Introduction
Esophageal cancer (EC), the eighth most common type
of cancer in the world, may be pathologically divided into two
major categories: Esophageal adenocarcinoma and esophageal squamous
cell carcinoma (ESCC) (1,2). In China, EC is highly prevalent, and is
the fourth-ranked cancer in terms of incidence (3). Due to the difficulties in early
diagnosis and poor treatment efficacy, the 5-year survival rate of
ESCC is considerably low, ranging from 15–25% (1–4). Thus far,
numerous studies have been conducted to attempt to clarify the
fundamental molecular mechanisms and biological behavior of
ESCC.
Abnormalities in the cell cycle are essential in the
process of human carcinogenesis, resulting in an increase in cell
proliferation and/or a reduction in the death of abnormal cells
(5). Several key proteins are
required to maintain the integrity of the normal cell cycle, and
aberrant expression of proteins such as cyclins A and B1 leads to
an abnormal cell cycle (5–8). Cyclin A, as an important checkpoint
mechanism in the G1-S transition of the cell cycle, is expressed
just prior to the start of DNA synthesis, while cyclin B1 acts as a
mitotic cyclin protein in the G2-M transition (9). It has been verified that the expression
of cyclin A and cyclin B1 is remarkably upregulated in human ESCC,
as opposed to neighboring normal tissues (10,11).
Therefore, cyclins A and B1 may be implicated in the tumorigenesis
and evolution of malignancies (9–13). Early
mitotic inhibitor-1 (Emi1), as a cell cycle regulator, governs the
progression to S phase and mitosis by stabilizing key
ubiquitination substrates of anaphase-promoting complex, including
cyclins A and B1 (14–16). It has been previously reported that
excess Emi1 added to Xenopus egg extracts prevents cyclins A
and B1 degradation, and is required for accumulation of cyclins A
and B1 (17). In addition,
upregulation of Emi1 messenger RNA exists in numerous malignant
tumors, and its overexpression produces mitotic defects, possibly
resulting in tumorigenesis (18–20).
Despite the frequent dysfunction of the cell cycle
machinery in human ESCC, the expression and clinical significance
of Emil protein in ESCC remain unclear. In the present study, Emi1
protein expression was determined by immunohistochemistry and
immunoblotting in ESCC, and the associations between Emi1 and
clinicopathological variables and prognosis were investigated. In
addition, ECA109 cells were transfected with Emi1 small interfering
(si)RNA vectors in vitro to investigate the functionality of
Emi1 as a potential therapeutic target for ESCC.
Materials and methods
Patients and tissue specimens
In the present study, 90 ESCC (55 males and 35
females) aged 31–80 years (mean, 60 years) were retrieved from the
archival files of the Department of Pathology of the Affiliated
Hospital of Nantong University (Nantong, China) from January 2000
to December 2004. None of the patients were treated with radiation,
chemotherapy or immunotherapy prior to operation. Upon signing
informed consent, patients were questioned regarding their
demographic characteristics. Histological differentiation was
divided into three grades, namely, grade I (well differentiated),
II (moderately differentiated) and III (poorly differentiated). The
90 patients examined were grouped into the above three grades (20
patients into grade I, 50 into grade II and 20 into grade III). In
addition, invasion of lymphatic and blood vessels was evaluated
microscopically.
Tissue specimens were treated as soon as surgical
removal was completed. For histological examination, all tumorous
and para-cancerous tissue portions were processed into 10% buffered
formalin-fixed, paraffin-embedded blocks. Protein expression was
analyzed in 8 tumorous and para-cancerous tissue samples stored at
−80°C.
Immunohistochemical analyses
The tissue sections were deparaffinized through a
graded ethanol series, and endogenous peroxidase activity was
blocked by immersion in 0.3% hydrogen peroxide
(H2O2). Next, the sections were treated in 10
mmol/l citrate buffer (pH 6.0; Beijing Zhongshan Jinqiao
Biotechnology Co., Ltd., Beijing, China), and heated to 121°C in a
pressure cooker for 20 min for antigen retrieval. Upon washing in
phosphate-buffered saline (PBS) (pH 7.2), 10% goat serum (Beijing
Zhongshan Jinqiao Biotechnology Co., Ltd.) was applied for 1 h at
room temperature to block nonspecific reactions. Then, the sections
were incubated for 12 h at 4°C with anti-Emi1 rabbit polyclonal
antibody (1:100; cat. no. sc-30182; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA), and anti-Ki-67 mouse monoclonal antibody (1:100;
clone 7B11; Zymed; cat. no. MA5-15690; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Negative control sections were also
processed in parallel with a nonspecific immunoglobulin (Ig)G (cat.
no. I5006-10MG; Sigma-Aldrich, St. Louis, MO, USA) applied at the
same concentration as the above primary antibodies. All sections
were treated using the peroxidase-antiperoxidase method (Dako,
Glostrup, Denmark). Upon washing in PBS, the peroxidase reaction
was visualized by incubating the slides with 3,3′-diaminobenzidine
tetrahydrochloride in 0.05 mol/l Tris buffer (pH 7.6) including
0.03% H2O2. Upon washing in water, the slides
were counterstained with hematoxylin, dehydrated in a graded
alcohol series and coverslipped.
Immunohistochemical evaluation
All the immunostained sections were assessed in a
blinded approach without knowing the patients' clinical and
pathological variables. Regarding Emil assessment, staining
intensity was evaluated using a four rating-level-scheme, where
scores ranging from 0 to 3 indicated negative, weak, medium and
strong staining, respectively. For extent of staining, a five
rating-level-scheme was employed. Thus, based on the total amount
of positive stained areas in the whole carcinoma region, the extent
of staining was evaluated with scores ranging from 0 to 4 as
follows: 0, 0%; 1, 1–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%. The
sum of intensity and extent scores was used as the final staining
score (0–7) for Emi1. Tumors were considered to be positive when
their final staining scores were ≥3 (21). In each specimen, five high-power
fields were randomly selected for Ki-67 assessment, together with
examination of nuclear staining. To determine the medium percentage
of immunostained cells among the total number of cells, >500
cells were counted. To avoid possible technical errors, staining
was performed twice, and similar results were achieved. All the
aforementioned evaluations were conducted independently by two
investigators with identical results.
