Introduction
Liver cancer has a high morbidity and mortality rate
(1), and is one of the most common
types of cancer in men. Hepatocellular carcinoma (HCC) is the major
histological subtype among primary liver cancer cases, accounting
for 70–85% of the total live cancer burden worldwide (2,3). Liver
cancer may be a result of multiple factors and it develops in a
multistep process. At present, there are no effective methods for
treatment. Although a number of genetic alterations have been
reported in the literature, including up- or down-regulated genes
(4–6),
the overall underlying mechanism remains unknown. It is widely
recognized that epigenetic alterations such as methylation and
acetylation contribute to carcinogenesis (7,8).
Therefore, further exploration of the gene epigenetic changes in
HCC are required.
Patatin glycoprotein is highly expressed in mature
potato tubers and is a non-specific, lipid acylhydrolase (9). Patatin-like phospholipase
domain-containing protein family (PNPLAs) has been identified in a
number of species varying from bacteria to human (10,11).
PNPLA7, also termed NTE-related 1 (NTE-R1) or NRE, is a member of
the PNPLAs family, which is conserved protein in mice, rats and
humans (12). It serves key roles in
triglyceride hydrolysis, energy metabolism, lipid droplet (LD), and
in regulation of adipocyte differentiation (10,13).
There is a marked difference between tumor cells and
normal cells in terms of their metabolic patterns, therefore, genes
that affect energy metabolism may be potential targets for tumor
treatment or diagnosis. It is well recognized that the liver is the
most important metabolism organ; whether PNPLA7 is deregulated in
HCC has not been previously reported. In the present study,
evidence is provided that PNPLA7 was down-regulated in HCC through
hypermethylation of its promoter. These findings indicate that
PNPLA7 is a potential biomarker for HCC diagnosis.
Materials and methods
Human tissues and cell lines
HCC tissue samples and the corresponding adjacent
non-cancerous tissues were obtained from 52 patients hospitalized
in Huashan Hospital (Shanghai, China). The study was approved by
the Human Research Review Committee of Huashan Hospital and written
informed consent was obtained from all patients. Each sample was
immersed in RNAlater (Ambion, Ahustin, TX, USA) and stored at −20°C
until use. HL-7702 (L02), Huh7, SMMC-7721, HCCLM-6, QGY-7703,
HepG2215, and HepG2 cell lines were purchased from the Cell Bank of
the Chinese Academy of Sciences (Shanghai, China). The cells were
cultured in Dulbecco's modified Eagle's medium with high glucose
(DMEM-h) (Gibco; Thermo Fisher Scientific, Inc., Gaithersburg, MD,
USA) supplemented with 10% fetal bovine serum (FBS), in a humid
atmosphere with 5% CO2 at 37°C.
DNA preparation, RNA extraction,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Genomic DNA of the cell lines was isolated by QIAamp
DNA Blood Mini Kit (Qiagen, Hilden, Germany), according to the
manufacturer's instructions. Total RNA was isolated using TRIzol
Reagent (Sigma-Aldrich, St. Louis, MO, USA). First-strand cDNA was
synthesized from 500 ng total RNA using the high-capacity cDNA
reverse transcription kit (Applied Biosystems, Foster City, CA,
USA). Quantitative PCR was performed using an Applied Biosystems
7900 Prism real-time PCR system and SYBR Premix Ex Taq (Takara,
Dalian, Japan), in accordance with the manufacturer's protocol.
Quantitative PCR primers were as follows: PNPLA7, F
5′-GGAAAAGCGTGATGGTTGC-3′ and R 5′-GAGCAGGTCCTTCTTGGCA-3′; and
GAPDH, F 5′-CAGGGCTGCTTTTAACTCTGGTAA-3′ and R
5′-ACTTGATTTTGGAGGGATCTCGCT-3′. Cycling conditions were as follows:
95°C for 3 min followed by 40 cycles of 95°C for 15 sec and 60°C
for 1 min.
Treatment of cells with 5-Aza-dC and
trichostatin A (TSA)
5-Aza-dC (Sigma-Aldrich) and TSA (Sigma-Aldrich)
were used for demethylation assay. HepG2 cells (5×105
cells/well) were seeded in 60 mm dishes. When the cells reached 30%
coverage, the demethylation agent 5-Aza-dC was added to the fresh
medium at a concentration of 3 µM. After 4 days, TSA was added at a
concentration of 0.5 µM. The cells were harvested on the 5th day
for the extraction of RNA and DNA. The control cells were incubated
without 5-aza-dC and TSA.
