients relapse, either locally or at distant sites,
resulting in a 5-year survival rate of 50% (1,2).
Chemotherapy and radiotherapy share common downstream effectors,
namely reactive oxygen species (ROS) (3). Although ROS toxicity for tumor cells is
well established, the activation of the oxidative stress pathway
also favors the development and spreading of certain tumors; thus,
oxidative stress exhibits a Janus-head effect in terms of cancer
progression (3,4).
In normal cells, glutathione (GSH) is one of the
main ROS scavenging molecules, and is important in the cellular
response to oxidation (4). GSH is
synthesized following a two-step reaction, by coupling three amino
acids, namely, cysteine, glutamine and glycine (5). Under normal conditions, the levels of
GSH depend on the efficiency of the first step of the synthesis
reaction, which is performed by the enzyme glutamate-cysteine
ligase (GCL) (5). GCL is composed of
two subunits, namely the catalytic (C) subunit and the modulator
(M) subunit (5). GCL activity only
requires the GCLC subunit, but it is strongly induced by the GCLM
subunit (6). These two GCL subunits
exhibit different pattern of expression within tissues, which
suggests an independent control of their expression (7). Notably, although only the expression of
GCLC is altered upon stimulation with hormones or drugs, the
expression of both subunits is induced following exposure of cells
to oxidative stress (8,9). The promoters of GCLC and GCLM harbor
binding sites for three transcription factors that have been
associated with the induction of the oxidative stress response
machinery (10–12). These transcription factors are nuclear
factor erythroid 2-related factor 2 (NRF2), nuclear factor (NF)-κB
and activator protein-1 (AP-1) (13).
Previous functional assays have reported the regulation of the
transcription of the GCL subunits genes by the transcription
factors NRF2 and AP-1 and by members of the NF-κB signaling pathway
(14,15). The NRF2 signaling pathway is a
prominent regulator of the cellular response to oxidative stress
(16). In the absence of oxidative
stress, Kelch-like erythroid cell-derived protein with cap'n'collar
homology-associated protein 1 (KEAP1) recruits NRF2, and the
KEAP1/NRF2 complex is then targeted to the proteasome (16). Oxidation of cysteine residues in KEAP1
prevents the formation of the complex (13). Upon stabilization of the complex, NRF2
is translocated to the nucleus, where it triggers the transcription
of the genes of phase II detoxifying enzymes, including the
aforementioned GCL subunits and heme oxygenase-1 (HO-1) (14,17).
Considering the role of GSH in ROS detoxification,
the present and other authors have previously attempted the
quantification of GSH within tumors, compared with normal tissues
(18,19). In agreement with previous studies
reporting the accumulation of GSH within various tumors, the
present authors have recently reported a higher ratio of reduced
vs. oxidized GSH in HN tumors, compared with the adjacent mucosa
(19). The aim of the present study
was to evaluate the expression of GCL, the rate-limiting enzyme of
GSH synthesis, in carcinoma tissues, compared with adjacent mucosa.
For that purpose, the messenger (m)RNA and protein expression
levels of the two GCL subunits and the mRNA levels of their
regulators were measured in biopsies of HN tumors that had not been
treated with radiotherapy or chemotherapy, in order to avoid any
potential interference with oxidative stress that may have been
induced by these therapies.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of André Vésale Hospital (Intermunicipal Public Health of
the Charleroi registration number OM008; Montigny-le-Tilleul,
Belgium) under Compliance Certification Board number B32520107991
and B325201111821.
Clinical data
Biopsy samples from carcinoma tissues and adjacent
normal tissues were collected from patients who had undergone
surgical resection of HNSCC at the André Vésale Hospital
(Montigny-le-Tilleul, Belgium) between 2011 and 2013 (Table I). Only patients who had not been
previously subjected to chemotherapy or radiotherapy were included
in the study. Cancer stages of the patients ranged from stage II to
IV (Table I), according to the
tumor-node-metastasis classification of malignant tumors (20). Patient's tumors were localized in the
oral cavity, hypopharynx and larynx, and ranged from poorly to well
differentiated (Table I).
 | Table I.Patient's clinical data. |
Table I.
Patient's clinical data.
| Gender | Age, years | Surgery date,
month/year | TNM stage | Localization | SSC
gradeb |
|---|
| M | 48 | 02/2013 | T4N2 | Larynx | III |
| M | 49 | 03/2013 | T2N0 | Mobile tongue | I |
| M | 72 | 06/2013 | T4N1 | Larynx | II |
| M | 55 | 07/2012 | T4N0 | Larynx | I |
| M | 57 | 07/2012 | T2N0 | Mobile tongue | I |
| M | 62 | 07/2012 | T2N2 | Oropharynx | I |
| M | 58 | 08/2012 | T4N0 | Larynx | I |
| M | 57 | 09/2011 | T4N2 | Mobile tongue | I |
| M | 85 | 09/2011 | T4N0 | Larynx | I |
| F | 84 | 10/2011 | T2N0 | Oropharynx | III |
| M | 66 | 11/2011 | T4N2 | Hypopharynx | III |
| M | 75 | 10/2012 | T4N0 | Mobile tongue | I |
| F | 73 | 10/2012 | T4N0 | Mobile tongue | II |
| M | 54 | 11/2012 | T4N2 | Larynx | I |
| F | 68 | 11/2012 | T2N1 | Mobile tongue | I |
| F | 63 | 12/2012 | T2N0 | Oropharynx | I |
| M | 78 | 01/2013 | T4N0 | Oropharynx | II |
| M | 50 | 01/2013 | T4N1 | Floor of the
mouth | III |
| M | 62 | 02/2013 | T2N0 | Mobile tongue | I |
| M | 58 | 05/2013 | T2N0 | Oropharynx | I |
| M | 72 | 06/2013 | T4N1 | Larynx | II |
| M | 54 | 04/2013 | T4N2 | Oropharynx | III |
| M | 58 | 04/2013 | T4N2 | Larynx | I |
| M | 59 | 09/2013 | T4N0 | Oropharynx | I |
| M | 54 | 10/2013 | T3N2 | Floor of the
mouth | I |
| M | 67 | 11/2013 | T4N0 | Larynx | I |
| M | 51 | 11/2013 | T4N2 | Hypopharynx | III |
| M | 50 | 12/2013 | T4N0 | Larynx | I |
| M | 63 | 03/2013 | T4N2 | Larynx | I |
| Fa | 75 | 07/2013 | T4N2 | Larynx | II |
| Fa | 89 | 08/2013 | T2N0 | Oropharynx | I |
| Ma | 63 | 05/2013 | T4N0 | Hypopharynx | II |
| Ma | 61 | 07/2013 | T4N1 | Larynx | I |
| Ma | 59 | 09/2013 | T4N1 | Larynx | I |
| Fa | 58 | 09/2013 | T2N0 | Oropharynx | I |
Sample collection
Fresh samples and formalin-fixed, paraffin-embedded
(FFPE) tissue sections of tumor and adjacent normal tissues were
collected from surgical resections of HNSCC.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Immediately following resection, samples for RNA
extraction were collected, frozen in liquid nitrogen and stored at
−80°C. Tissue samples were grinded with a mortar in a liquid
nitrogen bath (Bel-Art Products, Wayne, NJ, USA). RNA extraction
was performed using RNeasy Mini kit (Qiagen, Inc., Valencia, CA,
USA), according to the manufacturer's protocol, and including DNAse
treatment (Qiagen, Inc.).