Cell culture and cell cycle
analysis
The human ESCC cell line ECA109 was purchased from
the Chinese Academy of Sciences (Beijing, China) and cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.) with 10%
heat-inactivated fetal calf serum (Gibco; Thermo Fisher Scientific,
Inc.), 2 mM L-glutamine and 100 U/ml penicillin-streptomycin
mixture (Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5%
CO2. Cells were fixed in 70% ethanol for 1 h at 4°C, and
then incubated with 1 mg/ml RNase A for 30 min at 37°C for cell
cycle analysis. Next, cells were stained with propidium iodide (50
mg/ml; BD Biosciences, San Jose, CA, USA) and analyzed using a flow
cytometer (FACScan; BD Biosciences) and CellQuest Pro Acquisition
and Analysis software (BD Biosciences).
siRNA and transfection
The pSilencer 4.1-CMV Emi1-siRNA expression vectors
were constructed by incorporating the siRNA targeting nucleotide
residues AAGCACTAGAGACCAGTAGAC (Emi1-si1) and ACTTGCTGCCAGTTCTCA
(Emi1-si2) in the pSilencer 4.1-CMV vector (Thermo Fisher
Scientific, Inc.). ECA109 cells were seeded the day preceding
transfection using RPMI-1640 medium with 10% fetal calf serum but
without antibiotics. Transient transfection of Emi1-siRNA and
control siRNA vectors was conducted using Lipofectamine®
LTX & PLUS™ reagent (Thermo Fisher Scientific, Inc.) in
Opti-MEM® (Thermo Fisher Scientific, Inc.), as suggested
by the manufacturer. Cells were incubated with the pSilencer
vectors and Lipofectamine® LTX & PLUS™ reagent
complexes for 4 h at 37°C, and harvested 48 h post-transfection.
The experiments were repeated three times.
Cell Counting Kit (CCK)-8 assay
Cell proliferation was detected by the commercial
CCK-8 method (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan), according to the manufacturer's protocol. Shortly, cells
were seeded into 96-well cell culture cluster plates at a
concentration of 2×104 cells/well in volumes of 100 µl,
and cultured overnight. CCK-8 reagent was added to a subset of
wells containing cells under different treatments, and incubated
for 2 h at 37°C. The absorbance was next quantified at 450 nm with
an automated plate reader.
Western blot analysis
Tissues and cells were rapidly homogenized in a
homogenization buffer containing 1% Triton X-100, 1 M Tris-HCl (pH
7.5), 10% sodium dodecyl sulfate (SDS), 1% Nonidet ™ P-40, 10 µg/ml
leupeptin, 0.5% sodium deoxycholate, 10 µg/ml aprotinin, 0.5 M
ethylenediaminetetraacetic acid and 1 mM phenylmethylsulfonyl
fluoride, prior to be centrifuged at 10,000 g for 30 min to collect
the supernatant. Protein concentrations were measured with a
Bio-Rad protein assay (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). 2X SDS loading buffer was used to dilute the supernatant,
which was next boiled. Proteins were separated by
SDS-polyacrylamide gel electrophoresis, and then transferred to
polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked with 5% dried skimmed milk in
Tris-buffered saline and Tween 20, containing 20 mM Tris, 0.05%
Tween 20, and 150 mM NaCl. Following 2 h-incubation at room
temperature, the membranes were incubated overnight with the
following antibodies: Anti-Emi1 (1:500; cat. no. sc-30182),
anti-cyclin A (1:500; cat. no. sc-751), anti-cyclin B1 (1:500; cat.
no. sc-25764), anti-proliferating cell nuclear antigen (1:1,000;
cat. no. sc-56) and anti-glyceraldehyde 3-phosphate dehydrogenase
(1:1,000; cat. no. sc-25778). All the above primary antibodies were
purchased from Santa Cruz Biotechnology, Inc. Horseradish
peroxidase-linked IgG (cat. no. sc-2030; Santa Cruz Biotechnology,
Inc.) was used as a secondary antibody. The immunoreactive bands
were visualized by chemiluminescence (NEN Life Science Products,
Inc., Boston, MA, USA), and exposed to X-ray films, which were then
scanned using a Molecular Dynamics densitometer (GE Healthcare Life
Sciences, Chalfont, UK) and the Odyssey infrared imaging system
(LI-COR Biotechnology, Lincoln, NE, USA). The experiments were
repeated on three separate occasions.
Statistical analysis
The statistical software Stata version 11.0
(StataCorp LP, College Station, TX, USA) was used for statistical
analysis. The association between Emi1 protein expression and
clinicopathological factors was analyzed using the χ2
test. Survival curves were plotted using the Kaplan-Meier method,
and the log-rank test was employed for analysis. Multivariate
analysis was performed using Cox's proportional hazards model. The
risk rate and its 95% confidence interval were recorded for each
marker. P<0.05 was considered to indicate a statistically
significant difference.
