Introduction
Previous studies have demonstrated that alterations
in protein-coding genes are critical for the development of human
cancer (1,2). Other studies have indicated that
deregulation of non-coding genes, particularly microRNAs (miRNAs),
also contribute to tumorigenesis (3–6).
miRNAs belong to a class of phylogenetically
conserved non-coding RNAs that regulate an abundance of cellular
processes by binding to specific target messenger RNAs (7). Numerous studies indicate that miRNAs are
important in various biological processes, including the cell
cycle, development, cell proliferation, differentiation and
apoptosis (8–12). More than half of the miRNA genes are
located in chromosomes that have been observed to become amplified,
deleted or translocated during the development of cancer (13). In addition, deregulation of specific
miRNAs occurs in certain cancer types (14,15), and
this has been suggested to be associated with the prognosis of
cancer patients (16). Furthermore, a
previous study proposed that miRNAs may act as oncogenes or tumor
suppressor genes (6). These findings
highlight the importance of miRNAs in cancer development and
provide insight into the molecular mechanisms that cause
tumorigenesis.
Globally, breast cancer is the most common cause of
cancer-associated mortality in women, and accounted for ~1.38
million novel cases and 458,000 mortalities in 2008 (17). As with other solid tumors, multiple
genetic and epigenetic alterations in protein-coding genes have
been observed in breast cancer (18).
However, previous findings alone cannot explain the complexity of
breast cancer. Previous studies have demonstrated that dysfunction
of miRNAs contributes to breast cancer development (11,14). Zhang
et al reported that microRNA-520e (miR-520e) was associated
with hepatocarcinogenesis (19);
however, the biological significance of miR-520e in breast cancer
remains largely unknown. The present study demonstrated that
miR-520e is upregulated in breast cancer tissues and promotes
proliferation, invasion and migration, and inhibits the apoptosis
of cancer cells. The results of the present study suggest that
miR-520e may be an attractive target for cancer therapy.
Materials and methods
Human tissue specimens
The present study was approved by the Institutional
Review Board and Ethical Committee of Peking University Shenzhen
Hospital (Shenzhen, Guangdong, China). All patients provided
written informed consent prior to the study. In total, 21 paired
breast cancer and adjacent non-cancerous breast tissues were
obtained from patients that had undergone resection for breast
cancer. The patient characteristics are reported in Table I. The patients had no previous local
or systemic treatment prior to resection. No anticancer therapy was
administered to the patients prior to relapse. The tissue samples
were collected between 2012 and 2013 at the Peking University
Shenzhen Hospital, and were snap frozen in liquid nitrogen and
stored at −80°C until required. Tumor and non-cancerous tissue
samples were confirmed by histology. The histological grade was
determined based on the modified Scarff-Bloom-Richardson grading
system (20).
 | Table I.Clinical characteristics of 21
patients with breast cancer. |
Table I.
Clinical characteristics of 21
patients with breast cancer.
| Characteristics | Cases, n (%) |
|---|
| Age, years |
|
|
>50 | 10 (47.6) |
| ≤50 | 11 (52.4) |
| Histological
grade |
|
|
>I | 10 (47.6) |
|
I | 11 (52.4) |
| Tumor size |
|
| >5
cm | 6
(28.6) |
| ≤5
cm | 15 (71.4) |
| Tumor number |
|
|
>1 | 10 (47.6) |
|
1 | 11 (52.4) |
RNA oligoribonucleotides
All RNA oligoribonucleotides were purchased from
Genepharma, Ltd. (Shanghai, China). miRNA duplexes corresponding to
mature miR-520e were designed as previously described (21). The negative control (NC) RNA duplex
for the miRNA mimic was non-homologous to any human genome
sequence. The sequences were as follows: hsa-miR-520e mimic,
5′-AAAGUGCUUCCUUUUUAGGG-3′; and the hsa-miR-520e NC,
5′-UUCUCCGAACGUGUCACGUTT-3′.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) and primers
Total RNA was extracted from tissue cells using
Invitrogen TRIzol reagent (catalog no. 15596026; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. cDNA was synthesized from the extracted RNA using the
SYBR PrimeScript miRNA RT-PCR kit A (catalog no. RR716; Takara
Biotechnology, Co., Ltd., Dalian, China). qPCR was performed with
