Worldwide primary liver cancer, which includes
primary hepatocellular carcinoma (HCC), intrahepatic
cholangiocarcinoma (ICC) and fibrolamellar HCC, is the fifth most
common cancer and the third leading cause of cancer-associated
mortality (1). Currently, surgery and
liver transplantation are considered the optimal curative
treatments for the disease (2);
however, there is a significant shortage of organ donors, and
surgical complications, recurrence and metastasis are common
(3). Although epidemiological risk
factors, including hepatitis B virus (HBV) and hepatitis C virus
infection, have been identified, the molecular mechanisms
underlying primary liver cancer remain unclear (4). Therefore, elucidation of the molecular
pathogenesis of liver cancer and the development of effective
targeted therapies is urgently required (5).
The Notch signaling pathway is evolutionarily
conserved and controls numerous developmental processes, such as
cell fate determination, terminal differentiation and proliferation
(6). During embryonic development and
adulthood, intracellular Notch signaling is required for cell
specification, lineage commitment and maintenance of progenitor
cells (7), in particular in the
control of endothelial cell differentiation, arteriovenous
specification and vascular development (8).
In mammals, the canonical Notch pathway includes
four receptors (Notch1, 2, 3 and 4) and two ligand families [Jagged
(JAG) 1 and 2 and Delta-like-ligand (Dll) 1, 3 and 4]. The Notch
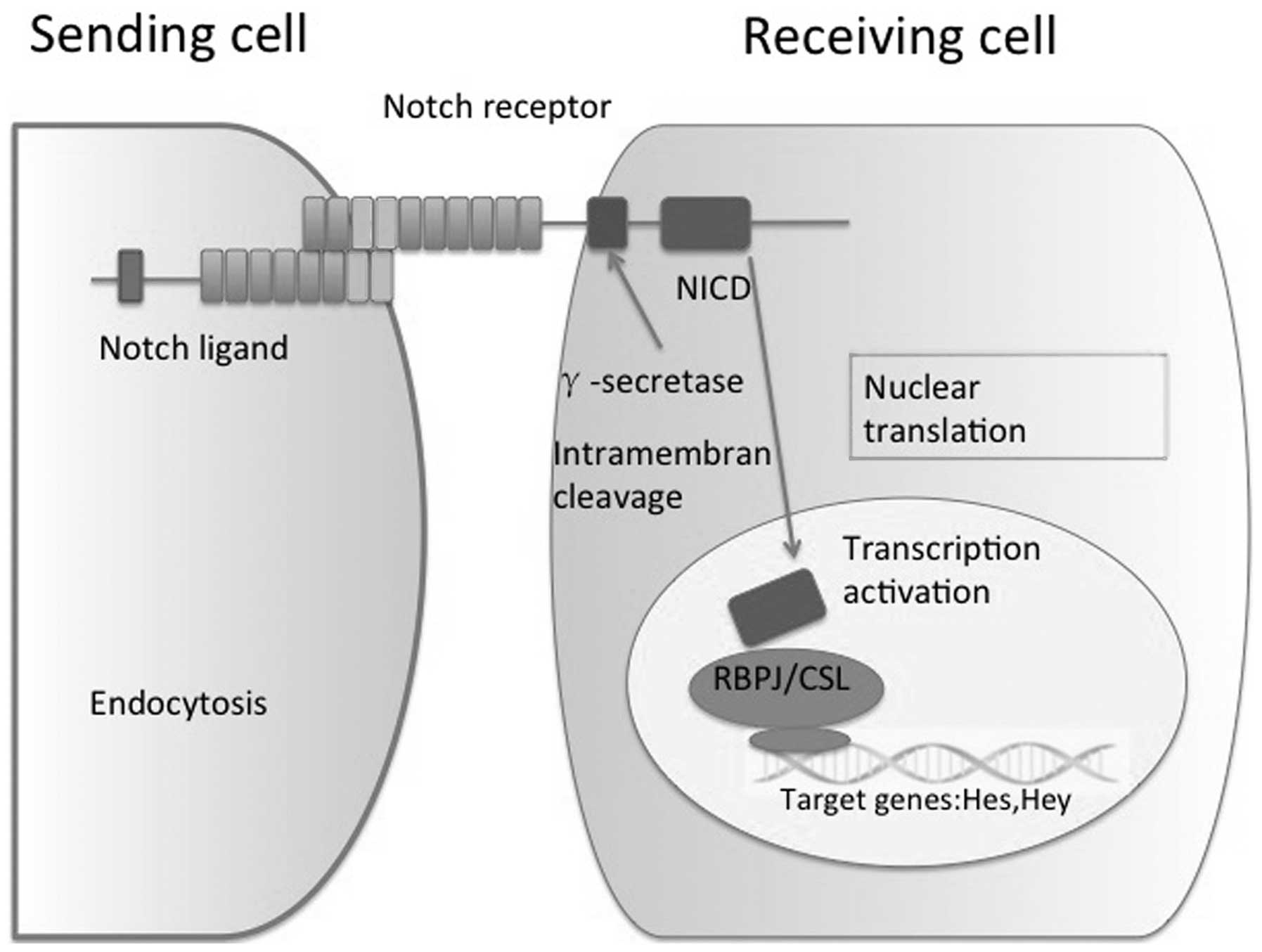
signaling pathway consists of ligand-induced activation of
receptors, proteolytic cleavage and subsequent translocation of the
notch intracellular domain (NICD) to the nucleus, where it
functions as a transcriptional regulator (9). Activation of Notch signaling requires
direct or indirect contact between cells expressing Notch ligands
or receptors, and transmitting and receiving cells are subsequently
modified by the interaction. Initially, cells express Notch
receptors and ligands, and as the interaction continues, one cell
becomes a transmitting cell by upregulating the expression of
ligands and downregulating of receptors, while the receiving cells
follow the opposite pattern (10).