Bisulfite-sequencing PCR (BSP)
analyses
The bisulfite conversion and PCR analyses were
performed as described previously (14). BSP primers used for PNPLA7 were F
5′-GTGTAGATTAAGGAGATGGTTT-3′ and R 5′-TACTTTTCCAAATTATCAAAATC-3′.
Cycling conditions were as follows: 94°C for 3 min; 40 cycles of
94°C for 15 sec, ~54°C for 20 sec (56°C for the 1st cycle, 54°C for
the 2nd cycle and 52°C for the remaining cycles) and 72°C for 30
sec; 72°C for 1 min; and 4°C thereafter. Then the PCR products were
subjected to TA cloning then sent to Invitrogen (Thermo Fisher
Scientific, Inc.) for first-generation sequencing (Sanger method).
Sequence data was analyzed using Chromas 2.23 (Technelysium Pty
Ltd., Brisbane, Australia) and CpGviewer 6.4 (http://dna.leeds.ac.uk/cpgviewer/).
Immunofluorescence
L02 cells (4×105/well) were seeded in a
20 mm glass bottom cell culture dish and cultured overnight. The
cells were washed with PBS and then fixed in 10% formaldehyde for
30 min at room temperature. After 3 washes with PBS, the cells were
treated with 0.5% Triton-X-100 on ice for 5 min, to permeabilize.
After a further 3 times of rinsing with PBS, the dish was blocked
with 1% Albumin from bovine serum and incubated with antibodies of