RT-qPCR was performed using total RNA. Complementary
DNA was synthesized with Transcriptor Reverse Transcriptase (Roche
Diagnostics, Indianapolis, IN, USA) using oligo(dT) primers
(Qiagen, Inc.), according to the manufacturer's protocol. RT-qPCR
was conducted with the primer sets presented in Table II (Sigma-Aldrich, St. Louis, MO,
USA), using SYBR Green I Master (Roche Diagnostics), according to
the manufacturer's protocol, in a LightCycler® 480
Instrument II (Roche Diagnostics). The cycle conditions were 95°C
for 5 min, followed by 50 cycles of 95°C for 15 sec, 60°C for 30
sec and 72°C for 30 sec. Relative expression (RE) of GCLM, GCLC,
NRF2, HO-1 and nuclear factor of kappa light polypeptide gene
enhancer in B-cells inhibitor, alpha (NFKBIA) was calculated using
succinate dehydrogenase complex flavoprotein subunit A and
ribosomal protein L27 as reference genes, according to the
following formula: RE=2Cq (reference)−Cq (target)
(21). Analyses of GCLC, NRF2, HO-1
and NFKBIA expression were restricted to 21, 24, 24 and 22
patients, respectively, since certain tissues samples collected for
RNA extraction were not sutible for qPCR analysis due to RNA
degradation. A no template control and no reverse transcriptase
control were performed to exclude extraneous nucleic acid
contamination and genomic DNA contamination, respectively.
 | Table II.List of the primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table II.
List of the primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Primer
sequence |
|---|
| SDHA |
|
|
Forward |
5′-CCCGAGGTTTTCACTTCACTGT-3′ |
|
Reverse |
5′-CCAGTTGTCCTCCTCCATGTTC-3′ |
| RPL27 |
|
|
Forward |
5′-ATCGCCAAGAGATCAAAGATAA-3′ |
|
Reverse |
5′-TCTGAAGACATCCTTATTGACG-3′ |
| NRF2 |
|
|
Forward |
5′-GCAAGTTTGGGAGGAGCTATTATC-3′ |
|
Reverse |
5′-AGTTTGGCTTCTGGACTTGGA-3′ |
| GCLM |
|
|
Forward |
5′-GAAGAGAGCATCTGGAGAACTAATGA-3′ |
|
Reverse |
5′-AGTTATGACACTGTCTTGCTTGTAGTCA-3′ |
| GCLC |
|
|
Forward |
5′-TTCCTGCACATCTACCACGC-3′ |
|
Reverse |
5′-TGTATTCCACCTCATCGCCC-3′ |
| HO-1 |
|
|
Forward |
5′-GCACTCAGGCAGAGGGTGATA-3′ |
|
Reverse |
5′-CTGGAGTGTGCCCAATGCTAT-3′ |
| NFKBIA |
|
|
Forward |
5′-CAATGCTCAGGAGCCCTGTAA-3′ |
|
Reverse |
5′-TCTGTTGACATCAGCCCCAC-3′ |
Immunohistochemistry (IHC)
IHC was performed on 5-µm paraffin-embedded, 10%
formalin-fixed tissue sections from 6 patients (Table I). Tissue sections were deparaffinized
during heat-induced antigen retrieval, which was conducted in
EnVision™ Flex Target Retrieval Solution High pH (catalog no.,
K8004; Dako, Glostrup, Denmark) for 10 min at 97°C, using the PT
Link apparatus (Dako), followed by a 20-min cool down period and
wash in Tris-buffered saline (Sigma-Aldrich). All subsequent steps
were performed using the EnVision™ FLEX/HRP kit (Dako) according to
the manufacturer's protocol, which includes the diaminobenzidine
(DAB) substrate. Polyclonal rabbit anti-GCLM (dilution, 1:40;
catalog no., HPA023696; Sigma-Aldrich) was incubated overnight at
4°C with the tissue slides for GCLM detection. Monoclonal mouse
anti-MIB-1 antibody (undiluted; catalog no., IR626; Dako) was
incubated for 30 min at room temperature with the tissue slides for
Ki-67 detection. Normal and tumor tissues were identified by
trained pathologists (University Hospital Center of Charleroi,
Charleroi and Institute of Pathology and Genetics, Gosselies,
Belgium). Quantification of the signal in the different cell types
was performed using 50 images captured on a Zeiss Axioplan
microscope, using the 40X objective (Carl Zeiss AG, Oberkochen,
Germany). Signal intensity was normalized using the white balance
function of Adobe Photoshop CS2 software (Adobe Systems, Inc., San
Jose, CA, USA) and the contrast enhancer of ImageJ software
(National Institutes of Health, Bethesda, MD, USA), set at 0.1%
saturated pixels. DAB signals were extracted using ImageJ and IHC
Profiler plugin (22). Relative
intensity was calculated as the mean gray value of the regions of
interest subtracted from the maximum intensity value. The intensity
of the GCLM signals was measured from the border to the center of
each lobule using ImageJ and its dedicated macro, which is
available at https://b2share.eudat.eu/record/149. In total, 60
lobules were analyzed as described for the different cell types,
except that the signal intensity was measured within concentric
selected areas of 10-µm width from the border to the center of the
selected lobule. The same procedure was applied to the
quantification of Ki-67-labeled nuclei within 90 lobules, except
for the following modification: The background was subtracted from
the DAB signal image, and the image was converted to a binary image
using the Rényi's entropy threshold (23) prior to nuclei count with the particle
analyzer function of ImageJ.