Results
The expression of Emi1 in human ESCC
tissue samples
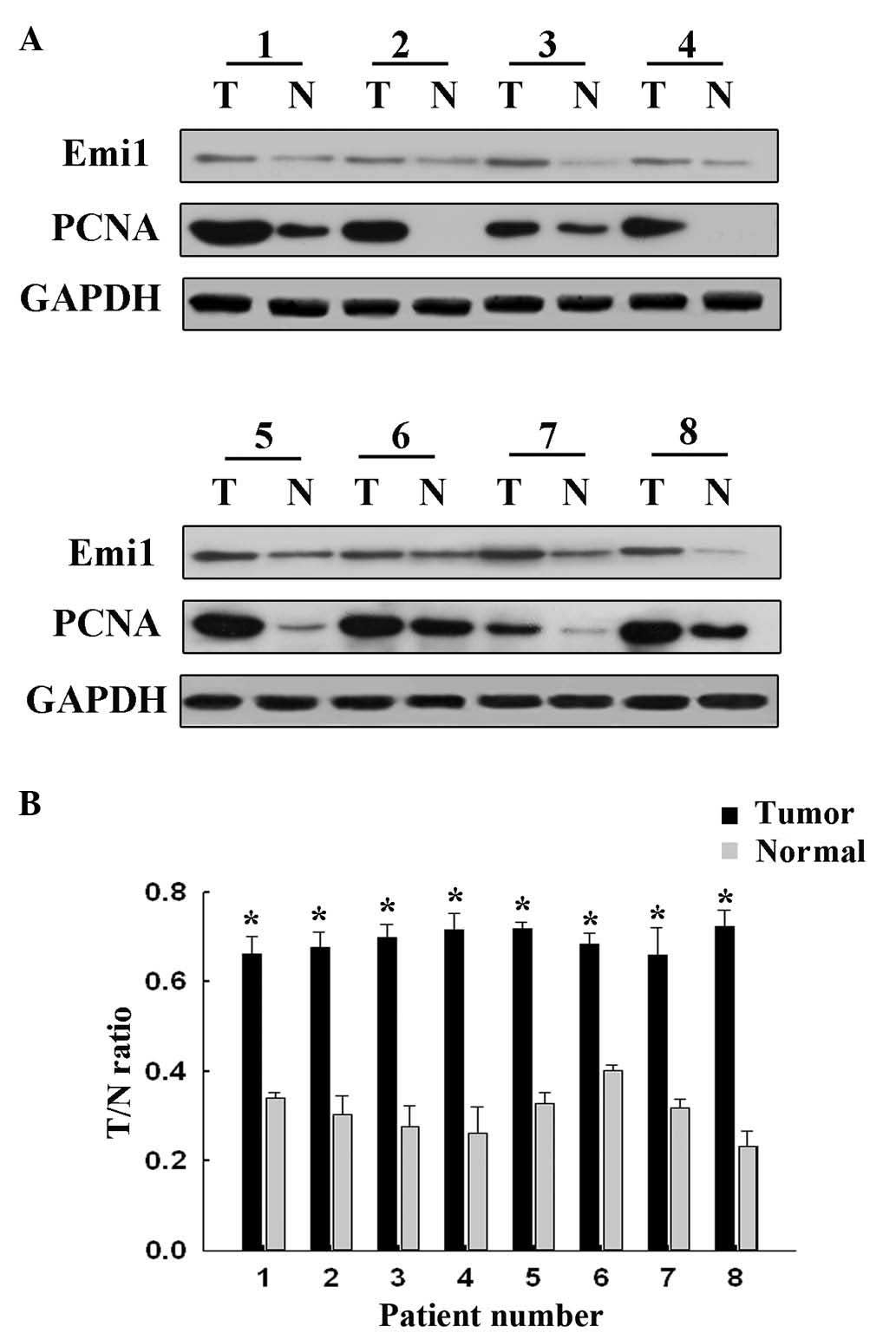
To reveal the role of Emi1 in ESCC, the expression
of Emi1 protein was detected by western blot analysis in 8 paired
frozen ESCC tumor tissues and para-cancerous tissues. The results
revealed that Emi1 expression was significantly increased in 6 of 8
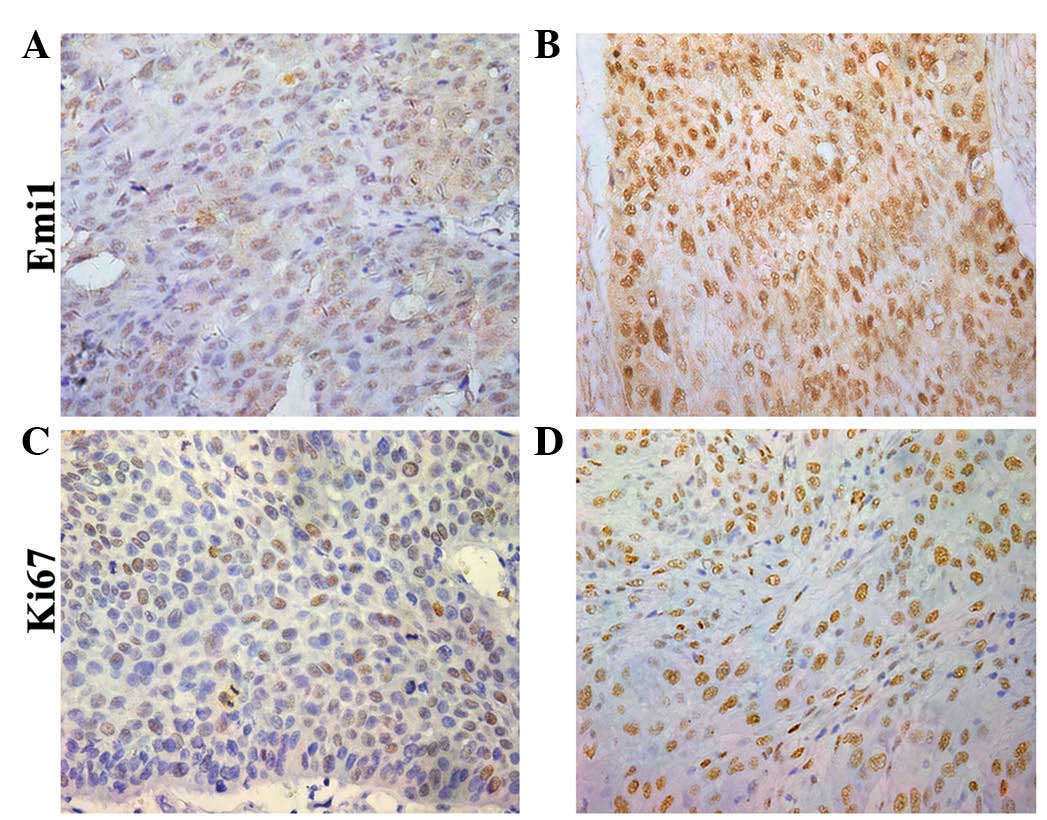
tumors, compared with para-cancerous tissues (P<0.05; Fig. 1). In addition, expression of Emi1 and
Ki-67 was simultaneously detected and further verified in 90 ESCC
samples by immunohistochemical staining. The results indicated that
Emi1 and Ki-67 proteins were overexpressed in ESCC specimens,
whereas in the matching para-cancerous tissue samples, their
expression was weak or absent (Fig.
