Introduction
Lung cancer is the leading cause of
cancer-associated mortalities worldwide, and adenocarcinoma is one
of the most common histological types of lung cancer in multiple
countries (1). In the recent past,
various major therapeutic advances have improved the prognosis of
specific subgroups of patients (2).
However, the 5-year survival rate of patients with lung cancer
remains <15% (3), and therefore,
novel therapeutic strategies are required for better management of
this disease.
MicroRNAs (miRNAs) are a class of small (18–25 nt),
non-coding, endogenous RNAs that have been implicated in a wide
range of cellular biological processes that involve the targeting
of messenger RNAs (mRNAs) for either degradation or inhibition of
translation (4,5). Aberrant expression of miRNAs has been
observed in lung cancer, including upregulation of miR-21,
miR-17-92, miR-155 and miR-367 (6–8) and
downregulation of let-7, miR-34b/c, miR-449a, miR-1 and miR-145
(8–12). This indicates that miRNAs can function
as tumor suppressors and oncogenes in tumor development.
miR-206 was first identified as a skeletal
muscle-specific miRNA that is involved in the process of skeletal
muscle differentiation (13). The
roles of miR-206 in breast cancer drew attention to the possibility
of its anti-cancer activity, since miR-206 is downregulated in
breast cancer, rhabdomyosarcoma, renal cell carcinoma, estrogen
receptor α-positive endometrioid adenocarcinoma, colorectal cancer,
gastric cancer and lung cancer (14–21). Wu
et al (22) noticed that the
serum levels of miR-206 were significantly upregulated in the early
stage of the tobacco-specific N-nitrosamine
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced rat lung
carcinogenesis, and downregulated at the late stage of lung
carcinogenesis, which indicates that miR-206 may be associated with
lung carcinogenesis. In addition, low expression of miR-206 was
demonstrated to be correlated with tumor invasion and metastasis in
individuals with lung cancer (20).
However, the regulatory effects and mechanisms of miR-206 in human
lung adenocarcinoma cell lines are not clear. In the present study,
the function and targets of miR-206 in human lung adenocarcinoma
cell lines were analyzed.
Materials and methods
Human tissue
Formalin-fixed and paraffin-embedded (FFPE) sections
of paired lung adenocarcinoma tissues and adjacent normal tissues
were obtained from patients who underwent radical resection for
lung cancer. The samples were collected between October and
December 2013 at The First Affiliated Hospital, School of Medicine,
Zhejiang University (Zhejiang, China). The patients provided signed
informed consent for participation in the study, which was approved
by the Ethics Committee of Zhejiang University. The demographic and
clinicopathological data are listed in Table I.
 | Table I.Clinical features of the 12 patients
with lung adenocarcinoma. |
Table I.
Clinical features of the 12 patients
with lung adenocarcinoma.
| Patient no. | Gender | Age (years) | Histological
type | Tobacco smoking
history | pTNM stage | Tumor grade | EGFR mutation |
|---|
| 1 | M | 47 | Adenocarcinoma | Yes | T2aN0M0 | IB | Exon 19 deletion |
| 2 | F | 82 | Adenocarcinoma | No | T2aN0M0 | IB | Unknown |
| 3 | F | 41 | Adenocarcinoma | No | T2aN1M1 | IV | Unknown |
| 4 | F | 50 | Adenocarcinoma | No | T2aN0M0 | IB | Unknown |
| 5 | F | 47 | Adenocarcinoma | No | T2aN0M0 | IB | Unknown |
| 6 | F | 69 | Adenocarcinoma | No | T1aN2M0 | IIIA | Exon 19 deletion |
| 7 | M | 46 | Adenocarcinoma | No | T2bN1M0 | IIB | Unknown |
| 8 | F | 73 | Adenocarcinoma | No | T2aN1M0 | IIA | No mutation |
| 9 | F | 50 | Adenocarcinoma | No | T1bN0M0 | IA | Unknown |
| 10 | F | 51 | Adenocarcinoma | No | T2bN3M0 | IIIB | No mutation |
| 11 | M | 37 | Adenocarcinoma | No | T1bN0M0 | IA | Unknown |
| 12 | M | 43 | Adenocarcinoma | No | T1bN2M0 | IIIA | Unknown |
Cell culture and reagents
The HCC827 and A549 cell lines were purchased from
the Committee on Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China). The PC-9 cell line was a gift from Dr
Caicun Zhou (Department of Oncology, Shanghai Pulmonary Hospital,
Shanghai, China) and was verified by short tandem repeats analysis.
HCC827 and A549 cells were cultured in RPMI-1640 (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) at 37°C and 5%
CO2. The PC-9 cells were maintained in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS.
Oligonucleotides and transfection
miR-206 mimics (double-stranded RNA
oligonucleotides) and a negative control (NC) duplex that lacks
significant homology to all known human sequences were purchased
from Shanghai GenePharma Co., Ltd. (Shanghai, China). Small
interfering RNA (siRNA) targeting MET (si-MET) and NC siRNA were
purchased from Shanghai GenePharma Co., Ltd. The sequences are
listed in Table II. The transfection
was performed using Lipofectamine 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
protocol.
 | Table II.Sequences of the oligonucleotides
used in the present study. |
Table II.