Prior to the NICD being transported to the nucleus, it is cleaved
by the γ-secretase complex. In the nucleus, NICD interacts with
C-repeat/DRE binding factor 1, a DNA-binding transcriptional
repressor also known as the recombination signal binding protein
for immunoglobulin Kappa J region (RBPJ), and converts it into a
transcriptional activator that induces the transcription of target
genes, including the family of Hes and Hey-associated transcription
factors. Furthermore, in the liver, Notch partially controls the
expression of Sox9, HNF1 Homeobox B and transforming growth
factor-β, which are key regulators in hepatic lineage commitment
(11,12) (Fig.
1).
RBPJ is a DNA-binding protein, also known as CSL,
which is a member of the Suppressor of Hairless (Drosophila
melanogaster) family of transcription factors that recognizes
the consensus sequence C(T)GTGGGAA. RBPJ predominantly acts as a
transcriptional repressor of promoters that possess RBPJ binding
sites via the recruitment of other co-repressors. The most
important function of RBPJ is to mediate signals from Notch
receptors. RBPJ is a common downstream transcription factor of
Notch receptors and its absence indicates a complete block of the
Notch signaling pathway (13).
The present review discusses the findings of recent
studies regarding Notch expression and summarizes Notch signaling
during HCC and ICC development.
Notch is a highly regulated signaling mechanism with
numerous specific features. In humans, mutations in Notch ligands
or receptors are associated with various diseases; for example,
JAG1 and Notch2 mutations are associated with Alagille syndrome
(14), and Notch3 mutations with
cerebral autosomal dominant arteriopathy with subcortical infarcts
and leukoencephalopathy (15). Human
genetic diseases and mutant mouse models have demonstrated the
importance of Notch signaling in the development and remodeling of
intrahepatic bile ducts (IHBD). Alagille syndrome (AGS) is a human
autosomal dominant disorder that is caused by mutations in the
Notch ligand JAG1, and less commonly in the Notch2 receptor
(16). The estimated prevalence of
AGS is 1 case per 70,000 live births worldwide (17). The disease is a multi-organ disorder
most commonly diagnosed by liver abnormalities, which lead to
hepatic bile duct paucity and cholestasis at birth (18). Cardiac, skeletal and ophthalmological
abnormalities, and less frequently renal or vascular deficiencies,
are also observed in patients with AGS (19). Numerous renal abnormalities are
observed in patients with AGS, including renovascular disease,
renal tubular acidosis, tubulointerstitial nephritis and renal
dysplasia/hypoplasia (20). Notably,
mice with haploinsufficiency for JAG1 exhibit no significant
phenotypic abnormalities, suggesting that additional modifier genes
contribute to the AGS phenotype observed in humans (21). Additionally, mutations in Notch3 are
associated with inherited vascular diseases, including degenerative
vascular disorder, cerebral autosomal dominant arteriopathy with
subcortical infarcts and leukoencephalopathy (22).
A number of Notch receptors have been identified in
mammals, including Notch1-4. The receptors are transmembrane
proteins with three domains: Extracellular Notch domain, a
transmembrane domain and NICD. A number of studies have
demonstrated that inhibition of the Notch signaling pathway induces
the downregulation of Notch receptors (34,35).
Notch1 regulates arteriovenous differentiation
during embryogenesis and in the hepatic endothelium of adult mice
(29). Homozygous disruption of the
Notch1 gene is fatal at embryonic day (ED) 10, suggesting that
Notch1 is essential for normal embryonic development. After ED10,
histological analyses revealed widespread cell death, which was
attributed to disorganized and delayed somitogenesis (30,31).
Notch1 is required for vascular homeostasis of hepatic sinusoids by
inducing quiescence and differentiation of liver sinusoidal
endothelial cells. Thus, disruption of the Notch1 pathway leads to
intussusceptive angiogenesis and nodular regenerative hyperplasia
(36).
Homozygous Notch2-deficient embryos exhibit
developmental retardation, widespread cell death and embryonic
mortality prior to ED11.5; however, normal somitogenesis is
observed compared with a Notch1 KO (37). Alagille syndrome is also associated
with Notch2 mutations (38).
Jeliazkova et al (39)
demonstrated that mice with a perinatal, liver-specific complete
elimination of Notch2 exhibited a marked reduction in the number of
mature bile ducts, an increased number of disorganized primitive
biliary-like structures, portal inflammation, portal tract
enlargement and fibrosis and biliary necrosis. Furthermore,
neonatal Notch2 KO mice are severely jaundiced, with livers that
exhibit no cytokeratin 19 positive ductal structures (40).
Young Notch3 KO mice are viable and fertile without
any apparent phenotypic abnormalities, whereas adult Notch3 KO mice
exhibit arterial defects due to abnormalities in differentiation
(41). In the liver, Notch3 regulates
the activation of hematopoietic stem cells and may exhibit an
anti-fibrogenic effect (42). Notch4
KO mice are viable and fertile, since during development Notch4
expression is restricted to vascular endothelial cells (42). However, Notch1/4 double KO mice
exhibit a more severe phenotype, presenting with extensive defects
in angiogenic vascular remodeling during embryonic development
compared with Notch1 KO mice (43).
Notch ligands include JAG1, JAG2, Dll1, Dll3 and
Dll4. Homozygous disruption of Notch ligands invariably affects the
liver. A previous study has demonstrated that deletion of JAG1 in
the portal vein mesenchyme lead in jaundice, liver failure and
small numbers of IHBDs (28).
Furthermore, JAG2 KO mice die perinatally due to craniofacial
defects, including fusion of the tongue with the palatal shelves,
and syndactyly of the fore and hind limbs (29). Homozygous inactivation of Dll1 causes
severe defects in somite patterning and the development of a
hyperplastic central nervous system (44). Following ED9, Dll1 KO mice become
hemorrhagic and die around ED11.5 (45). Dll3 is expressed in the presomitic
mesoderm and is localized to the rostral somatic compartments.