2).
Correlation of Emi1 protein expression
with clinicopathological variables in human ESCC tissues
The association between Emi1 expression and
clinicopathological variables was evaluated. For statistical
analysis of Emi1 expression, the ESCC tissue specimens were
classified into positive or negative groups, based on their final
staining scores. As presented in Table
I, Emi1 expression was correlated with histological
differentiation (P=0.032) and lymphatic metastasis (P=0.006), while
no correlation existed between Emi1 expression and other
prognostics factors, including age, gender, tumor diameter and
tumor depth. Furthermore, a positive correlation existed between
Emi1 and Ki-67 expression (which is indicative of proliferative
activity) in the majority of specimens (P=0.028).
 | Table I.Association between Emi1 protein
expression and clinicopathological features of esophageal squamous
cell carcinoma specimens. |
Table I.
Association between Emi1 protein
expression and clinicopathological features of esophageal squamous
cell carcinoma specimens.
|
|
| Emi1 expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Cases (n) | Negative (final
score, 0–2; n=32) | Positive (final score
3–7; n=58) | P-value |
|---|
| Age, years |
|
|
| 0.244 |
| ≤60 | 46 | 19 | 27 |
|
|
>60 | 44 | 13 | 31 |
|
| Gender |
|
|
| 0.120 |
| Male | 55 | 23 | 32 |
|
|
Female | 35 | 9 | 26 |
|
| Histological
differentiation |
|
|
| 0.032a |
| Well | 20 | 12 | 8 |
|
|
Moderately | 50 | 15 | 35 |
|
|
Poorly | 20 | 5 | 15 |
|
| Lymphatic
metastasis |
|
|
| 0.006a |
|
Positive | 23 | 9 | 34 |
|
|
Negative | 67 | 23 | 24 |
|
| Tumor diameter,
cm |
|
|
| 0.755 |
| ≤5 | 80 | 28 | 52 |
|
|
>5 | 10 | 4 | 6 |
|
| Tumor depth |
|
|
| 0.079 |
| T1 | 7 | 3 | 4 |
|
| T2 | 6 | 5 | 1 |
|
| T3 | 22 | 7 | 15 |
|
| T4 | 55 | 17 | 38 |
|
| Ki-67 expression,
% |
|
|
| 0.028a |
|
≤0.78 | 45 | 21 | 24 |
|
|
>0.78 | 45 | 11 | 34 |
|
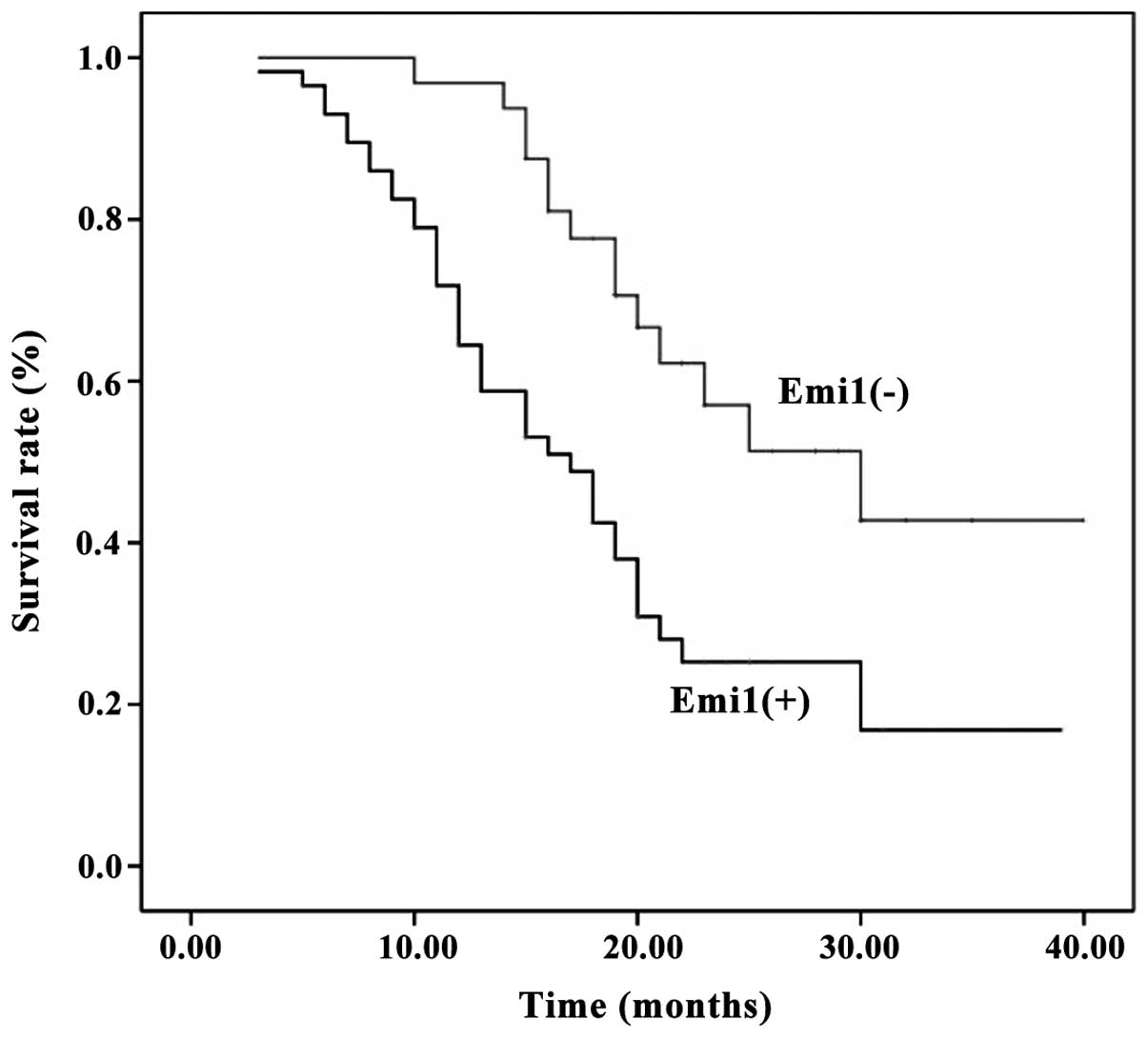
Prognostic significance of Emi1
expression in human ESCC samples
Survival information was available for all patients
at the end of clinical follow-up. Kaplan-Meier survival curves for
univariate analysis demonstrated that Emi1 protein overexpression
resulted in a poor survival rate (P<0.05) (Fig. 3). According to the Cox's proportional
hazards regression model, Emi1 expression, Ki67 expression and
lymphatic metastasis were independent prognostic factors of poor
prognosis in ESCC patients (Table
II).