Sequences of the oligonucleotides
used in the present study.
| Name | Sequence
(5′-3′) |
|---|
| miR-206 mimics |
UGGAAUGUAAGGAAGUGUGUGG |
|
|
ACACACUUCCUUACAUUCCAUU |
| si-MET |
GGAGGUGUUUGGAAAGAUAdTdT |
|
|
UAUCUUUCCAAACACCUCCdTdT |
| NC |
UUCUCCGAACGUGUCACGUTT |
|
|
ACGUGACACGUUCGGAGAATT |
| GAPDH F |
GGTCTCCTCTGACTTCAACA |
| GAPDH R |
AGCCAAATTCGTTGTCATAC |
| MET F |
GTTTGTCCACAGAGACTTGGCTG |
| MET R |
ATCCACTTCACTGGCAGCTTTG |
Cell viability assay
The day prior to transfection, 3,000 cells/well were
plated in 96-well plates. Upon overnight incubation, the cells were
transfected with miR-206 mimics or with the NC miRNA at 20 or 40-nM
concentration. After the culture period, 10 µl cell counting
solution (water-soluble tetrazolium-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was added to each well,
followed by an additional incubation for 2 h at 24 h
post-transfection, 1.5 h at 48 h post-transfection or 1 h at 72 h
post-transfection. The absorbance of the solution was measured
spectrophotometrically at 450 nm with an MRX II absorbance reader
(Dynex Technologies, Chantilly, VA, USA). Each experiment was
conducted three times in triplicate.
Cell migration assays
Transwell chambers (8-µm pore size; EMD Millipore,
Billerica, MA, USA) were used to determine the migration of tumor
cells. A total of 5×105 cells were resuspended in 0.2 ml
fresh medium without FBS 48 h after transfection, and then added to
the inserts. Next, 0.6 ml culture medium with 10% FBS was added to
the lower compartment. After 24 h, the non-migrating cells and
culture medium were removed using cotton buds, while the cells on
the lower surface of the inserts were fixed and stained with 0.1%
crystal violet. Four random fields per insert were counted. The
experiment was conducted three times in triplicate.
Cell apoptosis assay
Cell apoptosis was detected with an annexin
V-fluorescein isothiocyanate (FITC) apoptosis detection kit (BD
Biosciences, Franklin Lakes, NJ, USA). The cells were harvested 72
h after NC or miR-206 treatment. The cells were then washed twice
with pre-chilled phosphate-buffered saline and resuspended in 500
µl binding buffer at a concentration of 1×106 cells/ml.
Then, cells were stained with 5 µl annexin V-FITC and 5 µl
propidium iodide for 10 min in the dark. The apoptosis analysis was
performed by the BD FACS Verse flow cytometer with CellQuest Pro
6.0 software (BD Biosciences).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cultured cells with
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA
was reverse transcribed into complementary DNA (cDNA) using
PrimeScript™ RT reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China). miRNAs isolated from FFPE tissues were extracted
using the RNeasy FFPE kit (Qiagen GmbH, Hilden, Germany). miRNAs
that were isolated from the cultured cells were extracted with
RNAiso Plus (Takara Biotechnology Co., Ltd.) and reverse
transcribed into cDNA using an RT kit (Qiagen GmbH). PCR
amplification and detection were performed on a 7500 Fast Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.)
using miScript SYBR Green PCR kit (Qiagen GmbH). The amplification
protocol was as follows: Initial denaturation 95°C for 30 sec, and
then 40 cycles of denaturation at 95° for 5 sec, annealing and
extension at 60°C for 34 sec. The relative amounts of mRNA and
miRNA gene transcripts were normalized to the levels of
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and RNA, U6 small
nuclear 2 (RNU6-2). The sequences of the primers for GAPDH and MET
are listed in Table II. The primers
for RNU6-2 and miR-206 were purchased from Qiagen GmbH.
Vector construction and
dual-luciferase reporter assay
The 3′-untranslated regions (UTRs) and the mutant
3′-UTR of MET, which contained putative binding sites for miR-206,
were cloned downstream of the pmirGLO Dual-Luciferase miRNA Target
Expression vector (Promega Corporation, Madison, WI, USA). The
cells were plated in 24-well plates and were transfected with the
constructed luciferase vector (100 ng) and 40 nM miR-206 or NC. The
relative luciferase activity was measured 48 h after transfection
as described previously (23).
Immunoblotting
Following the various treatments, the cells were
harvested, lysed and quantified. Equivalent quantities (50 µg) of
protein were separated by 8% sodium dodecyl sulfate-polyacrylamide
gel electrophoresis and transferred onto polyvinylidene difluoride
membranes. Membranes were blocked for 1 h with 5% non-fat milk and
then incubated overnight with a primary rabbit polyclonal antibody
against MET (1:1,000 dilution; catalog no., 4560; Cell Signaling
Technology, Inc., Danvers, MA, USA) and a rabbit polyclonal
antibody against β-actin (1:2,000 dilution; catalog no., sc-130656;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Next, the
membranes were incubated with a chicken anti-rabbit horseradish
peroxidase-conjugated secondary antibody (catalog no., sc-516087,
Santa Cruz Biotechnology, Inc.) at a 1:2,000 dilution for 2 h.
Immunoreactive bands were detected with an enhanced
chemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.)
as described previously (23).
Statistical analysis
All data are presented as the mean ± standard
deviation of three independent experiments. All statistical
analyses were performed using SPSS 16.0 statistical software (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference according to Student's
t-test.