Homozygous disruption of Notch1 and Dll3 leads to severe
abnormalities in somitogenesis. Mutations in the human Dll3 homolog
result in recessive skeletal abnormalities in spondylocostal
dysostosis (46). Dll4 is essential
for embryonic vascular development and arterial specification and
is clearly upregulated in the tumor vessels of humans and mice;
Dll4 deficiency leads to severe vascular remodeling defects and
embryonic mortality (31).
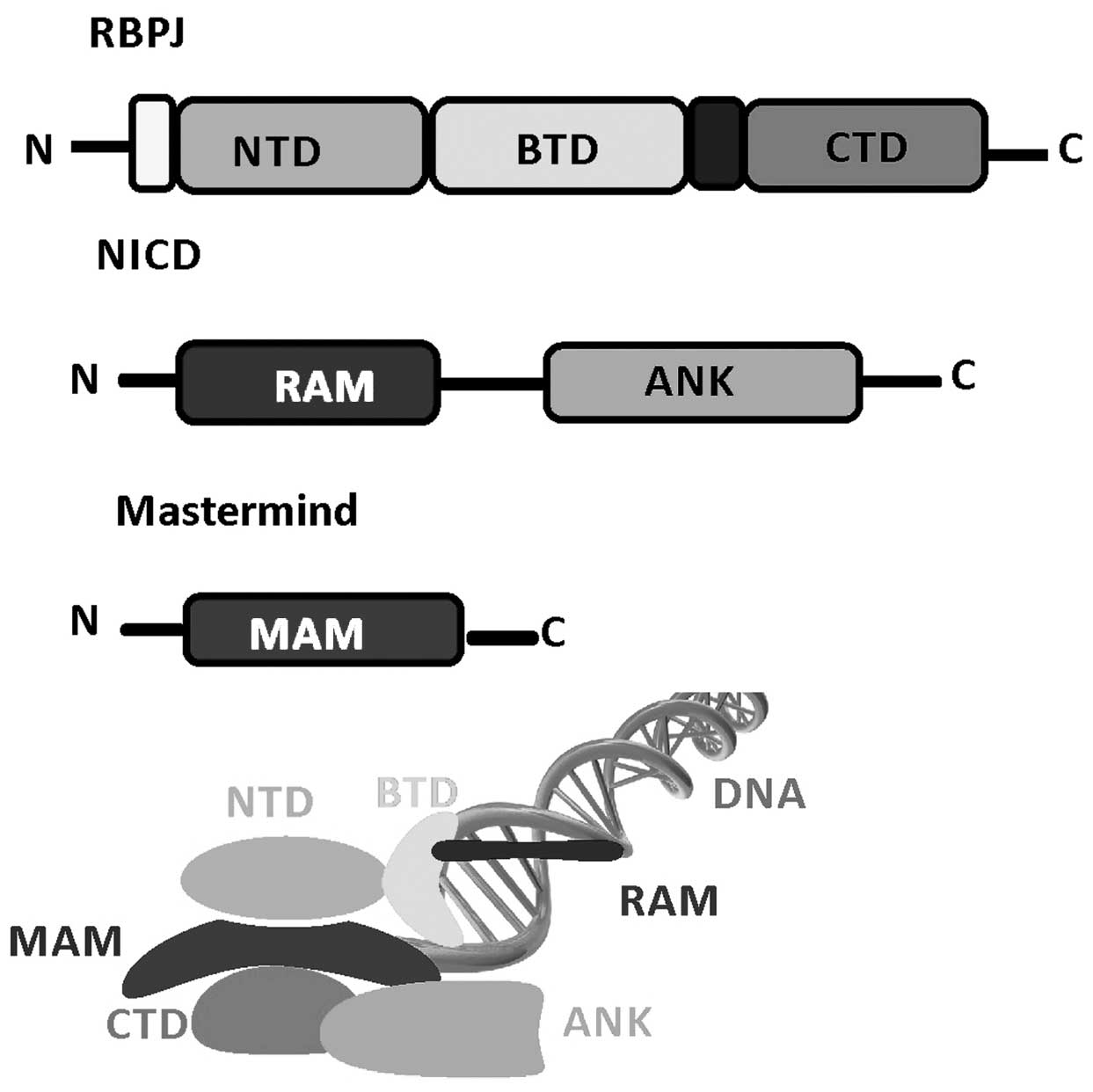
Structural studies of Notch transcription complexes
have identified the overall folds, domain organization and
interacting regions of RBPJ, NICD and MAM proteins (51–53). RBPJ
is composed of three domains: An N-terminal domain (NTD), a
β-trefoil domain (BTD) and a C-terminal domain (CTD). The NICD
binds RBPJ via a RBPJ-associated molecule and ankyrin (ANK) repeat
domains that interact with BTD and CTD. MAM forms a helix with a
distinctive bend, in which its N-terminal helical region forms a
tripartite complex with ANK and CTD, and its C-terminal helical
region binds the NTD of RBPJ. RBPJ binds the consensus DNA
sequence, CGTGGGAA with moderate affinity (~200 nm Kd) (54,55), which
is a similar site to that of the enhancer and promoter elements of
Notch target genes (56). The
structures of RBPJ and RBPJ-NICD MAM activator complexes, including
assembling at various target genes, have enabled detailed
biochemical and cellular studies. These transcriptionally active
ternary complexes bind to the promoter and enhancer elements of
Notch responsive genes, including hey1 and hes1 promoters (57) (Fig.
2).