 | Table II.Contribution of various potential
prognostic factors to survival in patients with esophageal squamous
cell carcinoma. |
Table II.
Contribution of various potential
prognostic factors to survival in patients with esophageal squamous
cell carcinoma.
| Variables | Hazard ratio | 95% Confidence
interval | P-value |
|---|
| Age | 1.143 | 0.655–1.995 | 0.638 |
| Gender | 0.898 | 0.483–1.668 | 0.733 |
| Histological
differentiation | 0.641 | 0.416–1.624 | 0.571 |
| Tumor diameter | 1.485 | 0.652–3.383 | 0.346 |
| Tumor depth | 1.171 | 0.800–1.713 | 0.416 |
| Lymphatic
metastasis | 0.822 | 0.421–0.976 | 0.018a |
| Emi1
expression | 1.967 | 1.024–3.782 | 0.042a |
| Ki-67
expression | 3.047 | 1.554–5.973 | 0.001a |
Emi1 is involved in ESCC cell
proliferation
To demonstrate whether Emi1 expression was cell
cycle-dependent in ESCC cells, the cell cycle was analyzed
following serum starvation and upon re-feeding with serum. ECA109
cells were arrested in the G1 phase by serum deprivation for 72 h,
and the percentage of cells in the G1 phase increased from 39.08 to
73.35% under these conditions (Fig.
4A). Upon serum addition, the cells were released from the G1
phase and reentered the S phase. As expected, the expression of
Emi1 increased as early as 4 h post-serum stimulation in ECA109
cells. Additionally, the expression of cyclins A and B was
upregulated (Fig. 4B and C). These
results indicate that Emi1 is important role in the regulation of
cell proliferation.