the SYBR PrimeScript miRNA RT-PCR kit B (catalog no. RR716; Takara
Biotechnology, Co., Ltd.) using a LightCycler® 480
Fluorescent Quantitative PCR system (Roche Diagnostics GmbH,
Mannheim, Germany). Briefly, single-stranded cDNA was synthesized
from 500 ng of total RNA in a 10 µl reaction volume using the SYBR
PrimeScript miRNA RT-PCR kit A. The reactions were incubated at
37°C for 60 min and were then inactivated by incubation at 85°C for
5 sec. RT-qPCR analysis for the expression of miR-520e and the
reference gene RNU6B was performed using SYBR PrimeScript miRNA
RT-PCR kit B. The temperature cycle profile for the RT-qPCR
reactions was as follows: 95°C for 30 sec; 40 cycles of 95°C for 5
sec; and 60°C for 20 sec. To verify the specificity of the PCR
product, a melting curve analysis was performed immediately
subsequent to amplification, as follows: Heating to 95°C for 20
sec; cooling to 60°C for 20 sec; and heating to 95°C with a
transition rate of 0.11°C/sec, while continuously collecting the
fluorescent signal. All reactions were performed on a LightCycler
480 and were run in triplicate. The cycle quantification (Cq)
values did not differ by more than 0.5 among the triplicates. The
levels of target genes were normalized to the levels of the
internal control genes to permit the calculation of the
2−ΔΔCq value. Cq is defined as the fractional cycle
number at which the fluorescence passes the fixed threshold. The
following primers were used: miR-520e forward,
5′-GAAAGTGCTTCCTTTTTAGGG-3′ and reverse, SYBR PrimeScript miRNA
RT-PCR kit universal primer; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-ACGCTTCACGAATTTGCGT-3′ (Thermo Fisher Scientific,
Inc.).
Cell culture and transfection
The human breast cancer MCF-7 and MDA-MB-231 cell
lines were purchased from the American Type Culture Collection
(Manassas, VA, USA). The cells lines were maintained in HyClone
Dulbecco's modified Eagle's medium (DMEM; catalog no. SH30022.01B;
GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10%
HyClone fetal bovine serum (FBS; catalog no. 10270-106; GE
Healthcare Life Sciences) and HyClone 100 U penicillin-streptomycin
(catalog no. SV30010; GE Healthcare Life Sciences) at 37°C in a
humidified atmosphere containing 5% CO2. RNA
oligoribonucleotides were reverse transfected using Invitrogen
Lipofectamine® 2000 (catalog no. 11668-019; Thermo
Fisher Scientific, Inc.). A final concentration of 100 nM RNA
duplex was used for each transfection. The RNA transfection
efficiency using this method is ~90%. All transfections were
performed according to the manufacturer's protocol.
Assessment of cell proliferation by
cell counting kit-8 (CCK-8) assay
CCK-8 assay was used to assess the proliferation of
MDA-MB-231 and MCF-7 subsequent to transfection with miR-520e
mimics. Briefly, MDA-MB-231 and MCF-7 cells were plated at
3.0×103 cells/well in a 96-well plate and transfected
with NC or miR-520e mimics at a final concentration of 100 nM.
Prior to harvesting the cells, 100 µl of spent medium was replaced
with an equal volume of fresh medium containing 10 µl CCK-8
(catalog no. CK04-500; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) and incubated for 1 h at 37°C. The light
absorbance of each well was measured at a wavelength of 450 nm
using an Enzyme Immunoassay Instrument (model no. 680; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Each assay was repeated in
triplicate.
Flow cytometric analysis of cell
apoptosis
The apoptotic rate of cells was determined using
Invitrogen Annexin V-fluorescein isothiocyanate (FITC) and
propidium iodide (PI) double staining (catalog no. V13241; Thermo
Fisher Scientific, Inc.). In total, ~10,000 MCF-7 and MDA-MB-231
cells were cultured in 5% CO2 in 6-well plates at 37°C.
The cells were transfected with the miR-520e mimic or NC when the
cells had reached ~50% confluency. Subsequent to 72 h, floating and
adherent cells were collected, washed twice with PBS (pH 7.4),
resuspended in 100 µl Annexin-binding buffer and incubated with 5
µl Annexin V-FITC and 3 µl PI (50 µg/ml). Subsequent to 20 min
incubation in the dark at room temperature, 400 µl Annexin-binding
buffer was added prior to flow cytometric analysis. The samples
were analyzed within 30 min of staining. Flow cytometry was
performed using the FACSCalibur and Cell Quest software (catalog
no. Navi105; Beckman Coulter, Inc., Miami, FL, USA).