Gain or loss of function mutations in the Notch
pathway have been identified in several types of cancers, including
neural stem cell tumors, lung carcinomas and prostate cancer
(68). Although Notch does not
directly lead to unregulated cell proliferation or genetic
alterations that are associated with tumor progression, it alters
the developmental state of cells and consequently maintains cells
in a proliferative or undifferentiated state (69). In chronic HBV infection (CHB),
repression of Notch receptors was demonstrated to lead to immune
dysfunction (70). The contribution
that a decreased expression of Notch receptors makes to ongoing
fibrosis, cirrhosis and HCC is unclear; however, repression of
Notch receptors in CHB has been suggested to repress immune
regulation, resulting in the inhibition of differentiation and
proliferation of effector cells leading to additional pathogenesis
of CHB (71). Notably, the
pro-mitogenic function of Notch was demonstrated in a model of
partial hepatectomy (72). The
pro-oncogenic function of Notch was also investigated by genome
wide analysis of HCC samples, which revealed that the Notch
coactivator MAML2 is a target of genetic alterations (73). Additionally, Notch signaling is
crucial for the differentiation of hepatocytes into biliary lineage
cells during the early stages of ICC developments. Gain and loss of
function studies have demonstrated that ICC develops via the
Notch-mediated differentiation of hepatocytes into biliary lineage
cells, and that the malignancy and progression of the tumor are
dependent on the intensity of Notch signaling in hepatocytes
(74). Notably, hepatitis-infected
hepatocytes may be converted into biliary lineage cells via Notch
signal activation and thus become the point of origin for ICC
(75). Therefore, suppression of
Notch signaling may present a novel strategy for the treatment of
ICC, since it may inhibit the conversion of hepatocytes into
biliary lineage cells during the early stages of ICC development
(76). Mice with liver-specific
constitutive activation of Notch1 intracellular domain (N1ICD) may
develop HCC when they reach adult age (77). Genomic profiling technology has
revealed that a Notch-specific gene expression signature reported
in mice overexpressing NICD was also present in a cluster of
patients with HCC (78). Histological
analyses of the mouse liver tissue exhibited similar features
compared with that of the HCC patients, which included the presence
of proliferating K-19 positive cells. Constitutive Notch2
overexpression causes HPCs to spontaneously develop into
dedifferentiated HCC cells (79). In
addition, Notch-induced malignant hepatocyte transformation is
associated with downregulation of hepatocyte-associated genes and
Sox9 expression (80). Fate-mapping
studies have demonstrated that clear-cell adenocarcinoma (CCA)
cells derived from hepatocytes may be converted to a biliary K-19
positive phenotype (81).
Accordingly, in CCA development, N1ICD association with protein
kinase B signaling in hepatocytes stimulated their malignant
dedifferentiation (82). The
stimulation of N1ICD expression by inflammatory mediators has also
been reported in human CCA, which additionally supports the role of
Notch in liver cancer (83) (Table II) (9,12,24,31,84–87).
Persistent activation of Notch signaling may lead to
oncogenesis depending on modifier factors, including the
inflammatory environment or the presence of other carcinogenic
conditions that may cause HCC or CCA (88). A Notch signature has been reported in
a subset of HCC patients and an overexpression of Notch receptors
has been reported in human CCAs, which is fundamentally required
for the development of targeted therapies (89). Silencing of the Notch pathway may
potentially inhibit Notch-driven tumor progression and interfere
with tumor aggressiveness, since Notch activation has been
associated with a more malignant phenotype (84). However, the identification of a
reliable tissue-specific biomarker of Notch inhibition is critical
for the application of Notch-targeted therapy. Notably, the hepatic
Notch target gene Sox9 is associated with a poor prognosis in liver
cancers (90). Therefore, the role of
Sox9 as a potential biomarker of Notch involvement and indication
for Notch-targeted treatment requires additional study.
An increased understanding of the mechanisms
involved in liver cancer proliferation and differentiation may aid
the development of therapeutic strategies for liver cancer. The
Notch pathway is emerging as a critical signaling pathway, which
regulates cell proliferation, differentiation and necrosis
associated with normal development, as well as stem cell renewal
and differentiation. In conclusion, loss or disruption of Notch
signaling may be a key contributing factor in bile ductular
disorders, sinusoidal capillarization and the neovascularization of
portal regions in the liver. To develop effective therapeutic
approaches for the treatment of liver cancer, additional studies
that investigate the Notch pathway are urgently required.
The present study was supported by the National
Science Foundation of China (grant nos. 81300340 and 81270515),
Shanghai Municipal Health Bureau Project (grant nos. 20124107 and
2011287) and Chinese Foundation for Prevention and Control of
Hepatitis (grant nos. WBN20100021 and CFHPC20131011).
|
1
|
Tinkle CL and Haas-Kogan D: Hepatocellular
carcinoma: natural history, current management, and emerging tools.
Biologics. 6:207–219. 2012.PubMed/NCBI
|
|
2
|
Jie L, Fan W, Weiqi D, Yingqun Z, Ling X,
Miao S, Ping C and Chuanyong G: The hippo-yes association protein
pathway in liver cancer. Gastroenterol Res Pract. 2013:1870702013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hackl C, Schlitt HJ, Kirchner GI, Knoppke
B and Loss M: Liver transplantation for malignancy: Current
treatment strategies and future perspectives. World J
Gastroenterol. 20:5331–5344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lam PH, Obirieze AC, Ortega G, Nwokeabia
I, Onyewu S, Purnell SD, Samimi MM, Weeks CB, Lee EL, Shokrani B,
et al: Characterization of hepatitis B and C among liver transplant
recipients with hepatocellular carcinoma: An analysis of the
Nationwide Inpatient Sample Database. Transplant Proc. 48:123–127.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: Globocan 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang F, He L, Dai WQ, Xu YP, Wu D, Lin CL,
Wu SM, Cheng P, Zhang Y, Shen M, et al: Salinomycin inhibits
proliferation and induces apoptosis of human hepatocellular
carcinoma cells in vitro and in vivo. PLoS One. 7:e506382012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dai W, Wang F, He L, Lin C, Wu S, Chen P,
Zhang Y, Shen M, Wu D, Wang C, et al: Genistein inhibits
hepatocellular carcinoma cell migration by reversing the
epithelial-mesenchymal transition: Partial mediation by the
transcription factor NFAT1. Mol Carcinog. 54:301–311. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roca C and Adams RH: Regulation of
vascular morphogenesis by notch signaling. Genes Dev. 21:2511–2524.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dou GR, Wang YC, Hu XB, Hou LH, Wang CM,
Xu JF, Wang YS, Liang YM, Yao LB, Yang AG and Han H: RBP-J, the
transcription factor downstream of Notch receptors, is essential
for the maintenance of vascular homeostasis in adult mice. FASEB J.