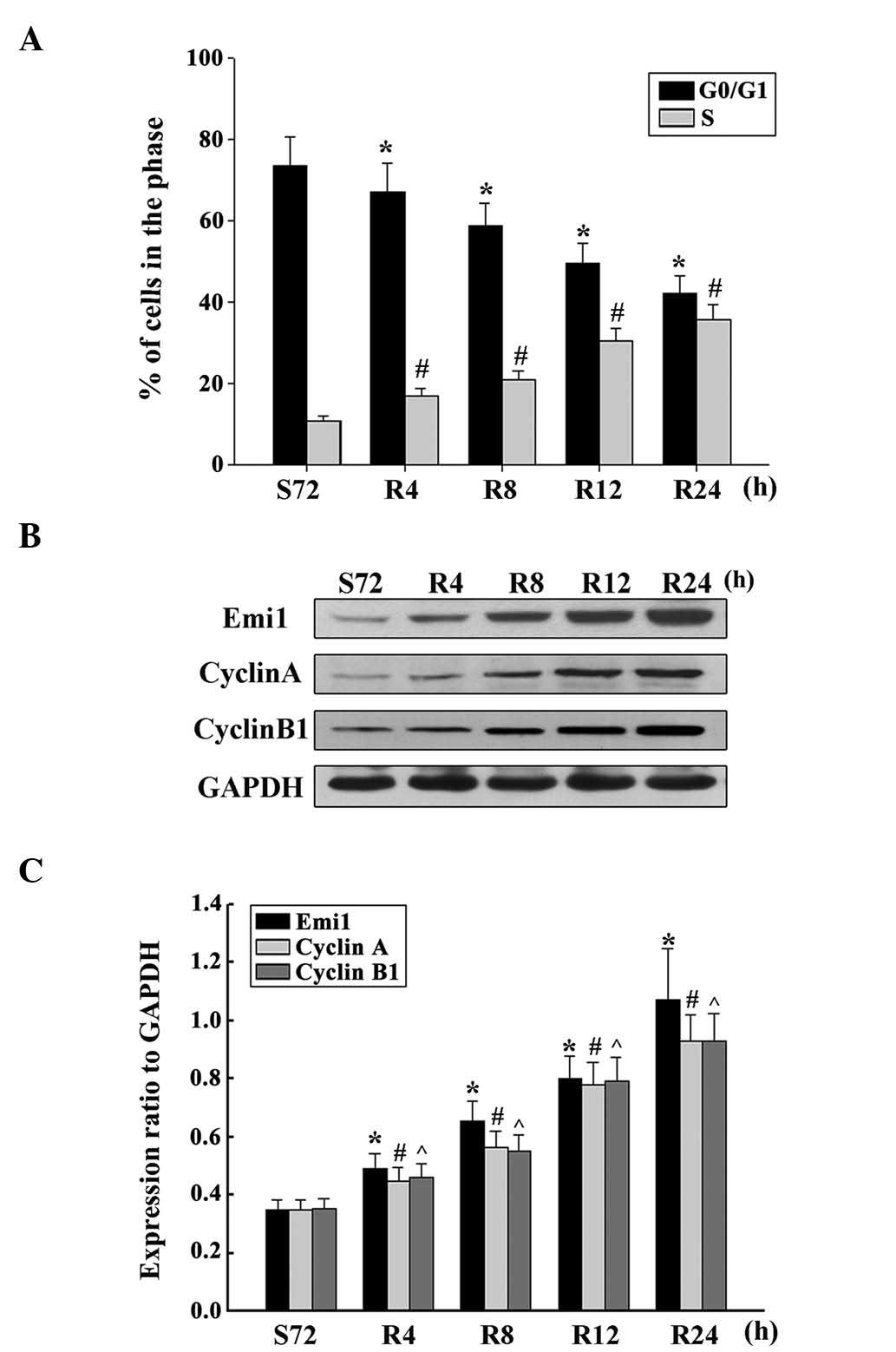 | Figure 4.Overexpression of Emi1 and cell
cycle-related molecules in proliferating esophageal squamous cell
carcinoma cells. (A) ECA109 cells were synchronized at G1, and
induced to progress into the cell cycle by serum addition at 0, 4,
8, 12 and 24 h. Upon cell cycle progression induction, the majority
of cells were in the S phase. Data represent the mean ± standard
deviation of three independent experiments *,#P<0.01
vs. control (S72 h). (B) ECA109 cells were serum starved for 72 h,
and following serum addition, cell lysates were prepared and
analyzed by western blotting using antibodies against Emi1, cyclin
A and cyclin B1. GAPDH was used as a control for protein loading
and integrity. (C) Ratio of Emi1, cyclin A and cyclin B1 protein
levels to those of GAPDH for each time point, as analyzed by
densitometry. Data represent the mean ± standard error of the mean
(n=3). *,#,^P<0.01, vs. control (S72 h). S, serum
starvation; R, serum addition; Emi1, early mitotic inhibitor-1;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase. |
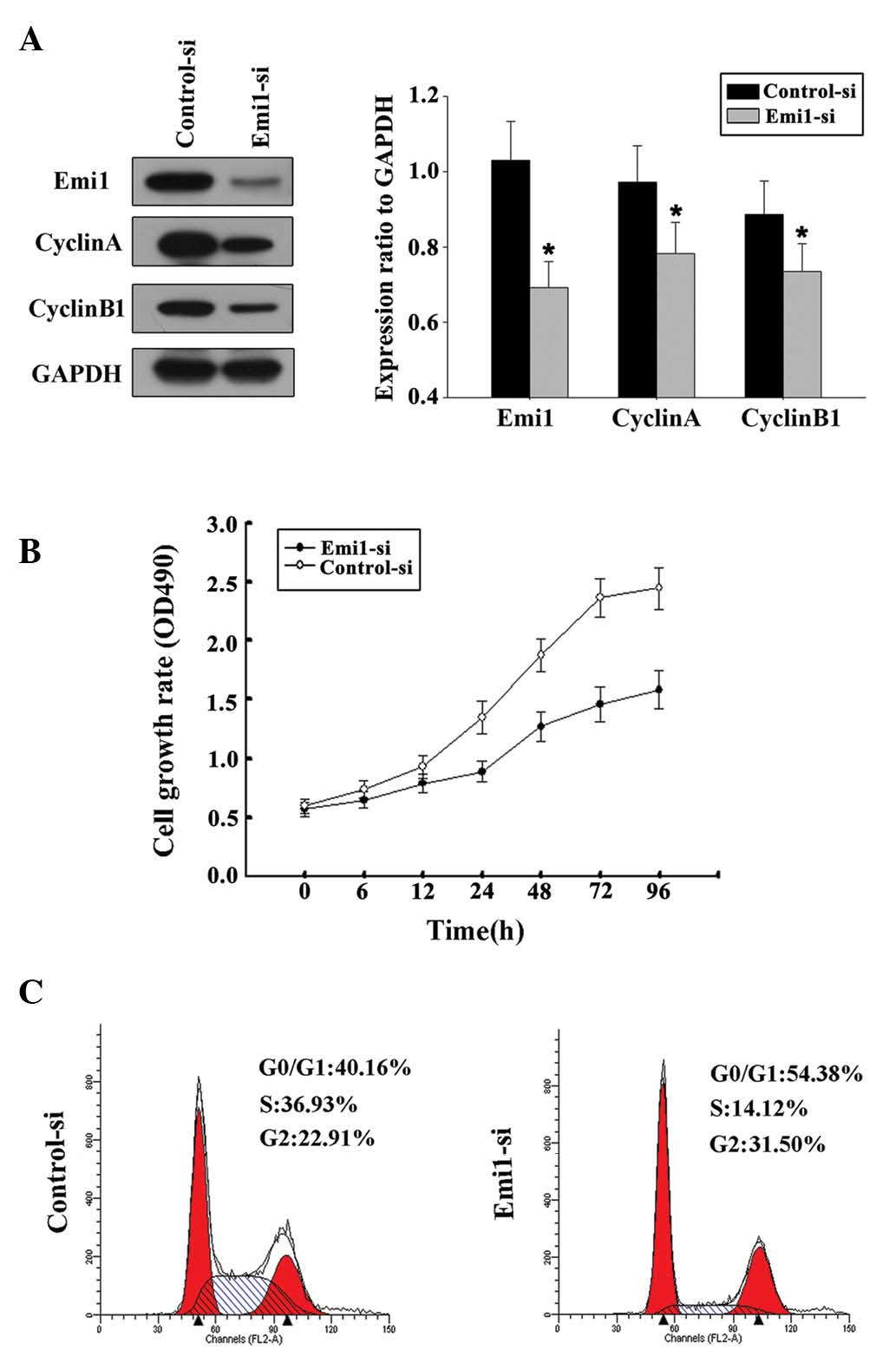
siRNA targeting Emi1 inhibits ESCC
cell proliferation
By transfecting ECA109 cells with Emil-siRNA or
control siRNA, the influence of Emil on ESCC cell proliferation was
further evaluated. In the present study, two siRNAs targeting Emi1
(Emi1-si1 and Emi1-si2) were tested, and the efficiency of Emi1
gene silencing was measured by immunoblotting. The results
demonstrated that Emi1-si1 exerted a better silencing effect.