In situ hybridization (ISH)
GCLM mRNA was detected in FFPE tissues using the ISH
kit RNAscope® 2.0 (Advanced Cell Diagnostics Inc.,
Hayward, CA, USA) and the Probe - Hs-GCLM, target, 1 (catalog no.,
411581; Advanced Cell Diagnostics Inc.), according to the
manufacturer's protocol.
Statistical analyses
Statistical analyses were performed using SigmaPlot
12 software (Systat Software, Inc., San Jose, CA, USA). RT-qPCR
data were analyzed using the Wilcoxon signed-rank test. Data
relative to IHC labeling in the different cell types were analyzed
using Kruskal-Wallis one-way analysis of variance (ANOVA) on ranks,
followed by Dunn's test as a post hoc procedure for pairwise
comparison. Statistical analysis of GCLM distribution was
restricted to 24 lobules that delivered data within 0–100 µm from
the lobule edge, while statistical analysis of Ki-67 distribution
was restricted to 34 lobules. Data were analyzed using repeated
measures ANOVA on ranks (Friedman's test), followed by Dunnett's
post hoc test vs. control.
Results
GCL mRNA levels in tumors
The mRNA expression levels of GCLM and GCLC were
evaluated in biopsy samples from carcinoma and adjacent tissues.
The mRNA expression levels of GCLM but not those of GCLC were
significantly increased in tumor samples, compared with normal
mucosa (P=0.029; Fig. 1A and B). The
role of the NRF2 and NF-κB signaling pathways in GCLM activation
was investigated in HNSCC tumors (Fig.
1C). The activation of the NRF2 signaling pathway was monitored
by measuring the mRNA levels of NRF2, which have been demonstrated
to be relevant for the activation of NRF2 in vivo (16). As the regulation of the NRF2 and NF-κB
signaling pathways involves post-translational modifications, the
expression levels of HO-1 and NFKBIA were used as a reporter of
NRF2 and NF-κB activity, respectively, since the HO-1 gene is under