22:1606–1617. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fortini ME: Notch signaling: The core
pathway and its posttranslational regulation. Dev Cell. 16:633–647.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Coffinier C, Gresh L, Fiette L, Tronche F,
Schütz G, Babinet C, Pontoglio M, Yaniv M and Barra J: Bile system
morphogenesis defects and liver dysfunction upon targeted deletion
of HNF1beta. Development. 129:1829–1838. 2002.PubMed/NCBI
|
|
12
|
Zong Y, Panikkar A, Xu J, Antoniou A,
Raynaud P, Lemaigre F and Stanger BZ: Notch signaling controls
liver development by regulating biliary differentiation.
Development. 136:1727–1739. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morell CM, Fiorotto R, Fabris L and
Strazzabosco M: Notch signalling beyond liver development: emerging
concepts in liver repair and oncogenesis. Clin Res Hepatol
Gastroenterol. 37:447–454. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahn KJ, Yoon JK, Kim GB, Kwon BS, Go JM,
Moon JS, Bae EJ and Noh CI: Alagille syndrome and a JAG1 mutation:
41 Cases of experience at a single center. Korean J Pediatr.
58:392–397. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Zuo Y, Sun W, Zhang W, Lv H, Huang
Y, Xiao J, Yuan Y and Wang Z: The genetic spectrum and the
evaluation of CADASIL screening scale in Chinese patients with
NOTCH3 mutations. J Neurol Sci. 354:63–69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oda T, Elkahloun AG, Pike BL, Okajima K,
Krantz ID, Genin A, Piccoli DA, Meltzer PS, Spinner NB, Collins FS
and Chandrasekharappa SC: Mutations in the human Jagged1 gene are
responsible for alagille syndrome. Nat Genet. 16:235–242. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brooks AS and Dooijes D: From gene to
disease: Arteriohepatic dysplasia or Alagille syndrome. Ned
Tijdschr Geneeskd. 147:1213–1215. 2003.PubMed/NCBI
|
|
18
|
McDaniell R, Warthen DM, Sanchez-Lara PA,
Pai A, Krantz ID, Piccoli DA and Spinner NB: NOTCH2 mutations cause
alagille syndrome, a heterogeneous disorder of the notch signaling
pathway. Am J Hum Genet. 79:169–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Habib R, Dommergues JP, Gubler MC,
Hadchouel M, Gautier M, Odievre M and Alagille D: Glomerular
mesangiolipidosis in alagille syndrome (arteriohepatic dysplasia).
Pediatr Nephrol. 1:455–464. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nyfeler Y, Kirch RD, Mantei N, Leone DP,
Radtke F, Suter U and Taylor V: Jagged1 signals in the postnatal
subventricular zone are required for neural stem cell self-renewal.
EMBO J. 24:3504–3515. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gridley T: Notch signaling in the
vasculature. Curr Top Dev Biol. 92:277–309. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng P, Dai W, Wang F, Lu J, Shen M, Chen
K, Li J, Zhang Y, Wang C, Yang J, et al: Ethyl pyruvate inhibits
proliferation and induces apoptosis of hepatocellular carcinoma via
regulation of the HMGB1-RAGE and AKT pathways. Biochem Biophys Res
Commun. 443:1162–1168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Croquelois A, Blindenbacher A, Terracciano
L, Wang X, Langer I, Radtke F and Heim MH: Inducible inactivation
of Notch1 causes nodular regenerative hyperplasia in mice.
Hepatology. 41:487–496. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dill MT, Rothweiler S, Djonov V, Hlushchuk
R, Tornillo L, Terracciano L, Meili-Butz S, Radtke F, Heim MH and
Semela D: Disruption of Notch1 induces vascular remodeling,
intussusceptive angiogenesis and angiosarcomas in livers of mice.
Gastroenterolog. 142:967–977. 2012. View Article : Google Scholar
|
|
25
|
Geisler F, Nagl F, Mazur PK, Lee M,
Zimber-Strobl U, Strobl LJ, Radtke F, Schmid RM and Siveke JT:
Liver-specific inactivation of Notch2, but not Notch1, compromises
intrahepatic bile duct development in mice. Hepatology. 48:607–616.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen JY, Feng L, Zhang HL, Li JC, Yang XW,
Cao XL, Liu L, Qin HY, Liang YM and Han H: Differential regulation
of bone marrow-derived endothelial progenitor cells and endothelial
outgrowth cells by the notch signaling pathway. PLoS One.