Decreased expression of cyclins A and B was detected in Emi-si1
(Fig. 5A). This result was in
agreement with a previous study that reported that Emi1 promoted
mitotic entry to enable accumulation of cyclins A and B1 (15). Flow cytometry confirmed that Emi1-si
could inhibit the cell cycle at the G1-S transition (Fig. 5B). Silencing of Emi1 led to a
significant inhibition of the rate of cell growth (Fig. 5C). These findings further suggested
that Emi1 may be involved in the regulation of the G1-S transition,
which could be responsible for the increased growth rate of ESCC
cells.
Discussion
Thanks to the advances in molecular and cellular
biology of tumors, it is well known that the occurrence of EC is
partly due to acquired alterations in oncogenes and tumor
suppressor genes (4). Cell
proliferation, differentiation and cell cycle control disorders are
important features in cancer (1).
Misregulation of the G1-S transition is an essential component of
the cellular transformation process in the cell cycle, and G1-S
regulatory defects have been reported in numerous types of human
malignancies (22–24).
Emi1 was firstly identified in a yeast two-hybrid
screen for F-box proteins using S-Phase kinase-associated protein 1
as bait (17). In mammalian cells,
Emi1 levels are regulated during the cell cycle, with its
transcription being induced at the G1-S transition under the
control of E2F, which is required to stabilize cyclins A and B, and
enables cells to initiate the S phase (18). A previous study indicated that Emi1 is
accumulated in ovarian clear cell carcinoma (25), and Liu et al (26) reported that Emi1 overexpression may be
a poor prognostic marker for breast carcinoma patients. These
findings suggested that the Emi1 gene may be involved in human cell
cycle disorders and may lead to oncogenesis.
To the best of our knowledge, Emi1 expression in
ESCC specimens has not been actively studied thus far. The present
study is the first to report that Emi1 protein is overexpressed in
human ESCC, and analyze a possible association between Emi1
expression and clinicopathological factors and prognosis of
patients with ESCC. In the present study, immunoblotting examined
the protein expression levels of Emi1 in ESCC specimens and
para-cancerous tissues. Furthermore, the expression of Emi1 was
investigated to confirm the participation of Emi1 in tumor
progression by immunohistochemical staining. High expression of
Emi1 as a useful marker of tumor proliferative activity (27,28) was
correlated with overexpression of Ki-67. Therefore, increased Emi1
levels may be closely associated with the pathogenesis of ESCC. In
addition, the association between Emi1 expression and
clinicopathological variables and patient prognosis was evaluated.
The results revealed that Emi1 expression was strongly correlated
with histological differentiation and lymphatic metastasis. The
results of survival analysis demonstrated that high expression of
Emi1 was strongly correlated with poor prognosis, while
multivariate analysis revealed that high expression of Emi1 was an
independent unfavorable prognostic factor. These findings indicated
that Emi1 may be a reliable factor of prognosis in patients with
EC.
The expression of Emi1 during cell cycle progression
was further detected in ESCC cells in vitro. The results
indicated that the expression of Emi1 was upregulated during the
G1-S phase transition. These results confirmed the association of
Emi1 expression with ESCC development. Furthermore, the present
data revealed that silencing Emi1 expression could suppress ECA109
cell proliferation. This observation is consistent with a previous
study in which Emi1 promoted mitotic entry and enabled accumulation
of cyclins A and B1 (15).
Hsu et al (18)
demonstrated that upregulation of Emi1 at the transcriptional level
occurred in various tumors. At the G1-S transition, Emi1 was
transcriptionally induced by the transcription factor E2F, which is
associated with cell cycle control (18). The E2F signaling pathway is frequently
activated in highly proliferative cells, and the central proteins
of the retinoblastoma (Rb)/E2F signaling pathway, including
p16INK4a, Rb and cyclin D, are frequently mutated in
cancer (29). This E2F activation is
expected to cause an increase in Emi1 levels.
In summary, the present study demonstrated that Emi1
protein expression was increased in ESCC, and positively correlated
with ESCC cell proliferation, indicating that Emi1 may play a key
role in ESCC and it is an independent candidate prognostic factor
for ESCC patients. However, further studies are required to clarify
the molecular mechanisms of Emi1 in the pathogenesis of ESCC.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pourhoseingholi MA, Vahedi M and
Baghestani AR: Burden of gastrointestinal cancer in Asia; an
overview. Gastroenterol Hepatol Bed Bench. 8:19–27. 2015.PubMed/NCBI
|
|
3
|
Zeng H, Zheng R, Guo Y, Zhang S, Zou X,
Wang N, Zhang L, Tang J, Chen J, Wei K, et al: Cancer survival in
China, 2003–2005: A population-based study. Int J Cancer.
136:1921–1930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lagergren J and Lagergren P: Recent
developments in esophageal adenocarcinoma. CA Cancer J Clin.