Transwell migration assay
A 24-well Boyden chamber with a 8 µm pore
polycarbonate membrane (catalog no. 3422; Corning, Inc., Corning,
NY, USA) was used to analyze the migration of MCF-7 and MDA-MB-231
cells. In total, 24 h subsequent to transfection with the RNA
duplex, tumor cells (4×104 MCF-7 cells or
3×104 MDA-MB-231 cells in 200 µl serum-free DMEM) were
added to the upper compartment of the Boyden chamber, while the
lower compartment was filled with 600 µl DMEM containing 10% FBS.
Subsequent to incubation at 37°C for indicated the times (24 h for
MDA-MB-231, 48 h for MCF-7), cells that had not migrated and
remained on the upper surface of the polycarbonate membrane were
removed with cotton swabs. The cells on the lower surface of the
membrane (those that had migrated) were fixed with formaldehyde,
stained with crystal violet and counted in 5 randomly selected
fields of view per well, under a light microscope at a
magnification of ×150. The cells were transfected in 3 separate
transfections and each was analyzed in triplicate.
Statistical analysis
The data was expressed as the mean ± standard error
of the mean for ≥3 independent experiments. Student's t-test
was employed to analyze the significance of differences between the
cancerous and non-cancerous tissue cells. For the comparison of
miR-520e expression levels between the matched breast cancer and
normal tissue samples, a Mann-Whitney U-test was used. All
statistical tests were two-tailed and P<0.05 was considered to
indicate a statistically significant difference. All analyses were
performed using SPSS software version 13.0 (SPSS, Inc., Chicago,
IL, USA).
Results
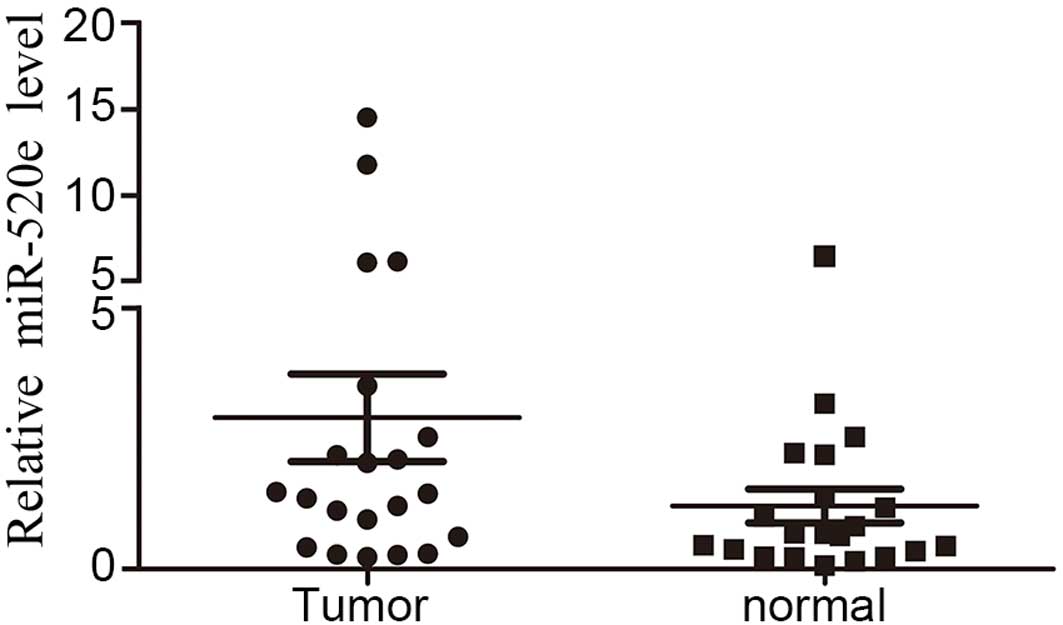
miR-520e expression is upregulated in
human breast cancer tissues
A RT-qPCR array was performed to analyze the
expression level of miR-520e in 21 paired breast cancer and
adjacent non-cancerous breast tissues. miR-520e was significantly
upregulated in breast cancer tissues compared with normal tissues
(Fig. 1), suggesting that miR-520e
overexpression in breast cancer cells is a frequent event and that
there is an association between miR-520e upregulation and breast
cancer tumorigenesis.