7:e436432012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krebs LT, Xue Y, Norton CR, Shutter JR,
Maguire M, Sundberg JP, Gallahan D, Closson V, Kitajewski J,
Callahan R, et al: Notch signaling is essential for vascular
morphogenesis in mice. Genes Dev. 14:1343–1352. 2000.PubMed/NCBI
|
|
28
|
Hofmann JJ, Zovein AC, Koh H, Radtke F,
Weinmaster G and Iruela-Arispe ML: Jagged1 in the portal vein
mesenchyme regulates intrahepatic bile duct development: Insights
into alagille syndrome. Development. 137:4061–4072. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiang R, Lan Y, Chapman HD, Shawber C,
Norton CR, Serreze DV, Weinmaster G and Gridley T: Defects in limb,
craniofacial and thymic development in Jagged2 mutant mice. Genes
Dev. 12:1046–1057. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Redeker C, Schuster-Gossler K, Kremmer E
and Gossler A: Normal development in mice over-expressing the
intracellular domain of DLL1 argues against reverse signaling by
DLL1 in vivo. PLoS One. 8:e790502013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Djokovic D, Trindade A, Gigante J, Badenes
M, Silva L, Liu R, Li X, Gong M, Krasnoperov V, Gill PS and Duarte
A: Combination of Dll4/Notch and Ephrin-B2/EphB4 targeted therapy
is highly effective in disrupting tumor angiogenesis. BMC Cancer.
10:6412010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Turnpenny PD, Whittock N, Duncan J,
Dunwoodie S, Kusumi K and Ellard S: Novel mutations in DLL3, a
somitogenesis gene encoding a ligand for the Notch signalling
pathway, cause a consistent pattern of abnormal vertebral
segmentation in spondylocostal dysostosis. J Med Genet. 40:333–339.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gale NW, Dominguez MG, Noguera I, Pan L,
Hughes V, Valenzuela DM, Murphy AJ, Adams NC, Lin HC, Holash J,
Thurston G and Yancopoulos GD: Haploinsufficiency of delta-like 4
ligand results in embryonic lethality due to major defects in
arterial and vascular development. Proc Natl Acad Sci USA.
101:15949–15954. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Traustadóttir GÁ, Jensen CH, Thomassen M,
Beck HC, Mortensen SB, Laborda J, Baladrón V, Sheikh SP and
Andersen DC: Evidence of non-canonical NOTCH signaling: Delta-like
1 homolog (DLK1) directly interacts with the NOTCH1 receptor in
mammals. Cell Signal. 28:246–254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hayashi T, Gust KM, Wyatt AW, Goriki A,
Jäger W, Awrey S, Li N, Oo HZ, Altamirano-Dimas M, Buttyan R, et
al: Not all NOTCH Is Created Equal: The Oncogenic Role of NOTCH2 in
Bladder Cancer and Its Implications for Targeted Therapy. Clin
Cancer Res. Jan 14–2016.(Epub ahead of print). View Article : Google Scholar
|
|
36
|
Conlon RA, Reaume AG and Rossant J: Notch1
is required for the coordinate segmentation of somites.
Development. 121:1533–1545. 1995.PubMed/NCBI
|
|
37
|
Swiatek PJ, Lindsell CE, del Amo FF,
Weinmaster G and Gridley T: Notch1 is essential for
postimplantation development in mice. Genes Dev. 8:707–719. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kamath BM, Bauer RC, Loomes KM, Chao G,
Gerfen J, Hutchinson A, Hardikar W, Hirschfield G, Jara P, Krantz
ID, et al: NOTCH2 mutations in Alagille syndrome. J Med Genet.
49:138–144. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jeliazkova P1, Jörs S, Lee M,
Zimber-Strobl U, Ferrer J, Schmid RM, Siveke JT and Geisler F:
Canonical Notch2 signaling determines biliary cell fates of
embryonic hepatoblasts and adult hepatocytes independent of Hes1.
Hepatology. 57:2469–2479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Falix FA, Aronson DC, Lamers WH and
Gaemers IC: Possible roles of DLK1 in the notch pathway during
development and disease. Biochim Biophys Acta. 1822:988–995. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Pippucci T, Maresca A, Magini P, Cenacchi
G, Donadio V, Palombo F, Papa V, Incensi A, Gasparre G, Valentino
ML, et al: Homozygous NOTCH3 null mutation and impaired NOTCH3
signaling in recessive early-onset arteriopathy and cavitating
leukoencephalopathy. EMBO Mol Med. 7:848–858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen YX, Weng ZH and Zhang SL: Notch3
regulates the activation of hepatic stellate cells. World J
Gastroenterol. 18:1397–1403. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Carlson TR, Yan Y, Wu X, Lam MT, Tang GL,
Beverly LJ, Messina LM, Capobianco AJ, Werb Z and Wang R:
Endothelial expression of constitutively active Notch4 elicits
reversible arteriovenous malformations in adult mice. Proc Natl
Acad Sci USA. 102:9884–9889. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rocha SF, Lopes SS, Gossler A and Henrique
D: Dll1 and Dll4 function sequentially in the retina and pV2 domain
of the spinal cord to regulate neurogenesis and create cell
diversity. Dev Biol. 328:54–65. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
de Hrabĕ Angelis M, McIntyre J II and
Gossler A: Maintenance of somite borders in mice requires the delta
homologue DII1. Nature. 386:717–721. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Maemura K, Yoshikawa H, Yokoyama K, Ueno
T, Kurose H, Uchiyama K and Otsuki Y: Delta-like 3 is silenced by
methylation and induces apoptosis in human hepatocellular
carcinoma. Int J Oncol. 42:817–822. 2013.PubMed/NCBI
|
|
47
|
Castel D, Mourikis P, Bartels SJ, Brinkman
AB, Tajbakhsh S and Stunnenberg HG: Dynamic binding of RBPJ is
determined by notch signaling status. Genes Dev. 27:1059–1071.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yuan Z, Friedmann DR, VanderWielen BD,
Collins KJ and Kovall RA: Characterization of CSL (CBF-1, Su (H),
Lag-1) mutants reveals differences in signaling mediated by Notch1
and Notch2. J Biol Chem. 287:34904–34916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kopan R and Ilagan MX: The canonical notch
signaling pathway: Unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Borggrefe T and Oswald F: The notch
signaling pathway: Transcriptional regulation at notch target
genes. Cell Mol Life Sci. 66:1631–1646. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kovall RA and Blacklow SC: Mechanistic
insights into notch receptor signaling from structural and
biochemical studies. Curr Top Dev Biol. 92:31–71. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Clayton T, Poe MM, Rallapalli S, Biawat P,
Savić MM, Rowlett JK, Gallos G, Emala CW, Kaczorowski CC, Stafford
DC, Arnold LA and Cook JM: A Review of the Updated Pharmacophore
for the Alpha 5 GABA(A) Benzodiazepine Receptor Model. Int J Med
Chem. 2015:4302482015.PubMed/NCBI
|
|
53
|
Zou JH, Xue TC, Sun C, Li Y, Liu BB, Sun
RX, Chen J, Ren ZG and Ye SL: Prognostic significance of Hes-1, a
downstream target of notch signaling in hepatocellular carcinoma
Asian Pac. J Cancer Prev. 16:3811–3816. 2015.