Results
miR-206 was frequently downregulated
in lung adenocarcinoma
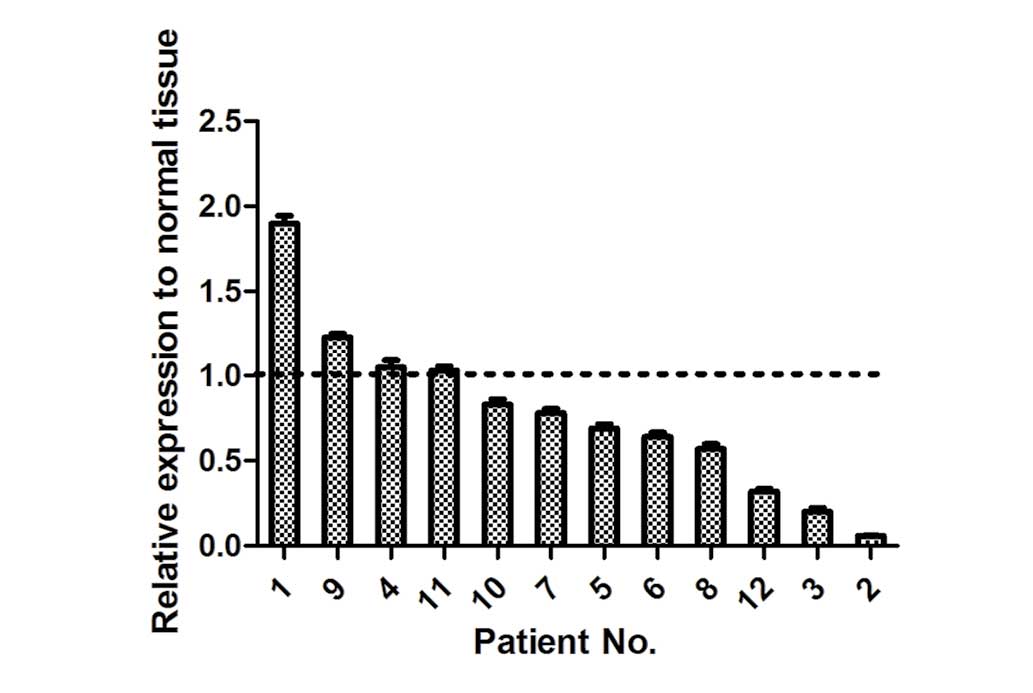
In the present study, the expression levels of
miR-206 were detected in 12 pairs of human lung adenocarcinoma
tissues and adjacent normal tissues. In total, 8 of 12 (75%) lung
adenocarcinoma tissues displayed a decreased expression of miR-206
compared with adjacent normal tissues (Fig. 1). This indicated that miR-206 is
frequently downregulated in human lung adenocarcinoma.
Overexpression of miR-206 inhibited
the viability of lung adenocarcinoma cells in vitro
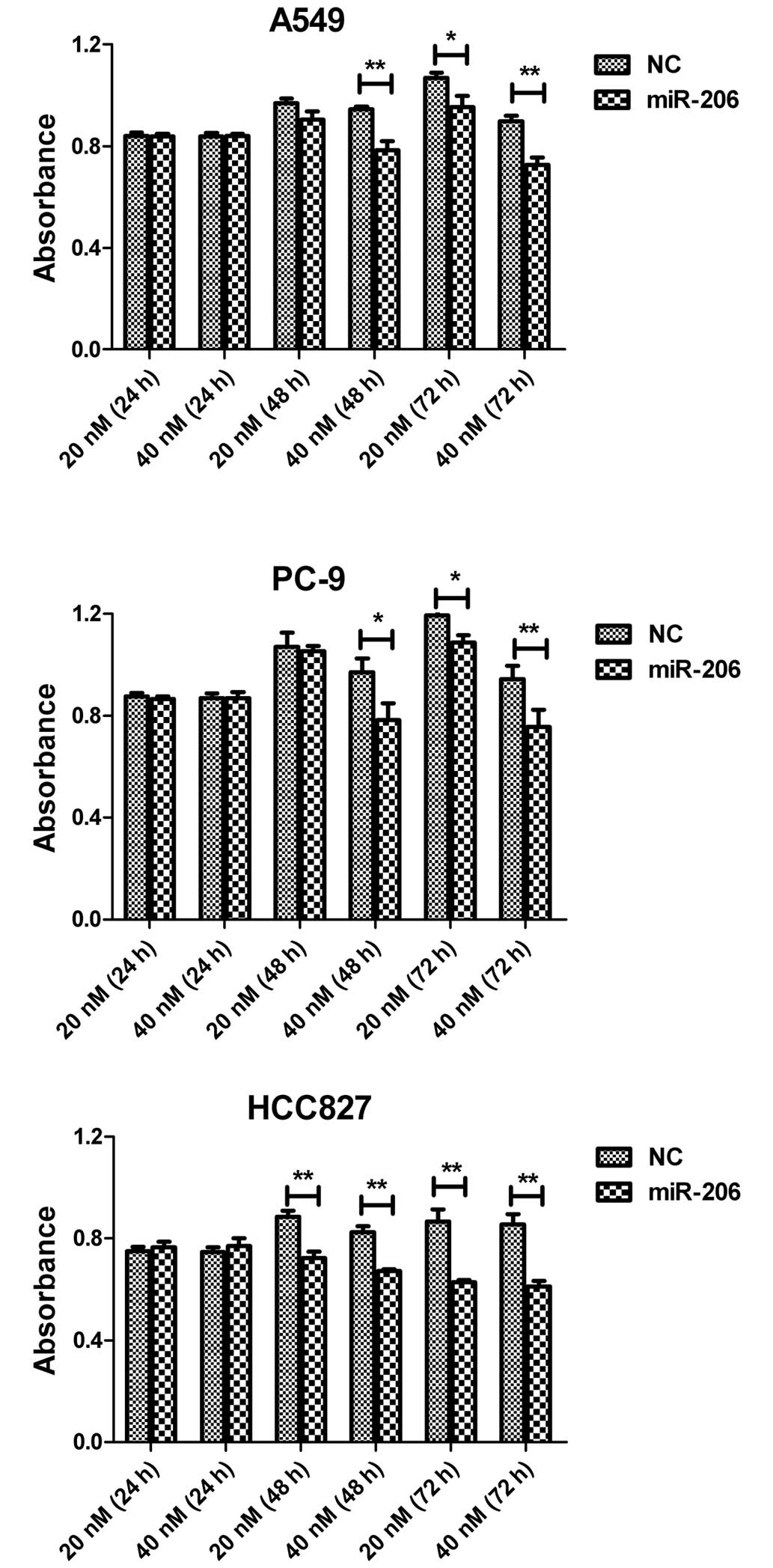
In order to determine if miR-206 could function as a
tumor suppressor in human lung adenocarcinoma cells, synthetic
miR-206 mimics and NC miRNA were independently transfected into
A549, PC-9 and HCC827 cells. The overexpression of miR-206 exerted
a potent inhibitory effect in a time- and dose-dependent manner.