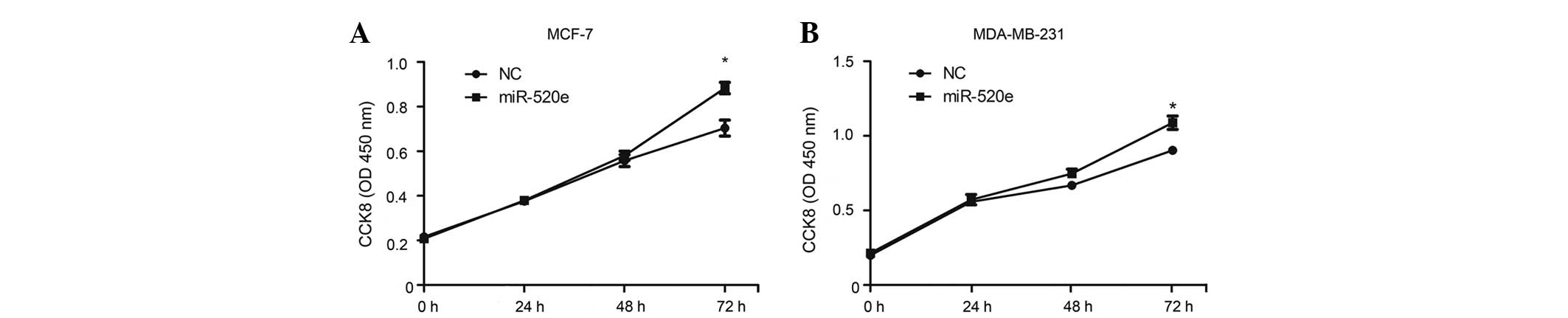
miR-520e promotes breast cancer cell
growth in vitro
The upregulation of miR-520e in breast cancer tissue
samples lead the present study to investigate whether miR-520e
functions as an oncogene. The effect of miR-520e restoration on the
proliferation of breast cancer cells was evaluated using a CCK-8
assay on the human breast cancer MCF-7 and MDA-MB-231 cell lines,
which were transfected with a control RNA duplex (NC) or miR-520e
duplex. miR-520e-transfected cells exhibited an increased
proliferation compared with NC transfectants (Fig. 2). These findings indicate a cell
growth-promoting role for miR-520e.
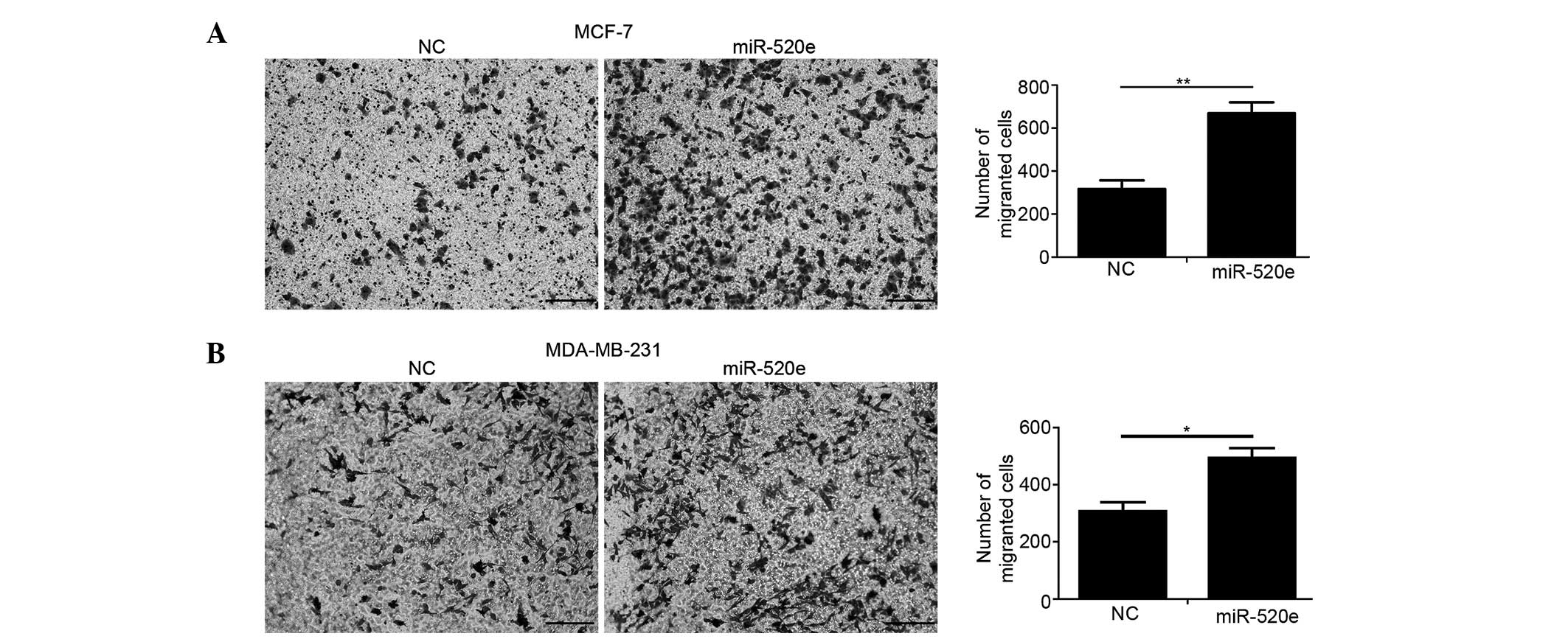
miR-520e promotes breast cancer cell
migration in vitro
The effect of miR-520e on breast cancer cell
migration was investigated using a Transwell assay. The restoration
of miR-520e substantially increased the number of MCF-7 and
MDA-MB-231 cells that invaded the lower chamber in the Boyden
chamber (Fig. 3).
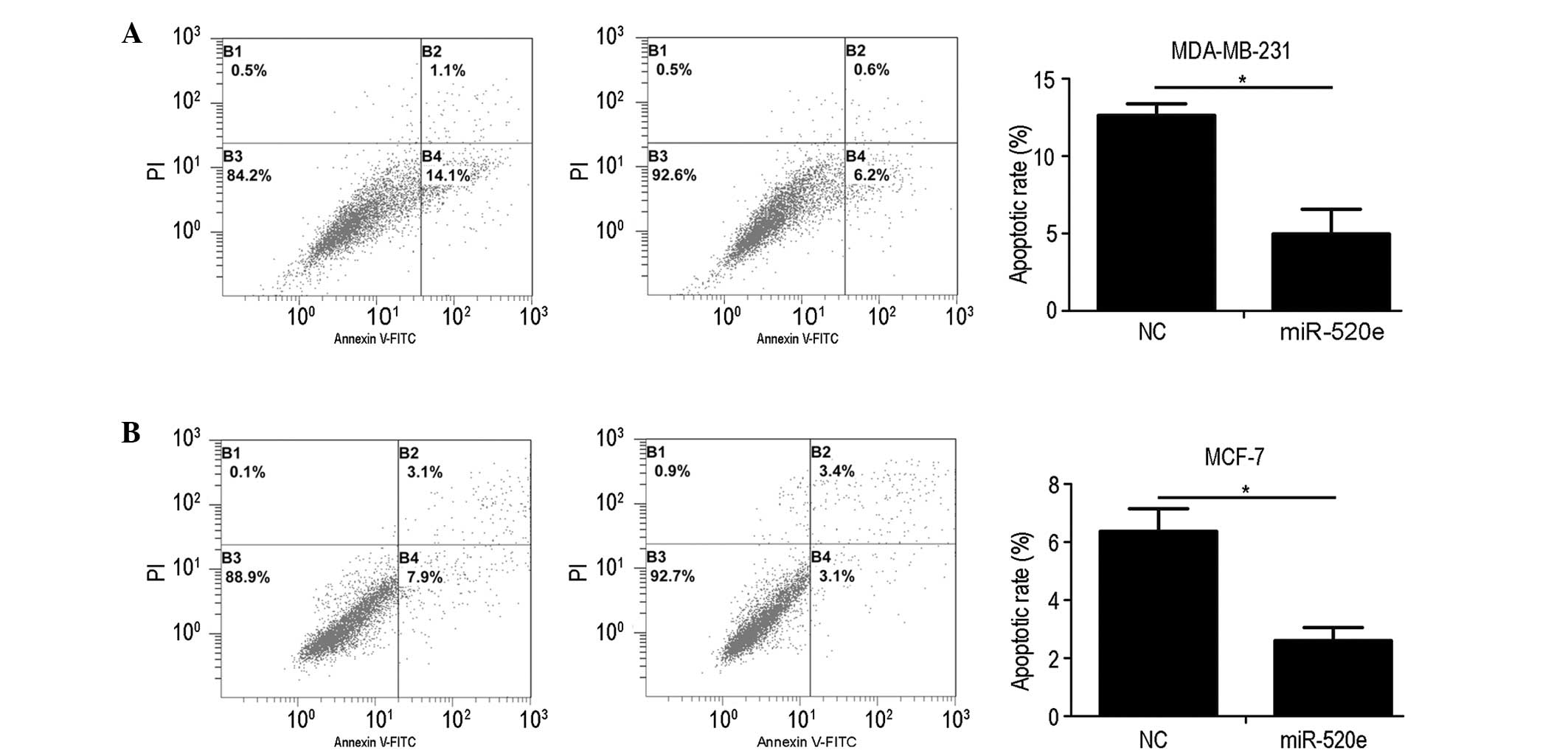
miR-520e suppresses breast cancer cell
apoptosis in vitro
The effect that miR-520e exerts on the apoptotic
rate of breast cancer cells was assessed using Annexin V-FITC and
PI double staining, followed by flow cytometric analysis. Fig. 4 demonstrates that the restoration of
miR-520e substantially decreased the percentage of MCF-7 and
MDA-MB-231 cells that underwent apoptosis. These results indicate
that miR-520e inhibits the apoptosis of breast cancer cells.
Discussion
Abnormal and uncontrolled cell proliferation is a