direct control of the transcription factor NRF2, while the
transcription of the NF-κB inhibitor NFKBIA has been demonstrated
to be a useful marker of NF-κB activation (17,24)
(Fig. 1C). The present results
indicated that the mRNA levels of NRF2 or HO-1 were not upregulated
in the tumor samples, compared with adjacent normal mucosa
(Fig. 1D and E), suggesting that the
activity of the NRF2 pathway was not altered in the tumors.
Regarding the NF-κB pathway, both tumors and adjacent mucosa
presented similar mRNA levels of NFKBIA (Fig. 1F).
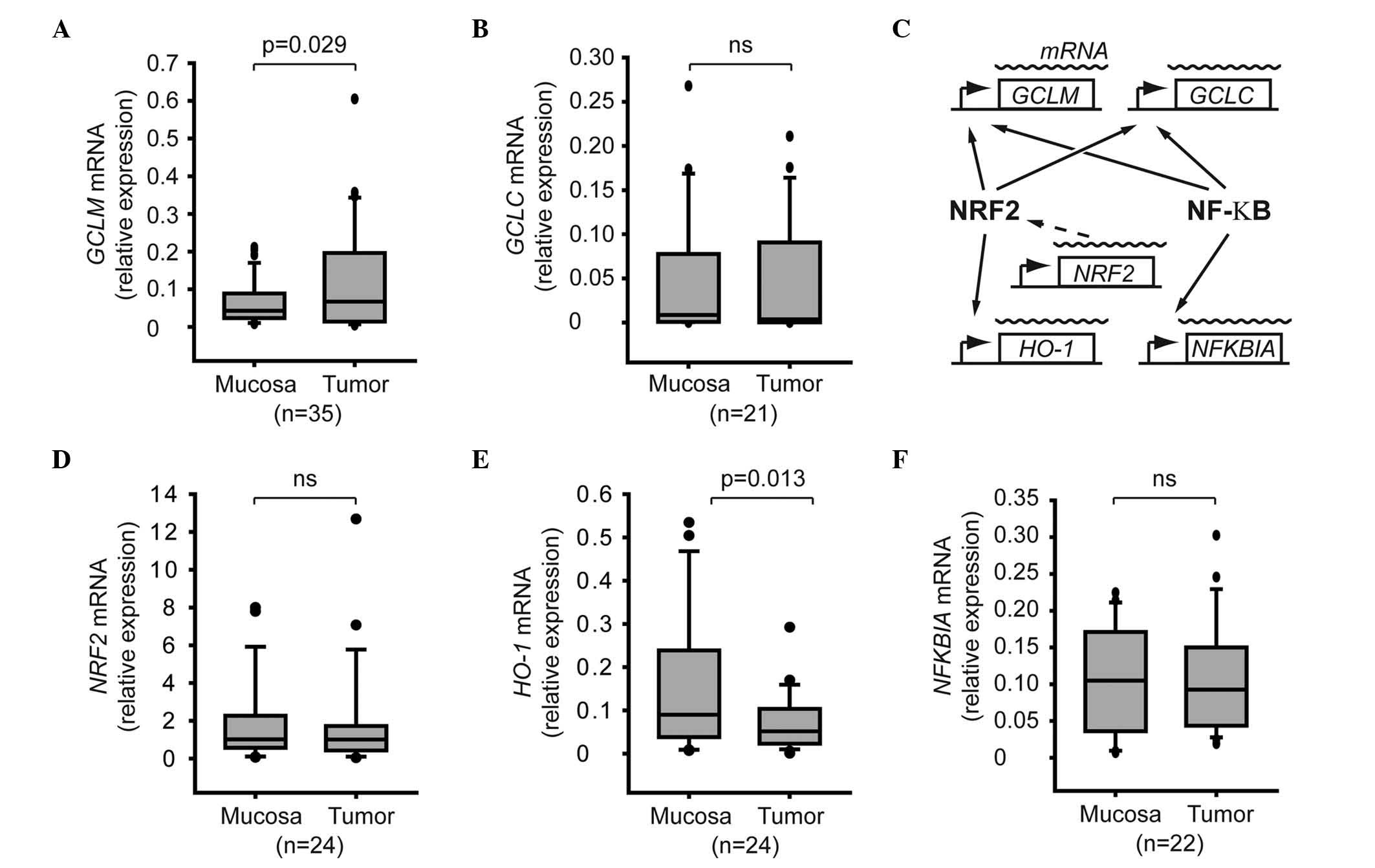 | Figure 1.Expression of GCL subunits and
regulators in tumor cells. Box plot graphs represent the relative
mRNA expression levels of (A) GCLM, (B) GCLC, (D) NRF2, (E) heme
oxygenase-1 and (F) nuclear factor of kappa light polypeptide gene
enhancer in B-cells inhibitor, alpha in normal mucosa and tumor
tissues. Total RNA was extracted from biopsy samples, and the
corresponding mRNA levels were quantified by reverse
transcription-quantitative polymerase chain reaction. The number of
patients included in each analysis is shown in brackets. (C)
Association between the genes of interest and the NRF2 and nuclear
factor-κB signaling pathways. SDHA, succinate dehydrogenase complex
flavoprotein subunit A; RPL27, ribosomal protein L27; NRF2, nuclear
factor erythroid 2-related factor 2; GCLM, glutamate-cysteine
ligase modulator subunit; GCLC, glutamate-cysteine ligase catalytic
subunit; HO-1, heme oxygenase-1; NFKBIA, nuclear factor of kappa
light polypeptide gene enhancer in B-cells inhibitor, alpha; mRNA,
messenger RNA; ns, not significant; NF-κB, nuclear factor-κB. |
GCLM localization in tumors
The identification of cell types expressing GCLM
mRNA within tumor samples was investigated at the mRNA and protein
level. For that purpose, IHC of GCLM protein expression was
performed on histological sections of tumors and adjacent mucosa.