Compared with the NC group, miR-206 caused an increased percentage
of inhibition of cell viability at a concentration of 40 nM in the
three cell lines tested at 48 or 72 h after transfection (Fig. 2). These data suggested that miR-206
negatively modulates the viability of lung adenocarcinoma cells.
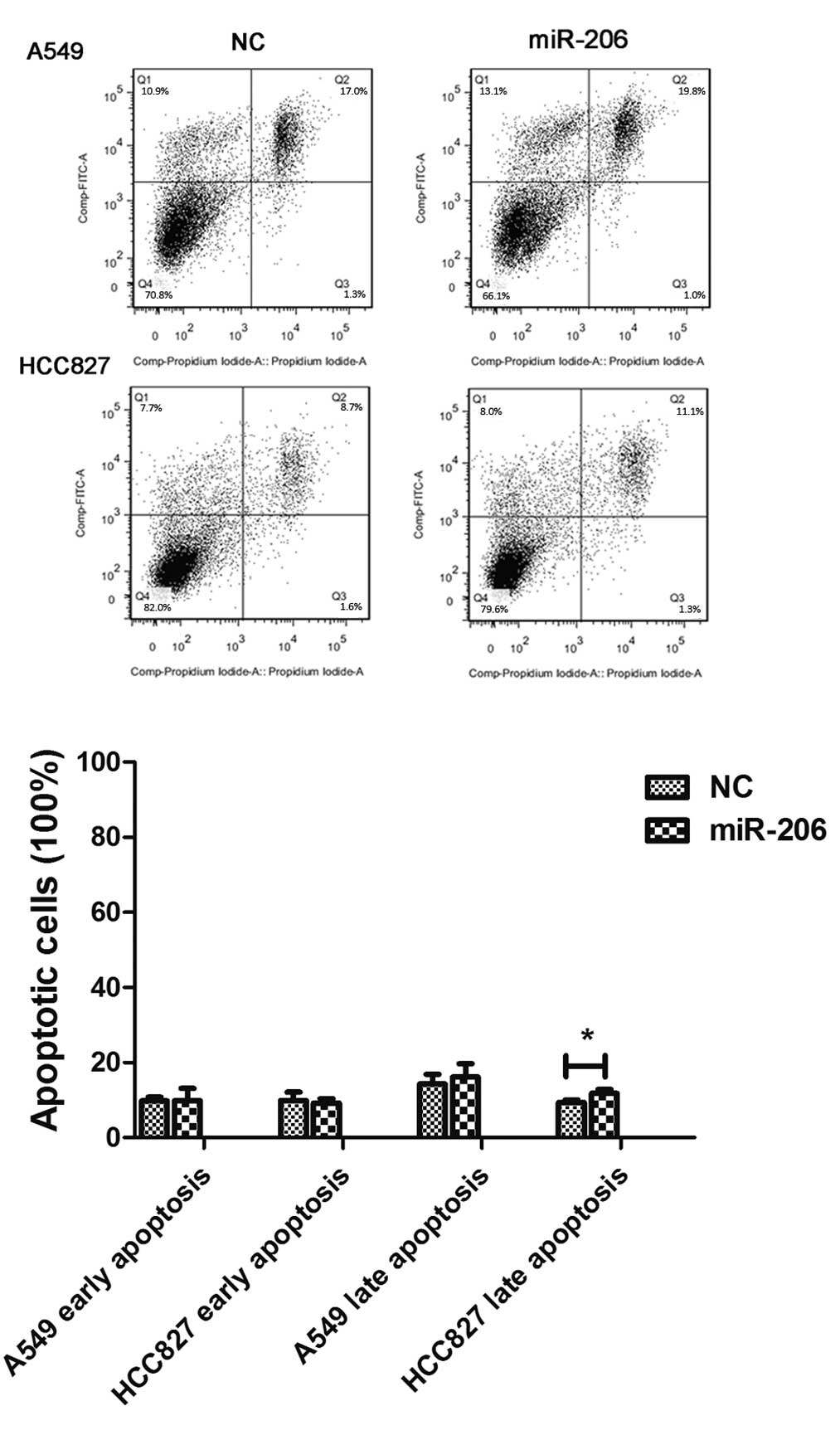
The apoptosis analysis by flow cytometry revealed that the early
apoptosis rate was not affected in A549 or HCC827 cells, whereas
the late apoptosis rate was higher in HCC827 cells following
miR-206 treatment (Fig. 3).