hallmark of cancer and is caused by the misregulation of various
crucial proteins and alterations in miRNA expression (22). miRNAs regulate numerous biological
processes, including disease progression and cell development,
proliferation, apoptosis and differentiation (8–12). Due to
the numerous biological processes that miRNAs are involved in, it
is not notable that the deregulation of miRNAs has been associated
with tumor progression in certain human cancers. Numerous miRNAs
have been observed in breast cancer development (23–27), by
functioning alone or in combination. Therefore, investigating
miRNAs that are aberrantly expressed in breast cancer may aid the
understanding of the mechanisms that cause breast cancer
carcinogenesis and progression. The present study demonstrated that
miR-520e was markedly upregulated in breast cancer tissues compared
with paired adjacent normal tissues. Furthermore, the ectopic
expression of miR-520e promoted breast cancer cell proliferation,
migration and invasion and inhibited cell apoptosis. The current
study suggests that miR-520e may act as a novel oncomir in breast
cancer and may be a potential therapeutic target.
Evading cell apoptosis is crucial for the malignant
transformation of a tumor and tumor progression. It aids cells to
avoid immune surveillance and survive in challenging tumor
environments, including those that are low in nutrition and hypoxic
(28). The present study demonstrated
that miR-520e could suppress the apoptosis of breast cancer cells.
Therefore, the upregulation of miR-520e may facilitate the survival
and development of breast cancer.