Within the normal epithelium, labeling was restricted to basal
cells, whose cytoplasm and nucleus were both labeled, with the
nuclei consistently presenting stronger labeling than the
cytoplasms (Fig. 2A). In the case of
pre-neoplastic lesions, dysplastic cells were labeled, with the
nuclei exhibiting a stronger signal than the cytoplasms (Fig. 2B). GCLM labeling of the tumors was
heterogeneous (Fig. 2C), but
similarly to the findings in epithelial and dysplastic cells, GCLM
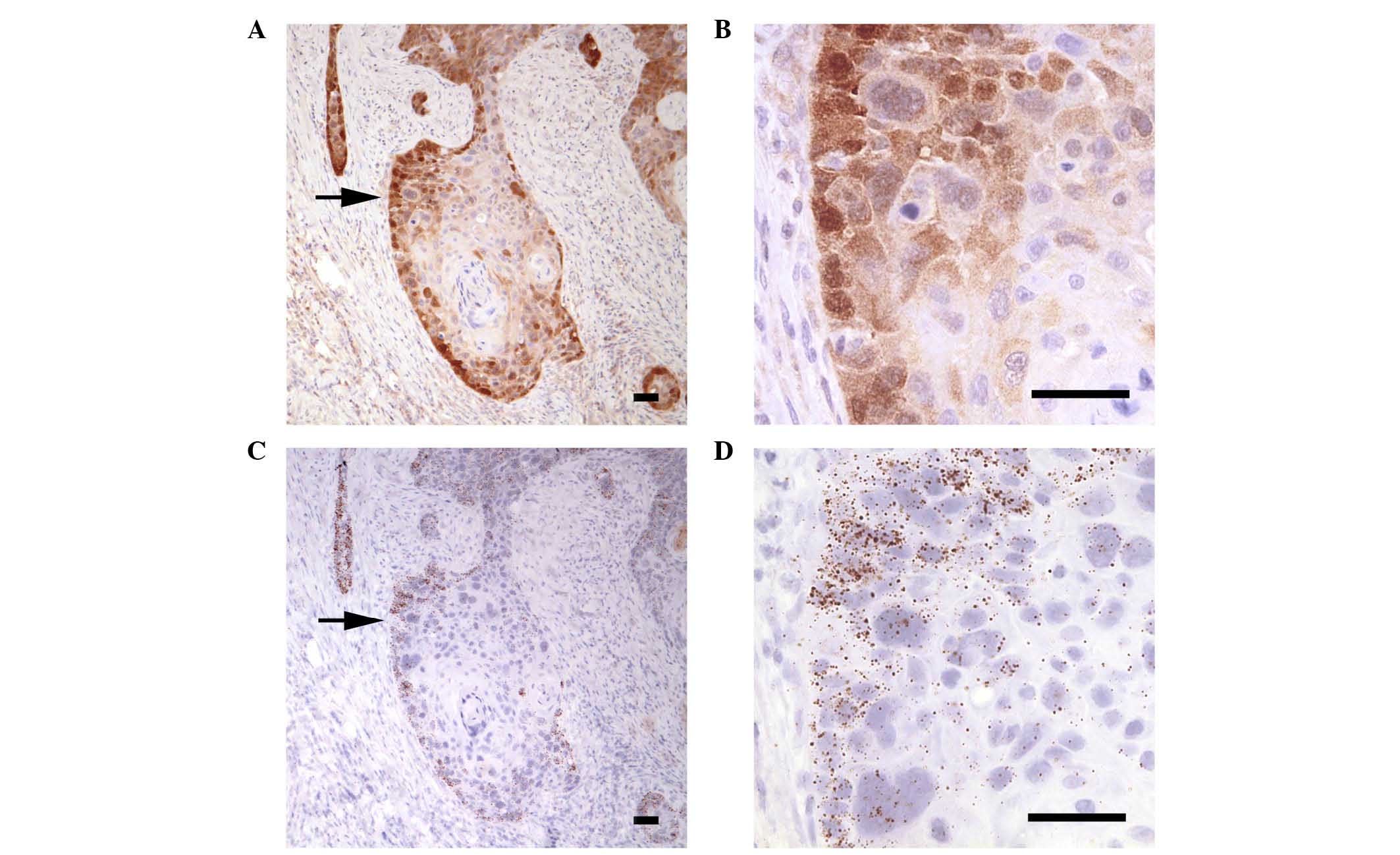
was detected in the cytoplasm and nucleus of tumor cells (Fig. 2C and D). Systematic analysis of
carcinoma lobules demonstrated that the mean GCLM labeling was
comparable in normal basal cells, dysplasia and tumor lobules
(Fig. 2E). The localization of GCLM
protein correlated with the areas where the corresponding mRNA was
detected, as indicated by the similar labeling patterns of the
protein (Fig. 3A and B) and mRNA
(Fig. 3C and D) expression in
sequential histological sections. In both cases, while the borders
of the tumor lobules were consistently labeled, the center
exhibited a range of strong to very weak protein and mRNA signals
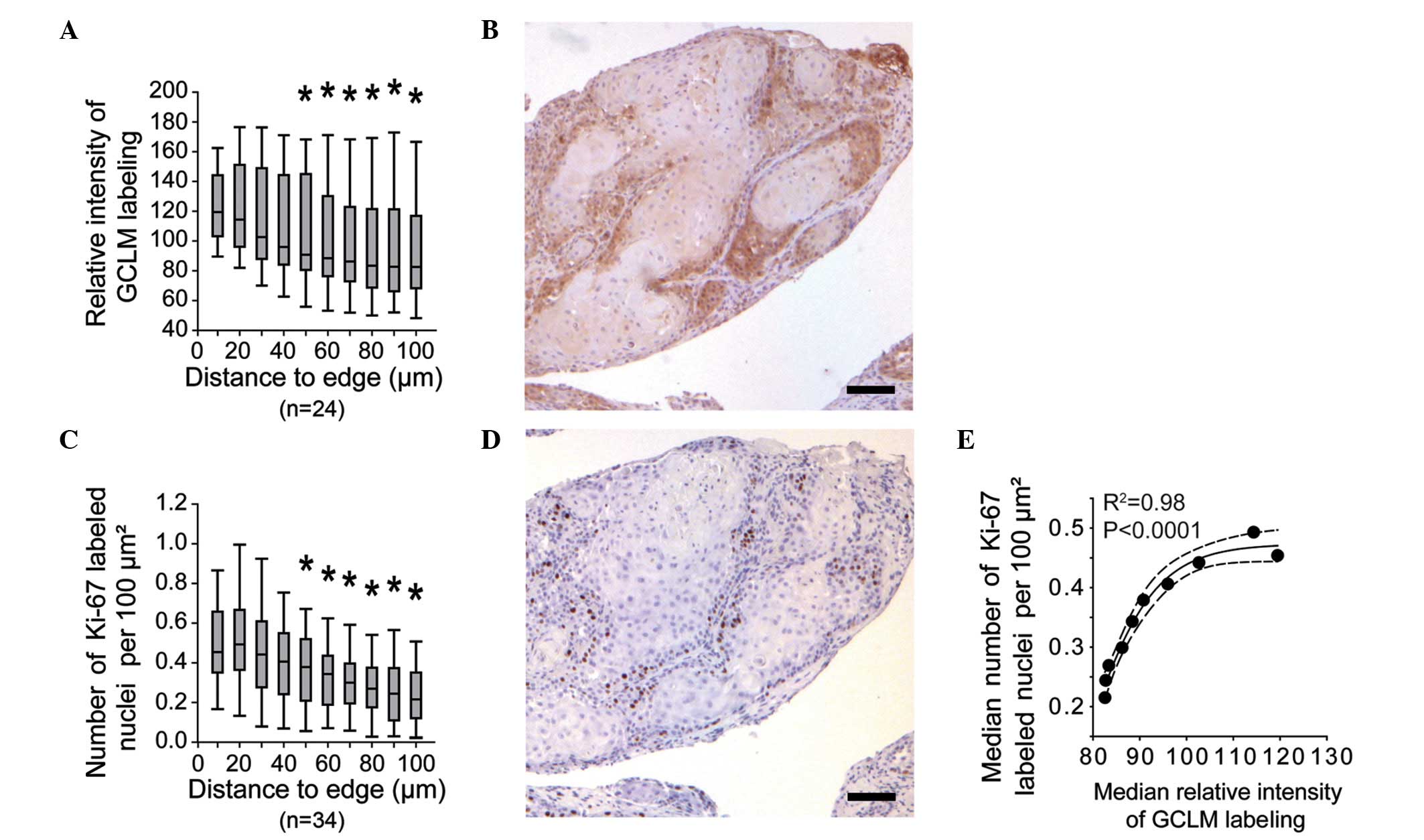
(Fig. 2C). Systematic measurement of
GCLM labeling within the tumor lobules revealed a significant
decrease in signal intensity from the periphery to regions located
≥50 µm from the lobule edge (Fig. 4A and
B). Based on previous studies reporting the peripheral
localization of proliferative cells within HNSCC lobules (25,26), the
relative density of Ki-67-labeled nuclei within the HNSCC lobules
was evaluated in the present study (Fig.
4C and D). The results revealed a consistent labeling of the
corresponding regions with anti-GCLM and anti-Ki-67 antibodies, as
illustrated by the correlation between the median values of both
signals (Fig. 4E).