rabbit polyclonal PNPLA7 (dilution, 1:200; cat no. ab121302; Abcam)
and subsequently with Alexa Fluor 488 Conjugate anti-rabbit IgG
(dilution, 1:1,000; cat no. 4412S; Cell Signaling Technology).
After 3 washes with PBS, Hoechst staining solution (1 µg/ml) (Life
Technologies, Inc., ThermoFisher Scientific, Inc.) was added to
completely cover the cells and the cells were incubated for 10 min
at room temperature in the dark. After a further 3 washes, the
cells were imaged with a LSM 710 confocal microscope (Carl Zeiss
AG, Oberkochen, Germany).
Statistical analysis
Student's t-test was performed to identify
statistical significance. Statistical analysis was performed using
SPSS software, version 20 (SPSS, Inc., Chicago, IL, USA).P<0.05
was considered to indicate a statistically significant difference.
GraphPad Prism 5 (GraphPad, San Diego, CA, USA) was used to draw
the figures.
Results
PNPLA7 was down-regulated in HCC
tissues and cell lines
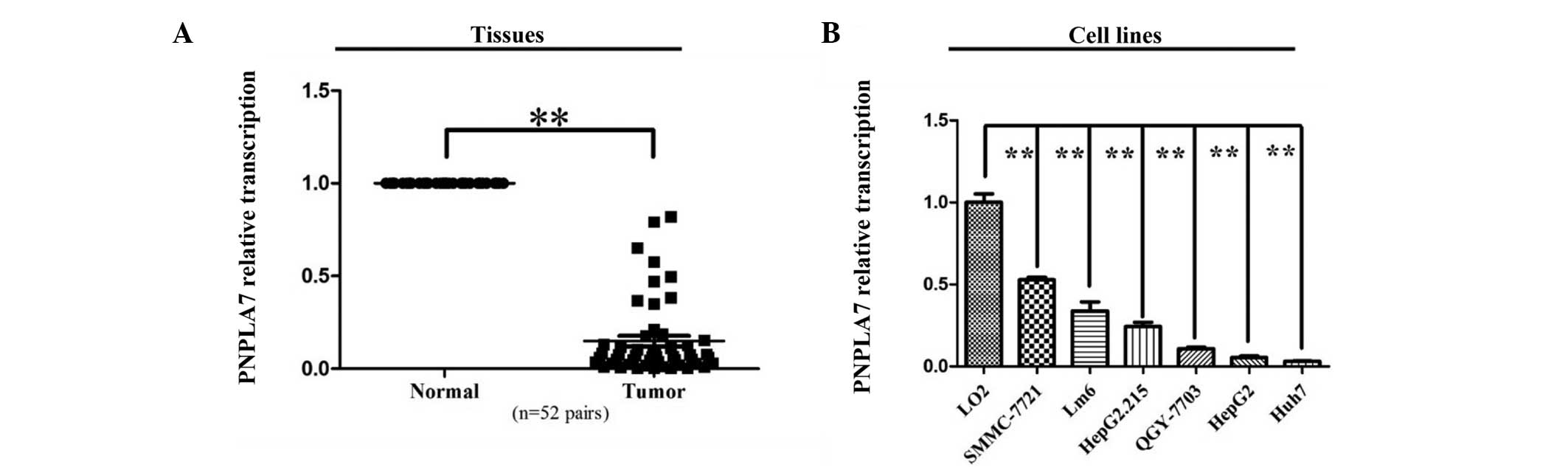
Transcriptional expression of PNPLA7 was evaluated
in 52 pairs of HCC samples by RT-PCR. After the expression levels
were normalized against GAPDH levels, the results showed that
PNPLA7 expression was dramatically decreasedin the HCC samples