63:232–248. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Johnson DG and Walker CL: Cyclins and cell
cycle checkpoints. Annu Rev Pharmacol Toxicol. 39:295–312. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wolgemuth DJ and Roberts SS: Regulating
mitosis and meiosis in the male germ line: Critical functions for
cyclins. Philos Trans R Soc Lond B Biol Sci. 365:1653–1662. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kronja I and Orr-Weaver TL: Translational
regulation of the cell cycle: When, where, how and why? Philos
Trans R Soc Lond B Biol Sci. 366:3638–3652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Fei M, Cheng C, Zhang D, Lu J, He
S, Zhao Y, Wang Y and Shen A: Jun activation domain-binding protein
1 negatively regulate p27 kip1 in non-Hodgkin's lymphomas. Cancer
Biol Ther. 7:460–467. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song Y, Zhao C, Dong L, Fu M, Xue L, Huang
Z, Tong T, Zhou Z, Chen A, Yang Z, et al: Overexpression of cyclin
B1 in human esophageal squamous cell carcinoma cells induces tumor
cell invasive growth and metastasis. Carcinogenesis. 29:307–315.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takeno S, Noguchi T, Kikuchi R, Uchida Y,
Yokoyama S and Müller W: Prognostic value of cyclin B1 in patients
with esophageal squamous cell carcinoma. Cancer. 94:2874–2881.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nozoe T, Korenaga D, Kabashima A, Ohga T,
Saeki H and Sugimachi K: Significance of cyclin B1 expression as an
independent prognostic indicator of patients with squamous cell
carcinoma of the esophagus. Clin Cancer Res. 8:817–822.
2002.PubMed/NCBI
|
|
12
|
Chetty R and Simelane S: p53 and cyclin A
protein expression in squamous carcinoma of the oesophagus. Pathol
Oncol Res. 5:193–196. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bernis C, Vigneron S, Burgess A, Labbé JC,
Fesquet D, Castro A and Lorca T: Pin1 stabilizes Emi1 during G2
phase by preventing its association with SCF(betatrcp). EMBO Rep.
8:91–98. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miller JJ, Summers MK, Hansen DV, Nachury
MV, Lehman NL, Loktev A and Jackson PK: Emi1 stably binds and
inhibits the anaphase-promoting complex/cyclosome as a
pseudosubstrate inhibitor. Genes Dev. 20:2410–2420. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Moshe Y, Bar-On O, Ganoth D and Hershko A:
Regulation of the action of early mitotic inhibitor 1 on the
anaphase-promoting complex/cyclosome by cyclin-dependent kinases. J
Biol Chem. 286:16647–16657. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Reimann JD, Gardner BE, Margottin-Goguet F
and Jackson PK: Emi1 regulates the anaphase-promoting complex by a
different mechanism than Mad2 proteins. Genes Dev. 15:3278–3285.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reimann JD, Freed E, Hsu JY, Kramer ER,
Peters JM and Jackson PK: Emi1 is a mitotic regulator that
interacts with Cdc20 and inhibits the anaphase promoting complex.
Cell. 105:645–655. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hsu JY, Reimann JD, Sørensen CS, Lukas J
and Jackson PK: E2F-dependent accumulation of hEmi1 regulates S
phase entry by inhibiting APC (Cdh1). Nat Cell Biol. 4:358–366.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Margottin-Goguet F, Hsu JY, Loktev A,
Hsieh HM, Reimann JD and Jackson PK: Prophase destruction of Emi1
by the SCF (betaTrCP/Slimb) ubiquitin ligase activates the anaphase
promoting complex to allow progression beyond prometaphase. Dev
Cell. 4:813–826. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao Y, Tang Q, Ni R, Huang X, Wang Y, Lu
C, Shen A, Wang Y, Li C, Yuan Q, et al: Early mitotic inhibitor-1,
an anaphase-promoting complex/cyclosome inhibitor, can control
tumor cell proliferation in hepatocellular carcinoma: Correlation
with Skp2 stability and degradation of p27(Kip1). Hum Pathol.
44:365–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Masunaga R, Kohno H, Dhar DK, Ohno S,
Shibakita M, Kinugasa S, Yoshimura H, Tachibana M, Kubota H and
Nagasue N: Cyclooxygenase-2 expression correlates with tumor
neovascularization and prognosis in human colorectal carcinoma
patients. Clin Cancer Res. 6:4064–4068. 2000.PubMed/NCBI
|
|
22
|
Roth JA and Cristiano RJ: Gene therapy for
cancer: What have we done and where are we going? J Natl Cancer
Inst. 89:21–39. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Roncalli M, Bosari S, Marchetti A,
Buttitta F, Bossi P, Graziani D, Peracchia A, Bonavina L, Viale G
and Coggi G: Cell cycle-related gene abnormalities and product
expression in esophageal carcinoma. Lab Invest. 78:1049–1057.
1998.PubMed/NCBI
|
|
25
|
Gütgemann I, Lehman NL, Jackson PK and
Longacre TA: Emi1 protein accumulation implicates misregulation of
the anaphase promoting complex/cyclosome pathway in ovarian clear
cell carcinoma. Mod Pathol. 21:445–454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu X, Wang H, Ma J, Xu J, Sheng C, Yang
S, Sun L and Ni Q: The expression and prognosis of Emi1 and Skp2 in
breast carcinoma: Associated with PI3K/Akt pathway and cell
proliferation. Med Oncol. 30:7352013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sheri A and Dowsett M: Developments in
Ki67 and other biomarkers for treatment decision making in breast
cancer. Ann Oncol. 23(Suppl 10): x219–x227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He WL, Li YH, Yang DJ, Song W, Chen XL,
Liu FK, Wang Z, Li W, Chen W, Chen CY, et al: Combined evaluation
of centromere protein H and Ki-67 as prognostic biomarker for
patients with gastric carcinoma. Eur J Surg Oncol. 39:141–149.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Harbour JW and Dean DC: The Rb/E2F
pathway: Expanding roles and emerging paradigms. Genes Dev.
14:2393–2409. 2000. View Article : Google Scholar : PubMed/NCBI
|