Tumor metastasis is responsible for the rapid
recurrence and poor prognosis of patients; therefore, the
identification of novel anti-metastasis therapeutics is imperative
(29). Previous studies using
clinical specimens and in vitro and in vivo models
have identified a limited number of miRNAs that promote metastasis,
including miR-200, miR-10b and miR-21 (30–33). The
present study revealed that miR-520e is associated with metastasis;
upregulation of miR-520e may facilitate the metastasis of breast
cancer cells and therefore the progression of breast cancer.
In summary, the present study investigated the
potential role of miR-520e in breast cancer development. The
findings of the current study suggest that upregulation of miR-520e
may favor malignant transformation and tumor progression of breast
cancer cells, and indicate that miR-520e possesses a potential
application in anti-cancer therapy.
Acknowledgements
The authors would like to thank Xiaowen Zhou and
Huajian Zhong for their advice on the study and manuscript, and
Xiaoqing Li and Hongfang Duan for their intellectual and technical
contributions. All are graduate students at the Central Laboratory
of Peking University Shenzhen Hospital. The present study was
supported by grants from the Medical Scientific Research Foundation
of Guangdong Province (grant no. A2011564), National Natural
Science Foundation of China (grant no. 31360212), Science and
Technology Development Fund Project of Shenzhen (grant no.
JC201005260209A) and Science and Technology Development Fund
Project of Shenzhen (grant no. JCYJ20130402114702122).