compared to normal controls in the tissue pairs studied (P<0.01,
Fig. 1A). PNPLA7 expression was also
significantly down-regulated in all six HCC cell lines compared to
the normal liver cell line, L02 (P<0.01, Fig. 1B).
Hypermethylation of PNPLA7 promoter
existed in HCC cell lines
DNA methylation is often correlated with
deregulation of variety of genes. The DNA methylation statue of the
region (chr9:140446407–140447247) was determined with a BSP-based
assay on 6 HCC cell lines and 1 normal liver epithelial cell line.
As presented in Fig. 2, the average
methylation rate in the 6 HCC cell lines (85.8%) was considerably
higher than that observed in the normal liver epithelial cell line
L02 (3.3%) (P<0.001), indicating that the promoter region was
hypermethylated in HCC.
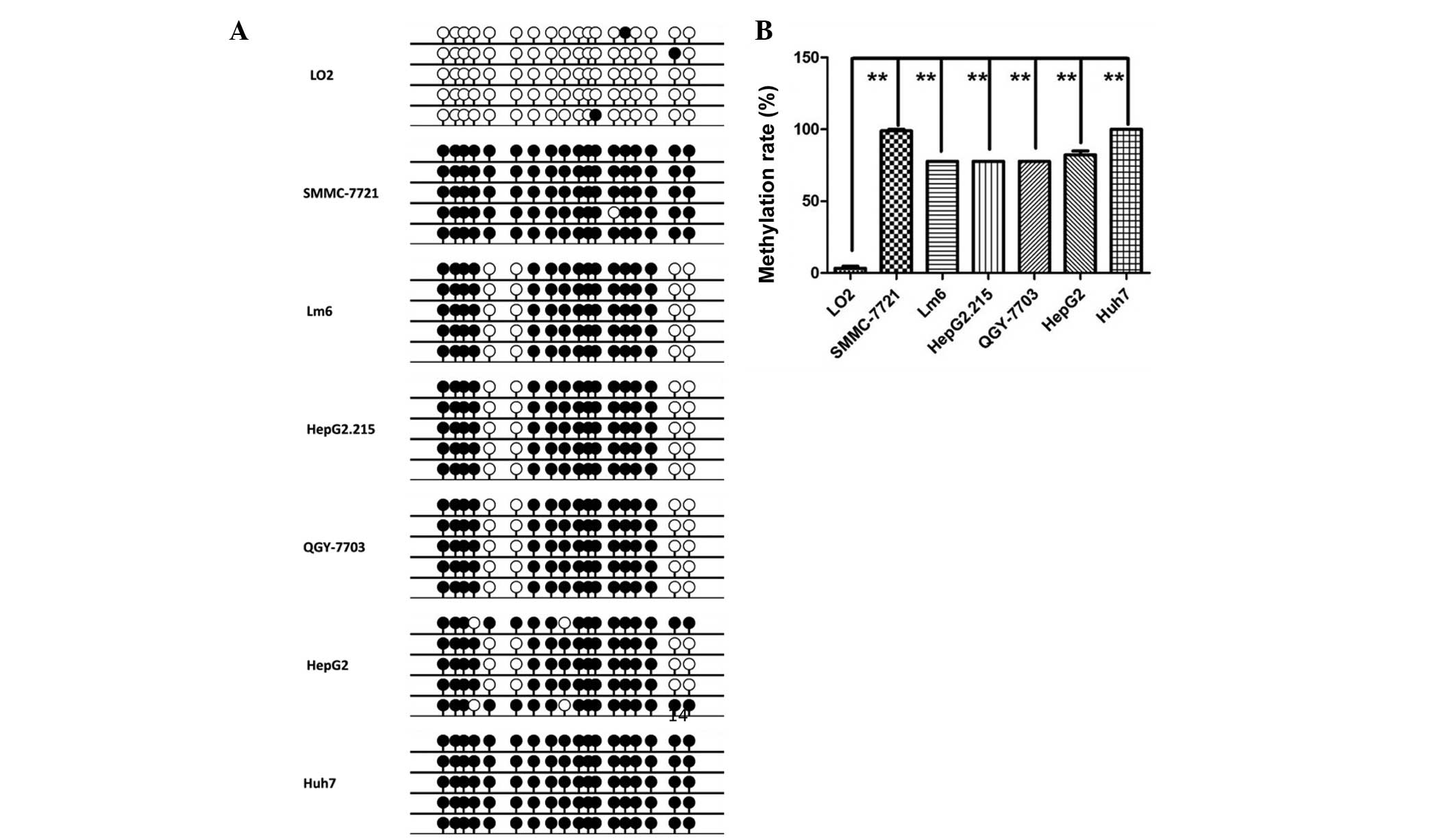 | Figure 2.Hypermethylation of PNPLA7 promoter
existed in HCC cell lines. (A) Methylation status of the fragment
was examined by BSP in HCC cell lines, SMMC-7721, Lm6, HepG2.215,
QGY-7703, HepG2, Huh7, and in normal liver cell line LO2.
Densitometric analysis of methylation rate of the tested fragment
in different cell lines based on BSP results. (B) Methylation rates