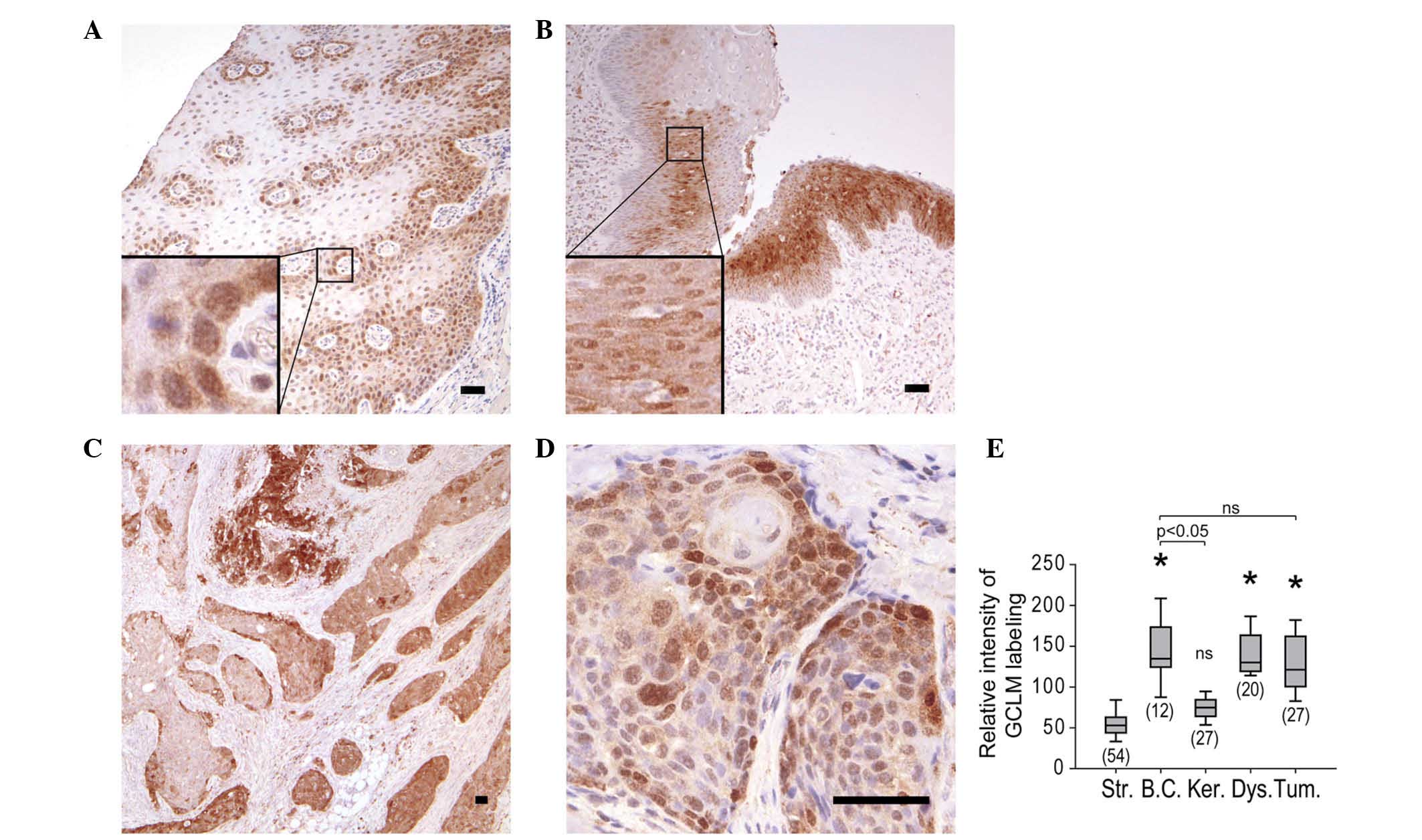 | Figure 2.IHC staining of GCLM in (A) normal
mucosa from resection margin (magnification, ×60), (B) dysplasia
within hemilarynx (magnification, ×60) and (C and D) carcinoma
(magnification C and D, ×30 and ×240, respectively). Boxes indicate
enlarged regions. Scale bars correspond to 50 µm. (E) Relative
intensity of IHC staining of GCLM in stroma, basal cells,
keratinocytes, dysplastic cells and tumor. The number of regions
analyzed for each tissue is shown in brackets. *P<0.05 vs.
stroma. Str, stroma; BC, basal cells; Ker, keratinocytes; Dys,
dysplastic cells; Tum, tumor; IHC, immunohistochemistry; GCLM,
glutamate-cysteine ligase modulator subunit; ns, not
significant. |
Discussion
Oxidative stress is the keystone of HN cancer
therapy, which requires the administration of radiotherapy and/or
chemotherapy for tumor treatment prior to or following surgical
resection (2). Both strategies rely
on the efficient induction of oxidative stress within the targeted
cells, but inducing an associated oxidative stress response that
will eventually salvage the cell (3,4). Among the
different salvage pathways, GSH is key in ROS detoxification, and
has been demonstrated to be important in tumor resistance to the
majority of chemotherapeutic drugs currently used against HN tumors
(27,28). By contrast, it is unclear whether the
levels of GSH alter the outcome of radiotherapy. While certain
studies have reported a correlation between the levels of GSH in
blood and the efficiency of SCC treatment, the levels of GSH within
the HN tumor itself do not appear to be associated with the degree
of radiosensitivity exhibited by the tumor (28,29). Thus,
it may be hypothesized that cell fate may not only depend on the
steady state levels of GSH, but also on the capability of the cell
to induce the appropriate response against ROS damage. In order to
evaluate this capability, the present study focused on the C and M
subunits of GCL, the rate-limiting enzyme of GSH synthesis
(5). While GCLC is sufficient to
perform the first step of GSH synthesis, GCLM is an essential
enhancer of GCLC activity, since it impairs the enzyme inhibition
by GSH and increases the affinity for glutamate (6).
The results of the present cross-sectional study
indicated that GCLM mRNA was more abundant in tumor biopsies than
in biopsies of adjacent tissues, whereas no significant differences
in GCLC mRNA levels were observed between tumor and normal tissues.
Although their expression is generally coordinated following
stimulation, the two GCL subunits present distinct patterns of
expression among different human tissues (5). This is partly due to the transcriptional
control of these genes (5). Both GCLM
and GCLC promoters contain the canonical antioxidant response
element sequence, which is targeted by the transcription factor
NRF2 (14). Upon oxidative stress,
the NRF2 pathway is the major trigger of the antioxidant response
(16). In addition, the two genes are
also regulated by the NF-κB pathway, which is another canonical
salvage pathway against oxidative stress (15). NF-κB signaling to GCL subunit
promoters is mediated by the AP-1 pathway (10). However, the induction of this pathway
was not evaluated in the present study, since the monitoring of the
AP-1 pathway was not amenable to mRNA quantification (10). Both NRF2 and the NF-κB are likely to
be activated in HN cancer, since increased expression of NRF2 in HN
tumors has been previously reported (30) and the dysregulation of the NF-κB
pathway has been demonstrated to influence the progression of HN
tumors (31). In the current study,
no significant changes in the expression of the NRF2 and NF-κB
genes were detected, thus precluding any conclusion on the
regulation of GCL by these pathways.