|
|
54
|
Friedmann DR and Kovall RA: Thermodynamic
and structural insights into CSL-DNA complexes. Protein Sci.
19:34–46. 2010.PubMed/NCBI
|
|
55
|
Dai W, Wang C, Wang F, Wang Y, Shen M,
Chen K, Cheng P, Zhang Y, Yang J, Zhu R, et al: Anti-miR-197
inhibits migration in HCC cells by targeting KAI 1/CD82. Biochem
Biophys Res Commun. 446:541–548. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang H, Zou J, Zhao B, Johannsen E,
Ashworth T, Wong H, Pear WS, Schug J, Blacklow SC, Arnett KL, et
al: Genome-wide analysis reveals conserved and divergent features
of Notch1/RBPJ binding in human and murine T-lymphoblastic leukemia
cells. Proc Natl Acad Sci USA. 108:14908–14913. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ridgway J, Zhang G, Wu Y, Stawicki S,
Liang WC, Chanthery Y, Kowalski J, Watts RJ, Callahan C, Kasman I,
et al: Inhibition of Dll4 signalling inhibits tumour growth by
deregulating angiogenesis. Nature. 444:1083–1087. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Sumazaki R, Shiojiri N, Isoyama S, Masu M,
Keino-Masu K, Osawa M, Nakauchi H, Kageyama R and Matsui A:
Conversion of biliary system to pancreatic tissue in Hes1-deficient
mice. Nat Genet. 36:83–87. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wolfe MS, Esler WP and Das C: Continuing
strategies for inhibiting alzheimer's gamma-secretase. J Mol
Neurosci. 19:83–87. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tchorz JS, Kinter J, Müller M, Tornillo L,
Heim MH and Bettler B: Notch2 signaling promotes biliary epithelial
cell fate specification and tubulogenesis during bile duct
development in mice. Hepatology. 50:871–879. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang QD, Xu MY, Cai XB, Qu Y, Li ZH and
Lu LG: Myofibroblastic transformation of rat hepatic stellate
cells: The role of Notch signaling and epithelial-mesenchymal
transition regulation. Eur Rev Med Pharmacol Sci. 19:4130–4138.
2015.PubMed/NCBI
|
|
62
|
Wen L, Liang C, Chen E, Chen W, Liang F,
Zhi X, Wei T, Xue F, Li G, Yang Q, Gong W, Feng X, Bai X and Liang
T: Regulation of Multi-drug Resistance in hepatocellular carcinoma
cells is TRPC6/Calcium Dependent. Sci Rep. 6:232692016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen QY, Jiao DM, Wang J, Hu H, Tang X,
Chen J, Mou H and Lu W: miR-206 regulates cisplatin resistance and
EMT in human lung adenocarcinoma cells partly by targeting MET.
Oncotarget. Mar 21–2016.(Epub ahead of print). doi:
10.18632/oncotarget.8229. View Article : Google Scholar
|
|
64
|
Lombardo Y, Faronato M, Filipovic A,
Vircillo V, Magnani L and Coombes RC: Nicastrin and Notch4 drive
endocrine therapy resistance and epithelial to mesenchymal
transition in MCF7 breast cancer cells. Breast Cancer Res.
16:R622014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Espinoza I, Pochampally R, Xing F, Watabe
K and Miele L: Notch signaling: Targeting cancer stem cells and
epithelial-to-mesenchymal transition. Onco Targets Ther.
6:1249–1259. 2013.PubMed/NCBI
|
|
66
|
Kang H, An HJ, Song JY, Kim TH, Heo JH,
Ahn DH and Kim G: Notch3 and Jagged2 contribute to gastric cancer
development and to glandular differentiation associated with MUC2
and MUC5AC expression. Histopathology. 61:576–586. 2012.PubMed/NCBI
|
|
67
|
Kodama Y, Hijikata M, Kageyama R,
Shimotohno K and Chiba T: The role of notch signaling in the
development of intrahepatic bile ducts. Gastroenterology.
127:1775–1786. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cheng HT, Kim M, Valerius MT, Surendran K,
Schuster-Gossler K, Gossler A, McMahon AP and Kopan R: Notch2, but
not Notch1, is required for proximal fate acquisition in the
mammalian nephron. Development. 134:801–811. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Loomes KM, Taichman DB, Glover CL,
Williams PT, Markowitz JE, Piccoli DA, Baldwin HS and Oakey RJ:
Characterization of notch receptor expression in the developing
mammalian heart and liver. Am J Med Genet. 112:181–189. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Pei J, Tang Z, Zang G and Yu Y: Blockage
of Notch1 signaling modulates the T-helper (Th)1/Th2 cell balance
in chronic hepatitis B patients. Hepatol Res. 40:799–805. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Nijjar SS, Crosby HA, Wallace L, Hubscher
SG and Strain AJ: Notch receptor expression in adult human liver: A
possible role in bile duct formation and hepatic
neovascularization. Hepatology. 34:1184–1192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Alvaro D, Bragazzi MC, Benedetti A, Fabris
L, Fava G, Invernizzi P, Marzioni M, Nuzzo G, Strazzabosco M and
Stroffolini T: Cholangiocarcinoma in Italy: A national survey on
clinical characteristics, diagnostic modalities and treatment.