in LO2, SMMC-7721, Lm6, HepG2.215, QGY-7703, HepG2 and Huh7 were
3.3, 98.9, 77.8, 77.8, 77.8, 82.2 and 100%, respectively.
**P<0.01. PNPL7, patatin-like phospholipase domain-containing
protein 7; HCC, hepatocellular carcinoma. |
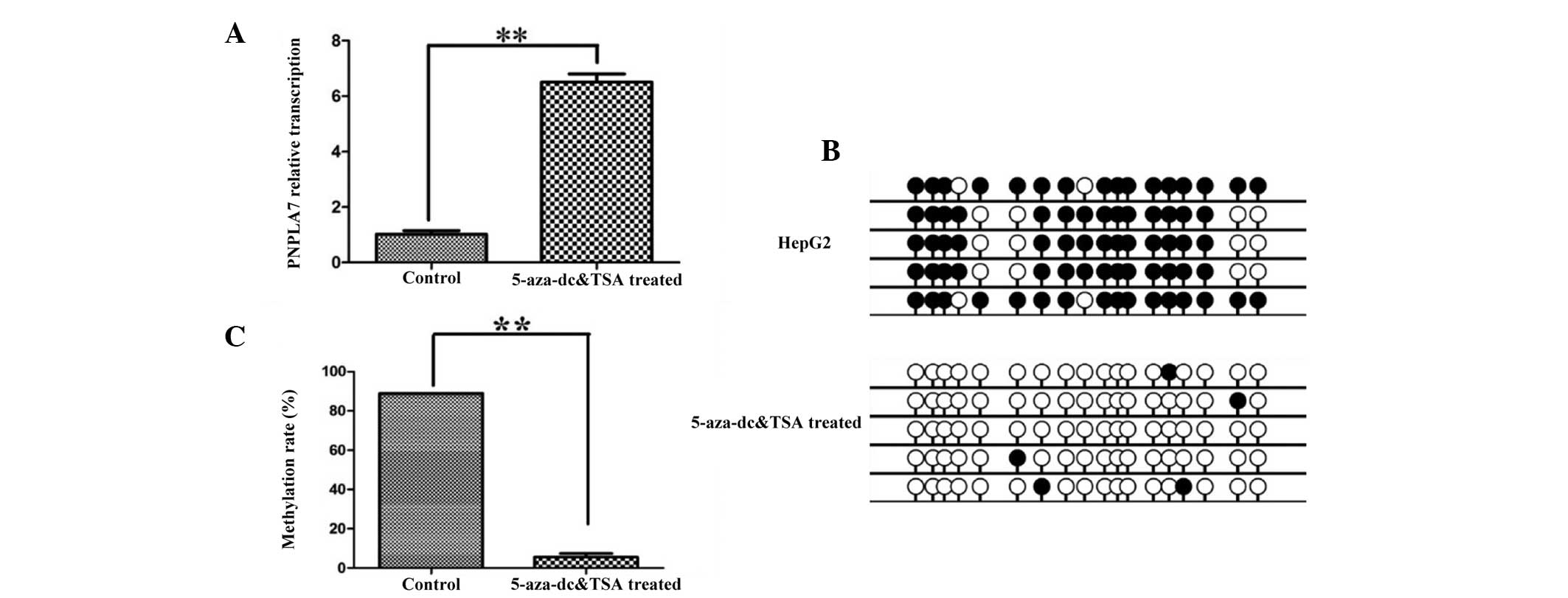
PNPLA7 expression was restored after
5-Aza-docy treatment
To confirm the correlation between PNPLA7 expression
and the methylation status of its promoter, a demethylation
experiment was performed. The HepG2 cell line was treated with DNA
methyltransferase inhibitor 5-Aza-dC and the histone deacetylase
inhibitor TSA. As shown in Fig. 3A,
the result of RT-qPCR assay revealed that the expression of PNPLA7
in HepG2 cell line was significantly up-regulated after treatment
with 5-Aza-dC and TSA (P<0.01). In addition, BSP results
(Fig. 3B) confirmed that the majority
of the methylated sites were demethylated following treatment, and
the corresponding methylation rate decreased by 93% (to 5.56%)
(Fig. 3C; P<0.01).
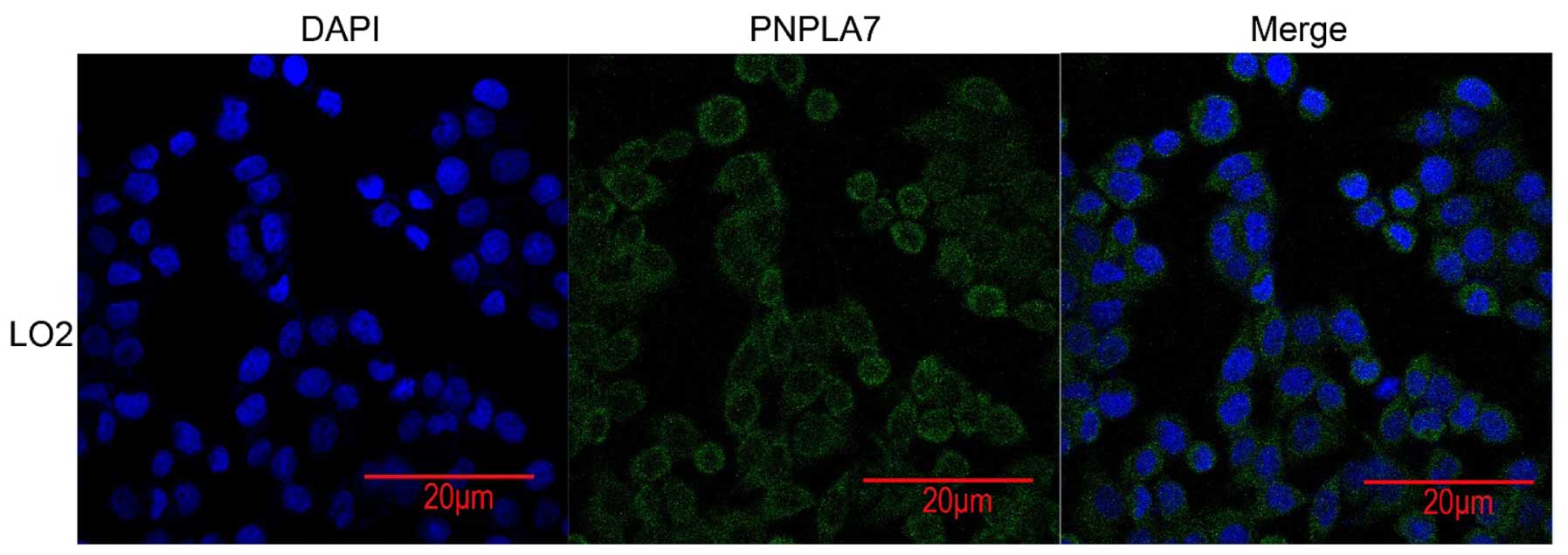
Subcellular location of PNPLA7 in HCC
cells
Confocal imaging was performed and indicated that
PNPLA7 was mainly located in plasma membrane and partly distributed
in cytoplasm (Fig. 4).