In addition, GCLM expression was restricted to basal
cells in normal pluristratified epithelium, while it was broadly
detected in dysplastic cells and non-differentiated tumor cells.
The present observations are consistent with the pattern of GCL
subunit expression in lung dysplasia, and confirmed earlier studies
reporting expression of GCL in HN tumors (32,33).
Despite the mechanisms involved are unclear, the marked increase in
GCLM expression in tumor biopsies may be responsible for the
increased GSH levels in HN tumors, compared with normal tissues,
observed in previous studies (18).
Thus, GCLM modulation appears to be sufficient to produce
significant changes in GSH synthesis (6). Under physiological conditions, GCL
activity is the result of the GCLC/GCLM ratio, which mostly depends
on the modulation of GCLM expression (34). In the present study, the expression of
GCLM was heterogeneous within tumor lobules, whereby the periphery
that was in close contact with the stroma exhibited the strongest
labeling for GCLM. Notably, these regions were identified as the
major sites of expression of Ki-67 (a well established cellular
proliferation marker), in accordance with previous reports
(25,26,35).
Therefore, the increased GCLM levels observed in the present study
may be associated with the proliferative state of tumor cells, thus
possibly linking cell proliferation with GSH levels (4). In the present study, the nuclear
localization of GCLM is reported, which is in contradiction with
the findings from previous studies conducted in Drosophila,
where only GCLC was detected in the nucleus (36,37).
However, the pattern of expression of GCLC reported in that study
hardly matched the distribution of GSH within mammalian dividing
cells (38). Thus, although GSH is
principally located in the nucleus of proliferating fibroblasts,
GCLC is mainly located into the cytosol of Drosophila cells
(36–38). Taken together, the importance of GCLM
for GCL activity and the reported localization of GCLM may explain
the high levels of GSH observed in the nucleus of proliferating
cells. The presence of enzymes involved in the synthesis of GSH
within the nucleus also explains the mechanism of GSH transport
into the nucleus (36,38).
In conclusion, the present study has demonstrated
that the expression levels of GCLM within dysplastic and tumor
cells derived from HN tumors are comparable with those observed in
basal epithelial cells. The association of cell proliferation and
GCL expression suggests that mechanisms involved in ensuring
protection against oxidative stress are associated with HN tumor
proliferation, which raises major concerns regarding individual
variations in tumor cell resistance toward chemotherapy and
radiotherapy among patients with HNSCC.
Acknowledgements
The present study was supported by the Scientific
Research Fund of the Intermunicipal Public Health of Charleroi,
University Hospital Center of Charleroi (Montigny-le-Tilleul,
Belgium), the Fund for Medical Research in Hainaut (Mons, Belgium),
the Fund for Cardiac Surgery (Brussels, Belgium) and the Institute
of Pathology and Genetics (Gosselies, Belgium). L.V. is Research
Director at the National Fund for Scientific Research (Brussels,
Belgium).
References
|
1
|
Cooper JS, Porter K, Mallin K, Hoffman HT,
Weber RS, Ang KK, Gay EG and Langer CJ: National Cancer Database
report on cancer of the head and neck: 10-year update. Head Neck.
31:748–758. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dandekar M and D'Cruz A: Organ
preservation strategies: Review of literature and their
applicability in developing nations. South Asian J Cancer.
3:147–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gupta SC, Hevia D, Patchva S, Park B, Koh
W and Aggarwal BB: Upsides and downsides of reactive oxygen species
for cancer: The roles of reactive oxygen species in tumorigenesis,
prevention and therapy. Antioxid Redox Signal. 16:1295–1322. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Traverso N, Ricciarelli R, Nitti M,
Marengo B, Furfaro AL, Pronzato MA, Marinari UM and Domenicotti C:
Role of glutathione in cancer progression and chemoresistance. Oxid
Med Cell Longev. 2013:9729132013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu SC: Glutathione synthesis. Biochim
Biophys Acta. 1830:3143–3153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Y, Shertzer HG, Schneider SN, Nebert
DW and Dalton TP: Glutamate cysteine ligase catalysis: Dependence
on ATP and modifier subunit for regulation of tissue glutathione
levels. J Biol Chem. 280:33766–33774. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dahl EL and Mulcahy RT: Cell-type specific
differences in glutamate cysteine ligase transcriptional regulation
demonstrate independent subunit control. Toxicol Sci. 61:265–272.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cai J, Huang ZZ and Lu SC: Differential
regulation of gamma-glutamylcysteine synthetase heavy and light
subunit gene expression. Biochem J. 326:167–172. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krzywanski DM, Dickinson DA, Iles KE,
Wigley AF, Franklin CC, Liu RM, Kavanagh TJ and Forman HJ: Variable
regulation of glutamate cysteine ligase subunit proteins affects
glutathione biosynthesis in response to oxidative stress. Arch
Biochem Biophys. 423:116–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang H, Magilnick N, Ou X and Lu SC:
Tumour necrosis factor alpha induces co-ordinated activation of rat
GSH synthetic enzymes via nuclear factor kappaB and activator
protein-1. Biochem J. 391:399–408. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wild AC, Moinova HR and Mulcahy RT:
Regulation of gamma-glutamylcysteine synthetase subunit gene
expression by the transcription factor Nrf2. J Biol Chem.
274:33627–33636. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang H, Wang J, Huang ZZ, Ou X and Lu SC:
Cloning and characterization of the 5′-flanking region of the rat
glutamate-cysteine ligase catalytic subunit. Biochem J.