Forced expression of miR-206 repressed
the migration of lung adenocarcinoma cells
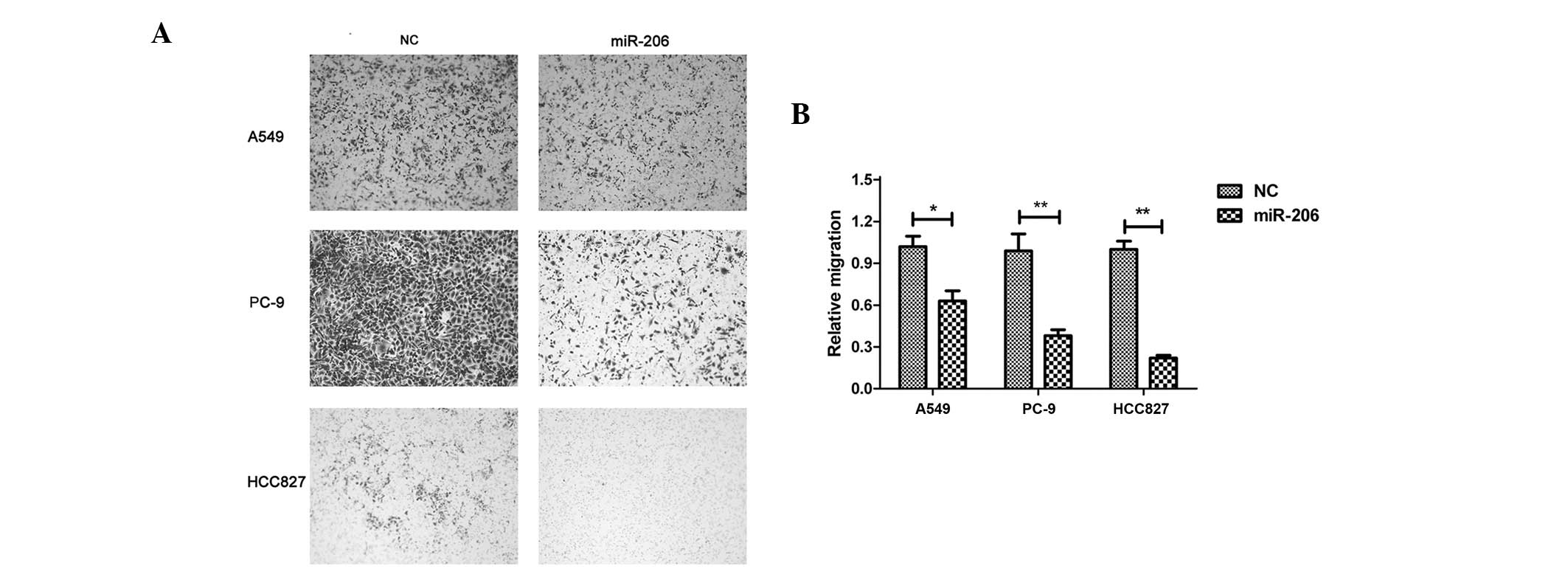
To better understand the function of miR-206 in lung
adenocarcinoma, the present study tested whether miR-206 could
affect the migration of lung adenocarcinoma cells. A549, PC-9 and
HCC827 cells were transfected with miR-206 mimics. The forced
expression of miR-206 caused significant inhibition of cell
migration compared with the control in all three cell lines
(Fig. 4).
Overexpression of miR-206
downregulated the expression of MET in lung adenocarcinoma cells
through targeting its 3′-UTR
The mRNA of MET, whose 3′-UTR contains
miR-206-binding sites, was previously demonstrated to be a specific
target of miR-206 in rhabdomyosarcoma (24). In the present study, it was observed
that MET was downregulated significantly at both the mRNA and
protein levels (Fig. 5A and B). A
luciferase reporter assay confirmed that MET was a direct target of
miR-206 in human lung adenocarcinoma cells (Fig. 5C and D). These results suggested a
targeted downregulation of MET by miR-206 in lung
adenocarcinoma.
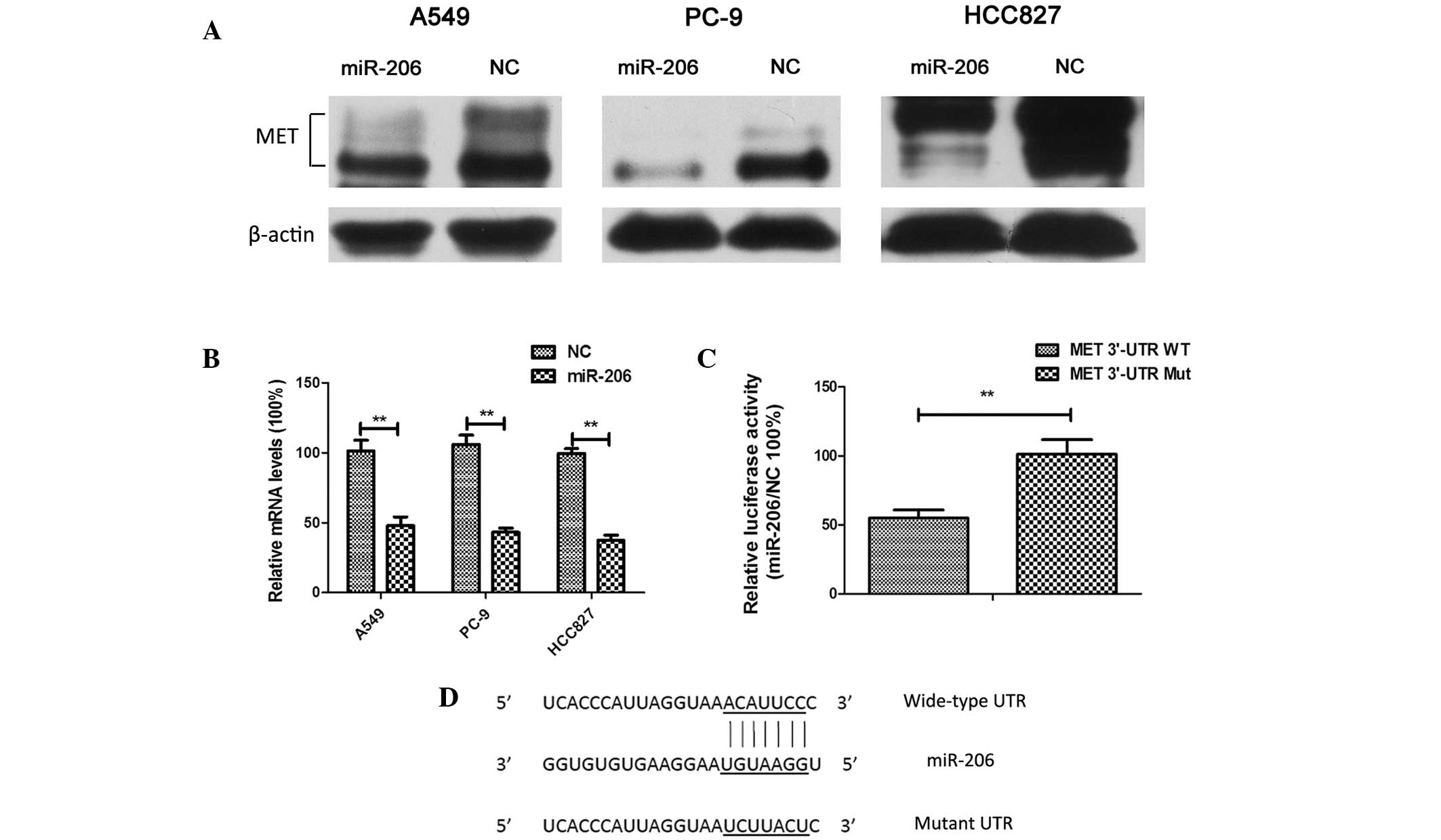 | Figure 5.Overexpression of miR-206 resulted in
the downregulation of MET expression in lung adenocarcinoma cells
through the targeting of its 3′-UTR. (A) A549, PC-9 and HCC827
cells were transfected with 40 nM NC or miR-206 mimics for 48 h.
The cell lysates were harvested, and the phosphorylation of the
indicated proteins was determined by western blotting. Three
independent experiments were performed. (B) A549, PC-9 and HCC827
cells were transfected with 40 nM NC or miR-206 mimics for 48 h.
Subsequently, RNA was extracted and reverse
transcription-quantitative polymerase chain reaction analysis was
conducted to determine the relative expression of MET. Data were
normalized to glyceraldehyde 3-phosphate dehydrogenase. Each
experiment was performed three times in triplicate. The results are
presented as the mean ±SD (**P<0.01). (C) Overexpression of
miR-206 suppressed pmirGLO-MET-3′-UTR-wild-type firefly luciferase
activity but exerted no effect on the mutant construct in A549
cells. Each experiment was performed three times in triplicate. The
values were normalized to the internal Renilla luciferase
control. The results are presented as the mean ± SD (**P<0.01).
(D) miR-206 target sites and substitution of nucleotides at the