Discussion
The present study indicated that deregulation of
PNPLA7 was associated with HCC and identified that hypermethylation
resulted in reduced expression levels of PNPLA7 in HCC.
PNPLAs are considered to be a divergent family, the
majority if which have a highly conserved orthologue in several
mammalian species (10). PNPLA family
members share a protein domain discovered initially in patatin,
which is a lipid hydrolase with an unusual folding topology that
differs for diverse substrates such as triacylglycerols,
phospholipids, and retinol esters (15).
PNPLA7 is 61% identical to PNPLA6 in amino acid
sequence and has the same domain structure, however, it cannot
substitute for PNPLA6 during embryonic development (10,12) and it
has a different tissue distribution, richest in lysosome,
mitochondrion, nucleus and vacuole, and was regulated by insulin
and glucose levels (16,17). Both these proteins are predicted to be
regulated by cyclic nucleotide as integral membrane proteins and
potent lysophospholipase activity, while they showed different
sensitivity to organophosphate inhibitors (18).
Other members of PNPLA family were reported to be
involved in lipid metabolism and chronic hepatitis C infection.
PNPLA3, particularly, was strongly associated with liver injury and
non-alcoholic fatty liver disease (NAFLD) (19–21).
However, there are few reports relating to PNPLA7 in human
diseases. Vrieze et al (22)
mapped ~85,000 rare nonsynonymous exonic single nucleotide
polymorphisms (SNPs) to 17 psychophysiological endophenotypes in
4,905 individuals and identified that PNPLA7 is associated with the
endophenotype pleasant difference startle, the difference in
startle magnitude between pleasant and neutral images. Therefore,
the present study aimed to explore whether PNPLA7 was also
associated with liver disease like PNPLA3. Here we observed that
PNPLA7 was down-regulated in HCC cell-lines and also in tissue
samples.
Promoter hypermethylation induced transcriptional
silencing has emerged recently as one important mechanism involved
in oncogenesis and cancer development (23,24). In
order to understand the molecular mechanism of the down-regulation
of PNPLA7 in HCC, the methylation status of CpG island of PNPLA7
promoter was analyzed and hypermethylation was confirmed in all HCC
cell lines. Then 5-Aza-dC, a demethylating agent, was used and this
treatment restored PNPLA7 expression in liver cancer cell lines.
The results revealed that promoter hypermethylation could result in
inhibition of PNPLA7 transcription in HCC.
In conclusion, the present study demonstrated that
PNPLA7 levels were dramatically down-regulated in both HCC cell
lines and tissues. It was also determined that DNA hypermethylation
was the mechanism of the down-regulation of PNPLA7 in HCC. These
results may offer a novel insight to identifying cancer markers and
understanding the mechanism of hepatocarcinogenesis.
Acknowledgements
The present study was supported by grants from the
Ministry of Science and Technology of China (grant no.