References
|
1
|
Chin K, DeVries S, Fridlyand J, Spellman
PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et
al: Genomic and transcriptional aberrations linked to breast cancer
pathophysiologies. Cancer Cell. 10:529–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stephens PJ, Tarpey PS, Davies H, Van Loo
P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell
GR, et al: Oslo Breast Cancer Consortium (OSBREAC): The landscape
of cancer genes and mutational processes in breast cancer. Nature.
486:400–404. 2012.PubMed/NCBI
|
|
3
|
Negrini M, Nicoloso MS and Calin GA:
MicroRNAs and cancer - new paradigms in molecular oncology. Curr
Opin Cell Biol. 21:470–479. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fang L, Deng Z, Shatseva T, Yang J, Peng
C, Du WW, Yee AJ, Ang LC, He C, Shan SW and Yang BB: MicroRNA
miR-93 promotes tumor growth and angiogenesis by targeting
integrin-β8. Oncogene. 30:806–821. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Díaz-López A, Moreno-Bueno G and Cano A:
Role of microRNA in epithelial to mesenchymal transition and
metastasis and clinical perspectives. Cancer Manag Res. 6:205–216.
2014.PubMed/NCBI
|
|
11
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Braun CJ, Zhang X, Savelyeva I, Wolff S,
Moll UM, Schepeler T, Ørntoft TF, Andersen CL and Dobbelstein M:
p53-Responsive micrornas 192 and 215 are capable of inducing cell
cycle arrest. Cancer Res. 68:10094–10104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Calin GA, Ferracin M, Cimmino A, Di Leva
G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et
al: A MicroRNA signature associated with prognosis and progression
in chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Grayson M: Breast cancer. Nature.
485:S492012. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Byler S, Goldgar S, Heerboth S, Leary M,
Housman G, Moulton K and Sarkar S: Genetic and epigenetic aspects
of breast cancer progression and therapy. Anticancer Res.
34:1071–1077. 2014.PubMed/NCBI
|
|
19
|
Zhang S, Shan C, Kong G, Du Y, Ye L and
Zhang X: MicroRNA-520e suppresses growth of hepatoma cells by
targeting the NF-κB-inducing kinase (NIK). Oncogene. 31:3607–3620.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lakhani SR, Elis IO, Schnitt SJ, Tan PH
and van de Vijver MJ: WHO Classification of Tumours of the Breast.
4th. IARC; Lyon: pp. 19–20. 2012
|
|
21
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu J, Mao Q, Liu Y, Hao X, Zhang S and
Zhang J: Analysis of miR-205 and miR-155 expression in the blood of
breast cancer patients. Chin J Cancer Res. 25:46–54.
2013.PubMed/NCBI
|
|
24
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Achari C, Winslow S, Ceder Y and Larsson
C: Expression of miR-34c induces G2/M cell cycle arrest in breast
cancer cells. BMC Cancer. 14:5382014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chao CH, Chang CC, Wu MJ, Ko HW, Wang D,
Hung MC, Yang JY and Chang CJ: MicroRNA-205 signaling regulates
mammary stem cell fate and tumorigenesis. J Clin Invest.
124:3093–3106. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feliciano A, Castellvi J, Artero-Castro A,
Leal JA, Romagosa C, Hernández-Losa J, Peg V, Fabra A, Vidal F,
Kondoh H, et al: miR-125b acts as a tumor suppressor in breast
tumorigenesis via its novel direct targets ENPEP, CK2-α, CCNJ, and
MEGF9. PLoS One. 8:e762472013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Talmadge JE and Fidler IJ: AACR centennial
series: The biology of cancer metastasis: Historical perspective.
Cancer Res. 70:5649–5669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma L: Role of miR-10b in breast cancer
metastasis. Breast Cancer Res. 12:2102010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song B, Wang C, Liu J, Wang X, Lv L, Wei
L, Xie L, Zheng Y and Song X: MicroRNA-21 regulates breast cancer
invasion partly by targeting tissue inhibitor of metalloproteinase
3 expression. J Exp Clin Cancer Res. 29:292010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Burk U, Schubert J, Wellner U, Schmalhofer
O, Vincan E, Spaderna S and Brabletz T: A reciprocal repression
between ZEB1 and members of the miR-200 family promotes EMT and
invasion in cancer cells. EMBO Rep. 9:582–589. 2008. View Article : Google Scholar : PubMed/NCBI
|