target sites in the MET 3′-UTR. miR, microRNA; NC, negative
control, WT, wild-type; Mut, mutant; UTR, untranslated region; SD,
standard deviation. |
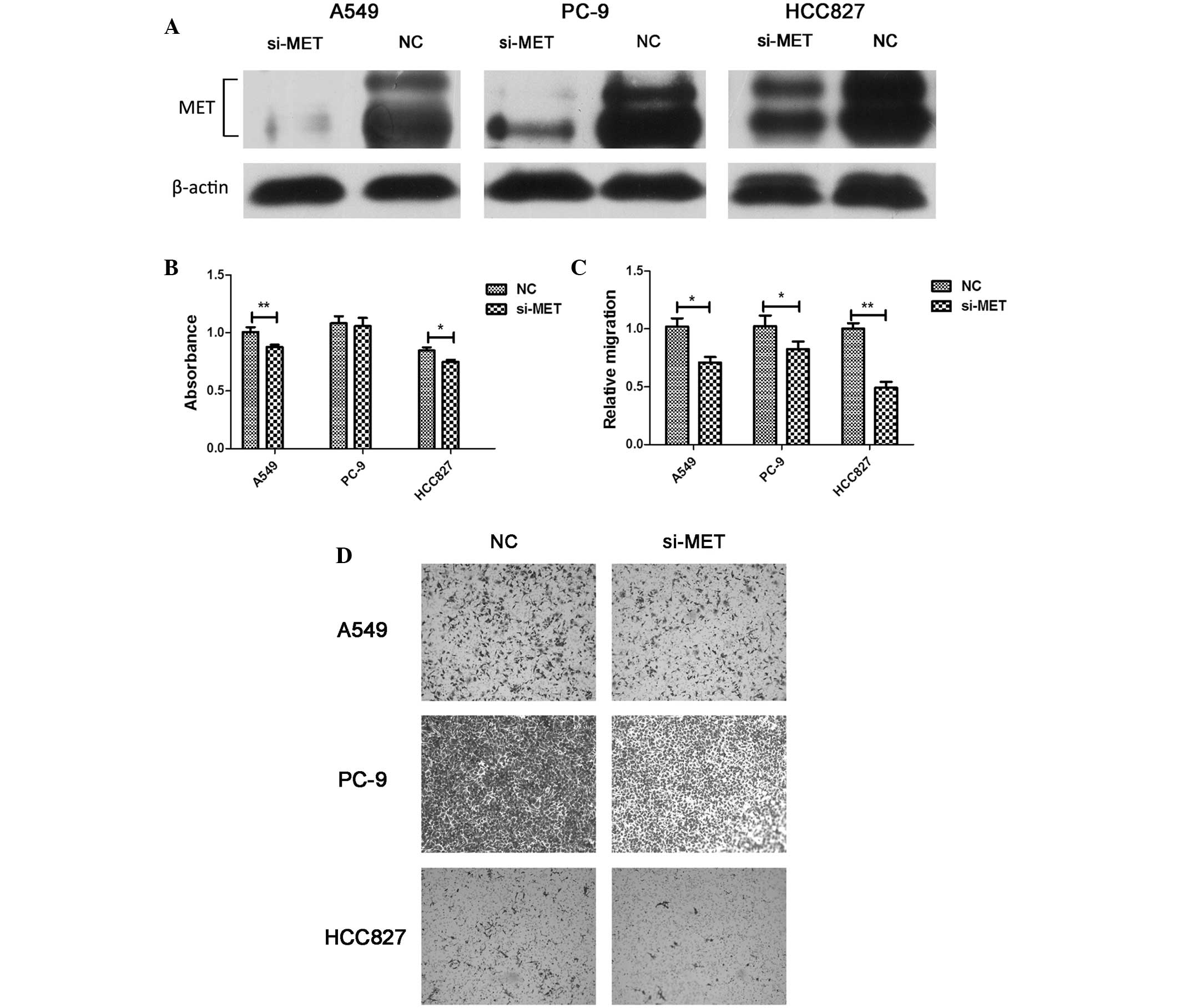
miR-206 triggered the inhibition of
cell viability and migration in lung adenocarcinoma cells in part
via targeting MET
To determine whether the downregulation of MET was
involved in the miR-206-mediated inhibition of cell viability and
migration, the physiological function of MET was evaluated by an
RNA interference approach. The knockdown of MET suppressed cell
viability and migration, but these effects were not as remarkable
as those observed upon miR-206 overexpression (Fig. 6). These findings indicated that the
anti-tumor effect of miR-206 is partly dependent on the targeted
regulation of MET.
Discussion
In recent years, the idea of targeted molecular
therapy has become one of the most attractive topics in the field
of human cancer treatment (3). Thus,
the identification of the molecular pathogenesis of cancer is
crucial for the development of effective therapeutic approaches.
Emerging evidence suggests that the dysregulation of miRNAs plays a
pivotal role in the pathogenesis of human cancer development
through the targeting of a variety of important molecules (25). Previous studies have demonstrated that
miR-206 is downregulated in lung cancer, which is correlated with
tumor invasion and metastasis (20).
However, the functional role of miR-206 in lung adenocarcinoma is
largely unknown. In the present study, the functional
characterization and the mechanism of action of miR-206 in human
lung adenocarcinoma were investigated. It was observed that tumor
tissues from patients with lung adenocarcinoma exhibited a decrease
in miR-206 expression. Furthermore, miR-206 expression caused
significant suppression of cell viability and migration in lung
adenocarcinoma cells in vitro. These results indicate that
miR-206 may serve as a tumor-suppressor gene in lung
adenocarcinoma.
MET is a receptor tyrosine kinase that is
dysregulated in multiple cancer types, including lung cancer
(26). Overexpression of MET protein
is detected in 36.0–72.3% of lung adenocarcinoma tissues, and is
associated with tumor metastasis and poor prognosis (27–29).
Silencing or inactivation of MET has been demonstrated to be
important for cell viability in vitro, as well as for cell
motility and migration (11,30). In rhabdomyosarcoma, MET was