2013CB945401) and the National Natural Science Foundation of China
(grant nos. 81125001 and 91129702).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leong TY and Leong AS: Epidemiology and
carcinogenesis of hepatocellular carcinoma. HPB (Oxford). 7:5–15.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buendia MA and Neuveut C: Hepatocellular
carcinoma. Cold Spring Harb Perspect Med. 5:a0214442015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lemmer ER, Friedman SL and Llovet JM:
Molecular diagnosis of chronic liver disease and hepatocellular
carcinoma: The potential of gene expression profiling. Semin Liver
Dis. 26:373–384. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Matter MS, Decaens T, Andersen JB and
Thorgeirsson SS: Targeting the mTOR pathway in hepatocellular
carcinoma: Current state and future trends. J Hepatol. 60:855–865.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Easwaran H, Tsai HC and Baylin SB: Cancer
epigenetics: Tumor heterogeneity, plasticity of stem-like states
and drug resistance. Mol Cell. 54:716–727. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma L, Chua MS, Andrisani O and So S:
Epigenetics in hepatocellular carcinoma: An update and future
therapy perspectives. World J Gastroenterol. 20:333–345. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rydel TJ, Williams JM, Krieger E, Moshiri
F, Stallings WC, Brown SM, Pershing JC, Purcell JP and Alibhai MF:
The crystal structure, mutagenesis, and activity studies reveal
that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic
dyad. Biochemistry. 42:6696–6708. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wilson PA, Gardner SD, Lambie NM, Commans
SA and Crowther DJ: Characterization of the human patatin-like
phospholipase family. J Lipid Res. 47:1940–1949. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Finn RD, Bateman A, Clements J, Coggill P,
Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J,
et al: Pfam: The protein families database. Nucleic Acids Res.
42(Database Issue): D222–D230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Winrow CJ, Hemming ML, Allen DM, Quistad
GB, Casida JE and Barlow C: Loss of neuropathy target esterase in
mice links organophosphate exposure to hyperactivity. Nat Genet.
33:477–485. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kienesberger PC, Lass A, Preiss-Landl K,
Wolinski H, Kohlwein SD, Zimmermann R and Zechner R: Identification
of an insulin-regulated lysophospholipase with homology to
neuropathy target esterase. J Biol Chem. 283:5908–5917. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herman JG and Baylin SB:
Methylation-specific PCR. Curr Protoc Hum Genet Chapter. 10:Unit
10.6. 2001. View Article : Google Scholar
|
|
15
|
Kienesberger PC, Oberer M, Lass A and
Zechner R: Mammalian patatin domain containing proteins: A family
with diverse lipolytic activities involved in multiple biological
functions. J Lipid Res. 50(Suppl): S63–S68. 2009.PubMed/NCBI
|
|
16
|
Chang PA, Long DX, Sun Q, Wang Q, Bu YQ
and Wu YJ: Identification and characterization of a splice variant
of the catalytic domain of mouse NTE-related esterase. Gene.
417:43–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Richardson RJ, Hein ND, Wijeyesakere SJ,
Fink JK and Makhaeva GF: Neuropathy target esterase (NTE): Overview
and future. Chem Biol Interact. 203:238–244. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang PA, Chen YY, Long DX, Qin WZ and Mou
XL: Degradation of mouse NTE-related esterase by macroautophagy and
the proteasome. Mol Biol Rep. 39:7125–7131. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kotronen A, Johansson LE, Johansson LM,
Roos C, Westerbacka J, Hamsten A, Bergholm R, Arkkila P, Arola J,
Kiviluoto T, et al: A common variant in PNPLA3, which encodes
adiponutrin, is associated with liver fat content in humans.
Diabetologia. 52:1056–1060. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sookoian S, Castaño GO, Burgueño AL,
Gianotti TF, Rosselli MS and Pirola CJ: A nonsynonymous gene
variant in the adiponutrin gene is associated with nonalcoholic
fatty liver disease severity. J Lipid Res. 50:2111–2116. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Valenti L, Al-Serri A, Daly AK, Galmozzi
E, Rametta R, Dongiovanni P, Nobili V, Mozzi E, Roviaro G, Vanni E,
et al: Homozygosity for the patatin-like
phospholipase-3/adiponutrin I148M polymorphism influences liver
fibrosis in patients with nonalcoholic fatty liver disease.
Hepatology. 51:1209–1217. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vrieze SI, Malone SM, Pankratz N,
Vaidyanathan U, Miller MB, Kang HM, McGue M, Abecasis G and Iacono
WG: Genetic associations of nonsynonymous exonic variants with
psychophysiological endophenotypes. Psychophysiology. 51:1300–1308.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jones PA and Laird PW: Cancer epigenetics
comes of age. Nat Genet. 21:163–167. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lopez-Serra P and Esteller M: DNA
methylation-associated silencing of tumor-suppressor microRNAs in
cancer. Oncogene. 31:1609–1622. 2012. View Article : Google Scholar : PubMed/NCBI
|