Results from the ‘Cholangiocarcinoma’ committee of the Italian
association for the study of liver disease. Dig Liver Dis.
43:60–65. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Lee JS, Heo J, Libbrecht L, Chu IS,
Kaposi-Novak P, Calvisi DF, Mikaelyan A, Roberts LR, Demetris AJ,
Sun Z, et al: A novel prognostic subtype of human hepatocellular
carcinoma derived from hepatic progenitor cells. Nat Med.
12:410–416. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
74
|
Gao J, Chen Y, Wu KC, Liu J, Zhao YQ, Pan
YL, Du R, Zheng GR, Xiong YM, Xu HL and Fan DM: RUNX3 directly
interacts with intracellular domain of Notch1 and suppresses notch
signaling in hepatocellular carcinoma cells. Exp Cell Res.
316:149–157. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sekiya S and Suzuki A: Intrahepatic
cholangiocarcinoma can arise from Notch-mediated conversion of
hepatocytes. J Clin Invest. 122:3914–3918. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Weng AP and Aster JC: Multiple niches for
notch in cancer: Context is everything. Curr Opin Genet De.
14:48–54. 2004. View Article : Google Scholar
|
|
77
|
Lu J, Zhou Y, Hu T, Zhang H, Shen M, Cheng
P, Dai W, Wang F, Chen K, Zhang Y, et al: Notch Signaling
Coordinates Progenitor Cell-Mediated Biliary Regeneration Following
Partial Hepatectomy. Sci Rep. 6:227542016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Espinoza I and Miele L: Notch inhibitors
for cancer treatment. Pharmacol Ther. 139:95–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hayashi Y, Osanai M and Lee GH: NOTCH2
signaling confers immature morphology and aggressiveness in human
hepatocellular carcinoma cells. Oncol Rep. 34:1650–1658.
2015.PubMed/NCBI
|
|
80
|
Kohn A, Rutkowski TP, Liu Z, Mirando AJ,
Zuscik MJ, O'Keefe RJ and Hilton MJ: Notch signaling controls
chondrocyte hypertrophy via indirect regulation of Sox9. Bone Res.
3:150212015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Fowlkes BJ and Robey EA: A reassessment of
the effect of activated Notch1 on CD4 and CD8 T cell development. J
Immunol. 169:1817–1821. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhu R, Yang J, Xu L, Dai W, Wang F, Shen
M, Zhang Y, Zhang H, Chen K, Cheng P, et al: Diagnostic performance
of des-γ-carboxy prothrombin for hepatocellular carcinoma: A
meta-analysis. Gastroenterol Res Pract. 2014:5293142014. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Nalesnik MA, Tseng G, Ding Y, Xiang GS,
Zheng ZL, Yu Y, Marsh JW, Michalopoulos GK and Luo JH: Gene
deletions and amplifications in human hepatocellular carcinomas:
Correlation with hepatocyte growth regulation. Am J Pathol.
180:1495–1508. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Villanueva A, Alsinet C, Yanger K, Hoshida
Y, Zong Y, Toffanin S, Rodriguez-Carunchio L, Solé M, Thung S,
Stanger BZ and Llovet JM: Notch signaling is activated in human
hepatocellular carcinoma and induces tumor formation in mice.
Gastroenterology. 143:1660–1669. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Fan B, Malato Y, Calvisi DF, Naqvi S,
Razumilava N, Ribback S, Gores GJ, Dombrowski F, Evert M, Chen X
and Willenbring H: Cholangiocarcinomas can originate from
hepatocytes in mice. J Clin Invest. 122:2911–2915. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Viatour P, Ehmer U, Saddic LA, Dorrell C,
Andersen JB, Lin C, Zmoos AF, Mazur PK, Schaffer BE, Ostermeier A,
et al: Notch signaling inhibits hepatocellular carcinoma following
inactivation of the RB pathway. J Exp Med. 208:1963–1976. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Vincent F, Bonnin P, Clemessy M, Contrerès
JO, Lamandé N, Gasc JM, Vilar J, Hainaud P, Tobelem G, Corvol P and
Dupuy E: Angiotensinogen delays angiogenesis and tumor growth of
hepatocarcinoma in transgenic mice. Cancer Res. 69:2853–2860. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang F, Dai W, Wang Y, Shen M, Chen K,
Cheng P, Zhang Y, Wang C, Li J, Zheng Y, et al: The synergistic in
vitro and in vivo antitumor effect of combination therapy with
salinomycin and 5-fluorouracil against hepatocellular carcinoma.
PLoS One. 9:e974142014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sekiya S and Suzuki A: Intrahepatic
cholangiocarcinoma can arise from notch-mediated conversion of
hepatocytes. J Clin Invest. 122:3914–3918. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ishimura N, Bronk SF and Gores GJ:
Inducible nitric oxide synthase up-regulates notch-1 in mouse
cholangiocytes: Implications for carcinogenesis. Gastroenterology.
128:1354–1368. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Roma J, Masià A, Reventós J, de Sánchez
Toledo J and Gallego S: Notch Pathway Inhibition Significantly
Reduces Rhabdomyosarcoma Invasiveness and Mobility. Vitro Clin
Cancer Res. 17:505–513. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Olsauskas-Kuprys R, Zlobin A and Osipo C:
Gamma secretase inhibitors of notch signaling. Onco Targets Ther.
6:943–955. 2013.PubMed/NCBI
|