demonstrated to be a direct target of miR-206 (15). In the present study, MET was
downregulated by miR-206 in lung adenocarcinoma cells at both the
mRNA and protein levels. The luciferase assay indicated that MET is
a target of miR-206, as the specific knockdown of MET by siRNA led
to a partial phenocopy of the inhibitory effect of cell viability
and migration inhibition that was observed with miR-206
overexpression. Together with the present findings, these results
suggested that MET acts as a potential oncogene and is involved in
the anti-tumor effects of miR-206.
In conclusion, the present study confirms that
miR-206 is frequently downregulated in lung adenocarcinoma. The
ectopic expression of miR-206 can exert an inhibitory effect on
cell viability and migration of lung adenocarcinoma cells, in part,
by targeting MET. These data suggest that miR-206 serves as a
potential tumor suppressor and may be developed as a novel
therapeutic strategy for patients with lung adenocarcinoma.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (Beijing, China; grant
no. 81101768), the State Scholarship Fund of China (Beijing, China;
grant no. 201308330145) and the Medicine and Health Sci-Tech
Project of Zhejiang Province (Hangzhou, China; grant no.
2015KYA074).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Research Network:
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Moreira AL and Eng J: Personalized therapy
for lung cancer. Chest. 146:1649–1657. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong
W, Liao Y and Du J: High expression of miR-21 and miR-155 predicts
recurrence and unfavourable survival in non-small cell lung cancer.
Eur J Cancer. 49:604–615. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hayashita Y, Osada H, Tatematsu Y, Yamada
H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y and
Takahashi T: A polycistronic microRNA cluster, miR-17-92, is
overexpressed in human lung cancers and enhances cell
proliferation. Cancer Res. 65:9628–9632. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Campayo M, Navarro A, Viñolas N, Diaz T,
Tejero R, Gimferrer JM, Molins L, Cabanas ML, Ramirez J, Monzo M
and Marrades R: Low miR-145 and high miR-367 are associated with
unfavourable prognosis in resected nonsmall cell lung cancer. Eur
Respir J. 41:1172–1178. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osada H and Takahashi T: let-7 and
miR-17-92: Small-sized major players in lung cancer development.