455:447–455. 2001. View Article : Google Scholar
|
|
13
|
Reuter S, Gupta SC, Chaturvedi MM and
Aggarwal BB: Oxidative stress, inflammation, and cancer: How are
they linked? Free Radic Biol Med. 49:1603–1616. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang H, Magilnick N, Lee C, Kalmaz D, Ou
X, Chan JY and Lu SC: Nrf1 and Nrf2 regulate rat glutamate-cysteine
ligase catalytic subunit transcription indirectly via NF-kappaB and
AP-1. Mol Cell Biol. 25:5933–5946. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng Z, Geh E, Chen L, Meng Q, Fan Y,
Sartor M, Shertzer HG, Liu ZG, Puga A and Xia Y: Inhibitor of
kappaB kinase beta regulates redox homeostasis by controlling the
constitutive levels of glutathione. Mol Pharmacol. 77:784–792.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Baird L and Dinkova-Kostova AT: The
cytoprotective role of the Keap1-Nrf2 pathway. Arch Toxicol.
85:241–272. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Na HK and Surh YJ: Oncogenic potential of
Nrf2 and its principal target protein heme oxygenase-1. Free Radic
Biol Med. 67:353–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gamcsik MP, Kasibhatla MS, Teeter SD and
Colvin OM: Glutathione levels in human tumors. Biomarkers.
17:671–691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dequanter D, Van de Velde M, Nuyens V,
Nagy N, Van Antwerpen P, Vanhamme L, Zouaoui Boudjeltia K,
Vanhaeverbeek M, Brohée D and Lothaire P: Assessment of oxidative
stress in tumors and histologically normal mucosa from patients
with head and neck squamous cell carcinoma: A preliminary study.
Eur J Cancer Prev. 22:558–560. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van der Schroeff MP and de Baatenburg Jong
RJ: Staging and prognosis in head and neck cancer. Oral Oncol.
4–5:356–360. 2009. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Varghese F, Bukhari AB, Malhotra R and De
A: IHC Profiler: An open source plugin for the quantitative
evaluation and automated scoring of immunohistochemistry images of
human tissue samples. PLoS One. 9:e968012014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sahoo P, Wilkins C and Yeager J: Threshold
selection using Renyi's entropy. Pattern Recognit. 30:71–84. 1997.
View Article : Google Scholar
|
|
24
|
Bottero V, Imbert V, Frelin C, Formento JL
and Peyron JF: Monitoring NF-kappa B transactivation potential via
real-time PCR quantification of I kappa B-alpha gene expression.
Mol Diagn. 7:187–194. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Edström SS, Gustafsson B, Stenman G, Lydén
E, Stein H and Westin T: Proliferative pattern of head and neck
cancer. Am J Surg. 162:412–416. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kearsley JH, Furlong KL, Cooke RA and
Waters MJ: An immunohistochemical assessment of cellular
proliferation markers in head and neck squamous cell cancers. Br J
Cancer. 61:821–827. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yellin SA, Davidson BJ, Pinto JT, Sacks
PG, Qiao C and Schantz SP: Relationship of glutathione and
glutathione- S-transferase to cisplatin sensitivity in human head
and neck squamous carcinoma cell lines. Cancer Lett. 85:223–232.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kato T, Duffey DC, Ondrey FG, Dong G, Chen
Z, Cook JA, Mitchell JB and Van Waes C: Cisplatin and radiation
sensitivity in human head and neck squamous carcinomas are
independently modulated by glutathione and transcription factor
NF-kappaB. Head Neck. 22:748–759. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bhattathiri VN, Sreelekha TT, Sebastian P,
Remani P, Chandini R, Vijayakumar T and Nair MK: Influence of
plasma GSH level on acute radiation mucositis of the oral cavity.
Int J Radiat Oncol Biol Phys. 29:383–386. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stacy DR, Ely K, Massion PP, Yarbrough WG,
Hallahan DE, Sekhar KR and Freeman ML: Increased expression of
nuclear factor E2 p45-related factor 2 (NRF2) in head and neck
squamous cell carcinomas. Head Neck. 28:813–818. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Loercher A, Lee TL, Ricker JL, Howard A,
Geoghegen J, Chen Z, Sunwoo JB, Sitcheran R, Chuang EY, Mitchell
JB, et al: Nuclear factor-kappaB is an important modulator of the
altered gene expression profile and malignant phenotype in squamous
cell carcinoma. Cancer Res. 64:6511–6523. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kaarteenaho-Wiik R and Kinnula VL:
Distribution of antioxidant enzymes in developing human lung,
respiratory distress syndrome, and bronchopulmonary dysplasia. J
Histochem Cytochem. 52:1231–1240. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nishimura T, Newkirk K, Sessions RB,
Andrews PA, Trock BJ, Rusmussen AA, Montogomery EA, Bischoff EK,
Hanigan MH and Cullen KJ: Association between expression of
glutathione-associated enzymes and response to platinum-based
chemotherapy in head and neck cancer. Chem Biol Interact.
111–112:187–198. 1998. View Article : Google Scholar
|
|
34
|
Lee JI, Kang J and Stipanuk MH:
Differential regulation of glutamate-cysteine ligase subunit
expression and increased holoenzyme formation in response to
cysteine deprivation. Biochem J. 393:181–190. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Watanabe S, Watanabe R, Oton-Leite AF,
Alencar RC, Oliveira JC, Leles CR, Batista AC and Mendonça EF:
Analysis of cell proliferation and pattern of invasion in oral
squamous cell carcinoma. J Oral Sci. 52:417–424. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Radyuk SN, Rebrin I, Luchak JM, Michalak
K, Klichko VI, Sohal RS and Orr WC: The catalytic subunit of
Drosophila glutamate-cysteine ligase is a nucleocytoplasmic
shuttling protein. J Biol Chem. 284:2266–2274. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Markovic J, Borrás C, Ortega A, Sastre J,
Viña J and Pallardó FV: Glutathione is recruited into the nucleus
in early phases of cell proliferation. J Biol Chem.
282:20416–20424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
García-Giménez JL, Markovic J, Dasí F,
Queval G, Schnaubelt D, Foyer CH and Pallardó FV: Nuclear
glutathione. Biochim Biophys Acta. 1830:3304–3316. 2013. View Article : Google Scholar : PubMed/NCBI
|