Cancer Sci. 102:9–17. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nadal E, Chen G, Gallegos M, Lin L,
Ferrer-Torres D, Truini A, Wang Z, Lin J, Reddy RM, Llatjos R, et
al: Epigenetic inactivation of microRNA-34b/c predicts poor
disease-free survival in early stage lung adenocarcinoma. Clin
Cancer Res. 19:6842–6852. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo W, Huang B, Li Z, Li H, Sun L, Zhang
Q, Qiu X and Wang E: MicroRNA-449a is downregulated in non-small
cell lung cancer and inhibits migration and invasion by targeting
c-Met. PLoS One. 8:e647592013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nasser MW, Datta J, Nuovo G, Kutay H,
Motiwala T, Majumder S, Wang B, Suster S, Jacob ST and Ghoshal K:
Downregulation of micro-RNA-1 (miR-1) in lung cancer. Suppression
of tumorigenic property of lung cancer cells and their
sensitization to doxorubicin-induced apoptosis by miR-1. J Biol
Chem. 283:33394–33405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rao PK, Kumar RM, Farkhondeh M,
Baskerville S and Lodish HF: Myogenic factors that regulate
expression of muscle-specific microRNAs. Proc Natl Acad Sci USA.
103:8721–8726. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kondo N, Toyama T, Sugiura H, Fujii Y and
Yamashita H: miR-206 Expression is downregulated in estrogen
receptor alpha-positive human breast cancer. Cancer Res.
68:5004–5008. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taulli R, Bersani F, Foglizzo V, Linari A,
Vigna E, Ladanyi M, Tuschl T and Ponzetto C: The muscle-specific
microRNA miR-206 blocks human rhabdomyosarcoma growth in
xenotransplanted mice by promoting myogenic differentiation. J Clin
Invest. 119:2366–2378. 2009.PubMed/NCBI
|
|
16
|
Hidaka H, Seki N, Yoshino H, Yamasaki T,
Yamada Y, Nohata N, Fuse M, Nakagawa M and Enokida H: Tumor
suppressive microRNA-1285 regulates novel molecular targets:
Aberrant expression and functional significance in renal cell
carcinoma. Oncotarget. 3:44–57. 2012.PubMed/NCBI
|
|
17
|
Chen X, Yan Q, Li S, Zhou L, Yang H, Yang
Y, Liu X and Wan X: Expression of the tumor suppressor miR-206 is
associated with cellular proliferative inhibition and impairs
invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett.
314:41–53. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vickers MM, Bar J, Gorn-Hondermann I,
Yarom N, Daneshmand M, Hanson JE, Addison CL, Asmis TR, Jonker DJ,
Maroun J, et al: Stage-dependent differential expression of
microRNAs in colorectal cancer: Potential role as markers of
metastatic disease. Clin Exp Metastasis. 29:123–132. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Liu X, Jin H, Guo X, Xia L, Chen
Z, Bai M, Liu J, Shang X, Wu K, et al: miR-206 inhibits gastric
cancer proliferation in part by repressing cyclinD2. Cancer Lett.
332:94–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Ling C, Bai Y and Zhao J:
MicroRNA-206 is associated with invasion and metastasis of lung
cancer. Anat Rec (Hoboken). 294:88–92. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Georgantas RW III, Streicher K, Luo X,
Greenlees L, Zhu W, Liu Z, Brohawn P, Morehouse C, Higgs BW,
Richman L, et al: MicroRNA-206 induces G1 arrest in melanoma by
inhibition of CDK4 and Cyclin D. Pigment Cell Melanoma Res.
27:275–286. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Yang T, Li X, Yang Q, Liu R, Huang
J, Li Y, Yang C and Jiang Y: Alteration of serum miR-206 and
miR-133b is associated with lung carcinogenesis induced by
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Toxicol Appl
Pharmacol. 267:238–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhou JY, Chen X, Zhao J, Bao Z, Chen X,
Zhang P, Liu ZF and Zhou JY: MicroRNA-34a overcomes HGF-mediated
gefitinib resistance in EGFR mutant lung cancer cells partly by
targeting MET. Cancer Lett. 351:265–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan D, Dong Xda E, Chen X, Wang L, Lu C,
Wang J, Qu J and Tu L: MicroRNA-1/206 targets c-Met and inhibits
rhabdomyosarcoma development. J Biol Chem. 284:29596–29604. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Skead G and Govender D: Gene of the month:
MET. J Clin Pathol. 68:405–409. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakamura Y, Niki T, Goto A, Morikawa T,
Miyazawa K, Nakajima J and Fukayama M: c-Met activation in lung
adenocarcinoma tissues: An immunohistochemical analysis. Cancer
Sci. 98:1006–1013. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma PC, Jagadeeswaran R, Jagadeesh S,
Tretiakova MS, Nallasura V, Fox EA, Hansen M, Schaefer E, Naoki K,
Lader A, et al: Functional expression and mutations of c-Met and
its therapeutic inhibition with SU11274 and small interfering RNA
in non-small cell lung cancer. Cancer Res. 65:1479–1488. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ichimura E, Maeshima A, Nakajima T and
Nakamura T: Expression of c-met/HGF receptor in human non-small
cell lung carcinomas in vitro and in vivo and its prognostic
significance. Jpn J Cancer Res. 87:1063–1069. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zillhardt M, Christensen JG and Lengyel E:
An orally available small-molecule inhibitor of c-Met, PF-2341066,
reduces tumor burden and metastasis in a preclinical model of
ovarian cancer metastasis. Neoplasia. 12:1–10. 2010. View Article : Google Scholar : PubMed/NCBI
|